Abstract
The flightless intertidal beetle genus Aegialites (family Salpingidae) is distributed along the Northern Pacific coasts, from California to Alaska and from Northern Japan to Kamchatka. Systematics of Aegialites and its phylogenetic relationships to other members of Salpingidae are unclear, and little genetic information is available. We here present the first complete mitochondrial genome of this genus, represented by Aegialites californicus (Motchoulsky, 1845) from Sonoma County, California, U.S.A. The complete mitochondrial genome of A. californicus is 15,899 bp long and comprises 13 protein-coding (PCG), two ribosomal RNA (rRNA) and 22 transfer RNA (tRNA) genes. The phylogenetic analysis places A.californicus as sister to other members of family Salpingidae. The mitochondrial genome sequence of A. californicus will contribute to future phylogenetic and taxonomic studies of genus Aegialites, family Salpingidae and superfamily Tenebrionoidea.
Introduction
The beetle genus Aegialites Mannerheim, 1853 (Salpingidae) comprises flightless beetles, which inhabit rocks and rock crevices in the supralittoral zone of rocky shores. They are recorded from a wide range of Northern Pacific coasts, from California to Alaska and from Northern Japan to Kamchatka (Zerche Citation2004). The supralittoral zone on rocky shores is a challenging environment with daily fluctuations in temperature, humidity and salinity as well as exposure to wave actions of different strength. The Aegialites beetles have therefore required special morphological adaptations like widely separated coxae, long tarsi and large claws.
Until recently, only four species have been recognized in the genus Aegialites (Spilman Citation1967). Zerche (Citation2004) conducted a revision of the genus based on morphology and increased the total number of species from four to thirty. Zerche (Citation2004) changed the previously accepted paradigm of few widespread and variable species in Aegialites to that of many locally distributed species. The validity of Zerche’s hypothesis has never been tested using molecular data. The only work that used molecular data in Aegialites (Hojito et al. Citation2010) compared different populations from Hokkaido using one mitochondrial gene (nad2).
To increase the number of genetic markers available, we here present the complete mitochondrial genome of A. californicus (Motchoulsky, 1845) from Sonoma County, California, assembled with genome skimming sequencing data. The mitochondrial genome can serve as a reference for further molecular work and help the research on taxonomic placement and systematics of the genus ().
Figure 1. Aegialites californicus, neotype (photo taken by Zerche Citation2004, pp 56) scale: 1 mm.

Material and methods
A living adult specimen of A. californicus was collected and directly placed in 100% ethanol. The specimen label data are as follows: U.S.A., California, Sonoma Co., 1.4 km SSE Carmet, 38°21.709’N 123°04.163’W WGS84, h = 0 m, accuracy 4 m, extent 2 m, splitting (crevices in) coastal/tidal rocks (5 m wide boulder) with chisel, V.I.Gusarov & M.N.Haugen leg., 23.xi.2019. The specimen was collected legally, as no special permits for collection or export was necessary as informed by the California Department of Fish and Wildlife. A voucher specimen was deposited in the insect collection at the Natural History Museum of the University in Oslo (https://www.nhm.uio.no/english/collections/zoological/insect/, Vladimir Gusarov, [email protected]) with specimen accession number ZMUN 10087480. The species identification was carried out by Vladimir Gusarov based on morphological examinations following the descriptions of Zerche (Citation2004). A. californicus, is morphologically most similar to the species A. farallonensis and A. rfuchsii, but is differentiated from them by being more metallically iridescent, having a more slender antenna and wide open anterior coxae toward the rear. Additionally, the pronotum of A. californicus is widest shortly just before the center, it is hardly narrower posteriorly than anteriorly, in A. farallonensis it is widest well before the center, much more narrowed posteriorly than anteriorly narrowed. The punctuation of the pronotum is quite fine in A. californicus and not stronger than the punctuation of the head; the interspaces on the pronotum measure about two to three times the diameter. In A. farallonensis the punctuation of the pronotum is stronger and much denser than in A. californicus, the punctuated interspaces measure about half the diameter, the punctuation is stronger than the punctuation of the head. In A. fuchsii the punctuation of the pronotum is somewhat stronger, the interspaces measure in places only a third of the diameter and are wrinkled.
