Abstract
Ammodendron bifolium, a rare deciduous shrub, is the only species of Ammodendron (Fabaceae) in China, which distributes in Huocheng county, Xinjiang. This study employed high-throughput sequencing technology to assemble the complete chloroplast genome sequence of A. bifolium. The entire length of chloroplast genome is 154,426 bp. It comprises 128 genes, which include 85 protein-coding genes, 35 tRNA genes, and 8 rRNA genes. The A. bifolium chloroplast genome has a GC content of 36.41%. Phylogenetic analysis strongly supported that A. bifolium is sister to the members of the Sophora genus. This study will provide the genetic information data for further phylogenetic studies of Ammodendron.
Introduction
Ammodendron bifolium (Pall.) Yakovlev, Citation1972, is a rare deciduous shrub and is the only species of Ammodendron (Fabaceae) in China (Yakovlev et al. 1996). The distribution of this species is only located in Tukai Desert in Huocheng county, Xinjiang, China. This species usually inhabits extremely arid fixed and semi-fixed deserts. Because of the narrow distribution, habitat destruction and the poor natural renewal ability of A. bifolium, it has been classified as a national second-class protected plant in the Information System of Chinese Rare and Endangered Plants (ISCRPE) (http://www.iplant.cn/rep/prot/Ammodendronbifolium). Meanwhile, due to its drought resistance, it can play an important ecological role as a windbreak and sand-fixation plant (Li et al. Citation2013a, Citation2013b) (). However, the chloroplast genome of A. bifolium has not been published yet. In this study, we generated the complete chloroplast genome sequence of A. bifolium based on the Illumina paired-end sequencing data, which would be beneficial for further protection and phylogenetic studies on this species.
Figure 1. Ammodendron bifolium: Vegetative morphology. A. Plant; B. Fruits (pods flat, long, and round lanceolate); C. Stipular thorns (the stipules are thorn-like, the leaflets are arranged in an opposite pattern, and both surfaces of the leaflets are covered with silvery-white sericeous hairs). Photo taken by Xiaojun Shi in Huocheng County.

Materials and methods
Fresh leaves of A. bifolium were collected from the Tukai Desert in Huocheng, Xinjiang Province, China (44°1′N, 80°44′E), and immediately dried using silica gel. The voucher specimen (No. SXJ160) is preserved in the Herbarium of Xinjiang Agricultural University (Contact: Xiaojun Shi, [email protected]). The collection of plant specimens complies with the Regulations on the Protection of Wild Plants of the People’s Republic of China. Following DNA extraction (Doyle and Doyle Citation1987), genome sequencing was conducted on the Illumina HiSeq Platform at Genepioneer Biotechnologies Inc, Nanjing, China. The sequencing yielded approximately 7.63 GB of clean data with an average sequencing depth of 2178.71× (Figure S1). The trimmed reads were mainly assembled using SPAdes (Bankevich et al. Citation2012). Subsequently, the assembled genome was annotated using CpGAVAS (Lohse et al. Citation2007) and visualized via OGDRAW (Liu et al. Citation2012). CPGview (Liu et al. Citation2023) was employed to generate cis-spliced and trans-spliced genes, as illustrated in Figures S2 and S3.
Figure 2. Chloroplast genome maps of Ammodendron bifolium. Genes situated on the inner side of the circle undergo transcription in a clockwise direction, while those on the outer side are transcribed in a counterclockwise direction. The inner circle, represented in dark gray, corresponds to the GC content, whereas the light gray denotes the content. Diverse colors symbolize different functional genes. The pronounced line on the large circle delineates the boundaries of the inverted repeat regions (IRa and IRb), segregating the genome into small single-copy (SSC) and large single-copy (LSC) regions.