The abdomen was dissected from the body with a sterile needle and tweezers, transferred to a new 1,5 mL Eppendorf tube with 100% ethanol before it was shipped on ice to StarSEQ, Mainz, Germany where genomic DNA extraction, library prep and sequencing was performed. A NEBNext® Ultra™ II FS DNA Library Prep kit (New England Biolabs) was utilized, and sequencing was done on the Illumina platform NextSeq 500 with paired end reads and 2 × 150 bp insert size.
Raw data were assembled using metaSPAdes (Nurk et al. Citation2017) and the linear contig produced by metaSPAdes was manually curated in Mega 7 (Kumar et al. Citation2016) and the 55 bp identical sequence overlap found in the area believed to be the control region was deleted to ensure the circularization of the mitochondrial genome. The mitochondrial sequence was then annotated using the MITOS2 web server (Donath et al. Citation2019). The annotation was manually edited so that the first gene feature of the sequence file was cox1. The read coverage depth of the Illumina reads was calculated using Bowtie2 (Langmead and Salzberg Citation2012) and SAMtools (Danecek et al. Citation2021), and the resulting coverage map was created using Matplotlib (Hunter Citation2007) (Supplementary Figure 1). The complete mitochondrial sequence of A. californicus was deposited at the National Center for Biotechnology Information GenBank under accession number OR168935. The circular mitochondrial genome map was generated using Chloroplot (Zheng et al. Citation2020).
To validate the phylogenetic position of A. californicus, its 13 protein coding genes, and those of 13 other species of Coleoptera were used to create a maximum-likelihood phylogenetic tree using IQ-TREE version 2.2.0 (Minh et al. Citation2020). Each gene from all 14 species was individually aligned in Mega 7 (Kumar et al. Citation2016) with the MUSCLE algorithm for codons (Edgar Citation2004), and a concatenated dataset of all 13 genes was created using FASconCAT-G_v1.05 (Kück and Longo Citation2014). The phylogenetic inference was run with 1000 bootstrap replicates and the option MFP + MERGE implementing the ModelFinder greedy partitioning strategy (Cameron Citation2014). The 13 sequences was selected non-randomly using NCBI blastn with cox1 of A. californicus as query with the following settings: taxID limited to superfamily Tenebrionoidea (TaxID 71527), sequence length 12,000-25,000, sequence similarity threshold 80-100%. From the resulting list at NCBI, 11 complete mitochondrial genomes was selected for download and two partial ones. Of the 13 mitochondrial genomes downloaded from NCBI, three other sequences come from the family Salpingidae (Salpingus planirostris, Lissodema cursor and Vincenzellus ruficollis).
Results
The mitochondrial genome of A. californicus is 15,899 bp and consists of 13 protein-coding genes (PCG), two ribosomal RNA (rRNA) genes, and 22 transfer RNA (tRNA) genes. The mitochondrial genome has an AT-bias, with an AT-content of 78% (A = 39%, T = 39%, C = 13%, G = 9%). The non-coding control region (also known as D-loop or origin of replication) could possibly be between trnI and rrnS (), but the annotation by MITOS2 was not certain in this respect, and hence not visualized in the genome plot. The gene order of the 13 PCGs is visualized in the genome plot in . All the 13 PCGs have a typical ATN (Met) start codon; five genes (atp6, cox3, nad4, nad4l and cob) have ATG and eight genes (atp8, cox1, cox2, nad1, nad2, nad3, nad5 and nad6) have ATA. All the PCGs have a typical TAN stop codon; six genes (atp6, cox3, nad2, nad4l, nad6 and cob) have TAA and three genes (atp8, nad1 and nad3) have TAG. Two genes (cox1 and cox2) have TGA and two genes (nad4 and nad5) are found to have TGG, so in these four genes, TAA stop codon is completed by the addition of 3′ A residues to the mRNA.
Figure 2. Mitochondrial genome map of A. californicus generated by Chloroplot (Zheng et al. Citation2020). Overlapping features are excluded for a better graphical representation of the architecture of the genome. The dark grey areas inside the map represents GC-content, genes on the outside of the map are transcribed counterclockwise, and genes on the inside are transcribed clockwise as informed by the arrows in the figure.
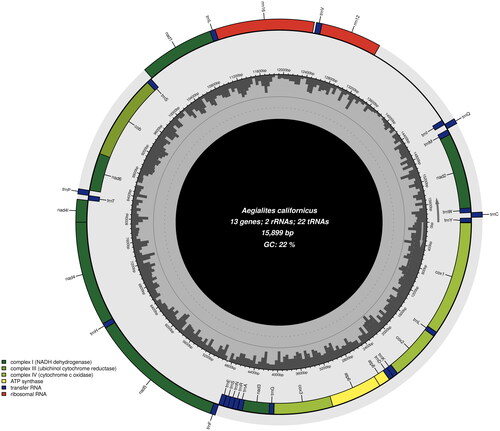
The 22 tRNA genes in A. californicus are interspersed throughout the coding region and range from 63 bp (trnT, trnW, trnS and trnH) to 71 bp (trnK). An overlap of seven bp was detected between the protein coding genes atp6 and atp8 and one bp overlap between atp6 and cox3 and between nad6 and cob. Additional overlaps between t-RNA genes and PCGs were discovered; the longest being 42 bp between cox2 and trnL. The ribosomal RNA genes, rrnL (also known as l-rRNA or 16S rRNA) and rrnS (s-rRNA, 12S) were 1278 and 772 bp long, respectively.
Based on ModelFinder in IQtree and the Bayesian information criterion (BIC) the best fitting partition scheme and the corresponding substitution models comprised three partitions with the following genes grouped together: atp6, cob, cox1, cox2, cox3: mtART + I+R3; atp8, nad2, nad3, nad6: mtART + F+R4; nad1, nad4, nad4l, nad5: mtART + F+R4.
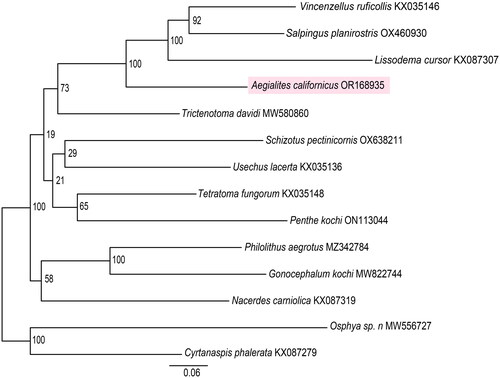
The resulting tree based on 13 mitochondrial genes places A. californicus as sister to the clade composed of V. ruficollis, S. planirostris and L. cursor, the three other representatives of family Salpingidae, with a bootstrap support of 100 (). The four members of Salpingidae are monophyletic in this dataset and they are found closest to T. davidi from family Trictenotomidae.
Figure 3. Maximum-likelihood tree for Aegialites californicus (highlighted) and 13 additional insect taxa from superfamily Tenebrionoidea derived from the following records: Vincenzellus ruficollis KX035146 (unpublished), Salpingus planirostris OX460930 (unpublished), Lissodema cursor KX087307 (unpublished), trictenotoma davidi MW580860 (Sheng et al. Citation2021), schizotus pectinicornis OX638211 (unpublished), usechus lacerta KX035136 (unpublished), tetratoma fungorum KX035148 (unpublished), penthe kochi ON113044 (unpublished), philolithus aegrotus MZ342784 (Smith et al. Citation2021), gonocephalum kochi MW822744 (unpublished), nacerdes carniolica KX087319 (unpublished), osphya sp. n. MW556727 (Liu et al. Citation2023) and cyrtanaspis phalerata KX087279 (unpublished). The GenBank accession numbers for the sequences are indicated next to the species names. Bootstrap values are found close to the nodes and a scale bar indicating the mean number of nucleotide substitutions per site is located at the bottom.
Discussion and conclusions
In this study we assembled and described the characteristics of the mitochondrial genome of A. californicus. The mitochondrial genome is found to be 15,899 bp and consists of 13 protein-coding genes (PCG), two ribosomal RNA (rRNA) genes, and 22 transfer RNA (tRNA) genes, with gene order found to be identical as in the ancestral insect genome (Cameron Citation2014). The phylogentic analysis confirms the placement of A. californicus as sister to three representatives of family Salpingidae. The four salpingids are found to be monophyltic and found closest to T. davidi as expected according to Hu et al. (Citation2020). Lastly, this study provides genetic data for further analyses and studies of genus Aegialites, family Salpingidae and superfamily Tenebrionoidea.
Ethical approval
No ethical issues were involved in this study. As informed by California Department of Fish and Wildlife in Fish and Game Code (FGC) sections 1002, 1002.5 and 1003, trough Section 650, Title 14, California Code of Regulations (CCR), no special Scientific Collection Permit (SCP) was necessary to obtain, because beetles of this genus is not listed on the California Terrestrial and Vernal Pool invertebrates of Conservation Priority list. Information on the voucher specimen and who identified it were included in the manuscript.
Author contributions
MNH, VG and THS designed the project, MNH conducted the data analysis, MNH wrote the manuscript and MNH, VG and THS revised and read the final version of the manuscript to be published.
Supplemental Material
Download MS Word (12.8 KB)Supplemental Material
Download JPEG Image (1 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI GenBank database at https://www.ncbi.nlm.nih.gov with the accession number OR168935, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Associated BioProject, SRA, and BioSample accession numbers are PRJNA985217, SRR25009919 and SAMN35791155, respectively.
Additional information
Funding
References
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59(1):95–117. doi:10.1146/annurev-ento-011613-162007.
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, et al. 2021. Twelve years of SAMtools and BCFtools. Gigascience. 10(2):giab008. doi:10.1093/gigascience/giab008.
- Donath A, Jühling F, Al-Arab M, Bernhart SH, Reinhardt F, Stadler PF, Middendorf M, Bernt M. 2019. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 47(20):10543–10552. doi:10.1093/nar/gkz833.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. doi:10.1093/nar/gkh340.
- Hojito S, Kobayashi N, Katakura H. 2010. Population structure of Aegialites beetles (Coleoptera, Salpingidae) on the coasts of Hokkaido, Northern Japan. Zoolog Sci. 27(9):723–728. doi:10.2108/zsj.27.723.
- Hu F-S, Pollock DA, Telnov D. 2020. Comparative morphology of immature Trictenotoma formosana Kriesche, 1919 and systematic position of the Trictenotomidae (Coleoptera, Tenebrionoidea). European J Taxon. 640:1–22. doi:10.5852/ejt.2020.640.
- Hunter JD. 2007. Matplotlib: a 2D graphics environment. Comput Sci Eng. 9(3):90–95. doi:10.1109/MCSE.2007.55.
- Kück P, Longo G. 2014. FASconCAT-G: extensive functions for multiple sequence alignment preparations concerning phylogenetic studies. Front Zool. 11(1):81. doi:10.1186/s12983-014-0081-x.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. doi:10.1093/molbev/msw054.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359. doi:10.1038/nmeth.1923.
- Liu H, Yuan L, Wang P, Pan Z, Tong J, Wu G, Yang Y. 2023. First record of osphya (Melandryidae: osphyinae) from Chinese mainland based on morphological evidence and mitochondrial genome-based phylogeny of tenebrionoidea. Diversity. 15(2):282. doi:10.3390/d15020282.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534. doi:10.1093/molbev/msaa015.
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27(5):824–834. doi:10.1101/gr.213959.116.
- Sheng L, Zhou T, Shi Z, Pan X, Weng X, Ma J, Wu S. 2021. The complete mitochondrial genome of Trictenotoma davidi Deyrolle, 1875 (Coleoptera: Trictenotomidae). Mitochondrial DNA B Resour. 6(7):2026–2027. doi:10.1080/23802359.2021.1942266.
- Smith AD, Kamiński MJ, Kanda K, Sweet AD, Betancourt JL, Holmgren CA, Hempel E, Alberti F, Hofreiter M. 2021. Recovery and analysis of ancient beetle DNA from subfossil packrat middens using high-throughput sequencing. Sci Rep. 11(1):12635. doi:10.1038/s41598-021-91896-8.
- Spilman TJ. 1967. The heteromerous intertidal beetles (Coleoptera: salipingidae: aegialitinae). Pacific Insects. 9(1):1–21.
- Zerche L. 2004. Revision der Gattung Aegialites Mannerheim (Coleoptera: salpingidae: aegialitinae). Stuttgarter Beiträge Zur Naturkunde, Serie A (Biologie). 666:1–116.
- Zheng S, Poczai P, Hyvönen J, Tang J, Amiryousefi A. 2020. Chloroplot: an online program for the versatile plotting of organelle genomes. Front Genet. 11:576124. doi:10.3389/fgene.2020.576124.
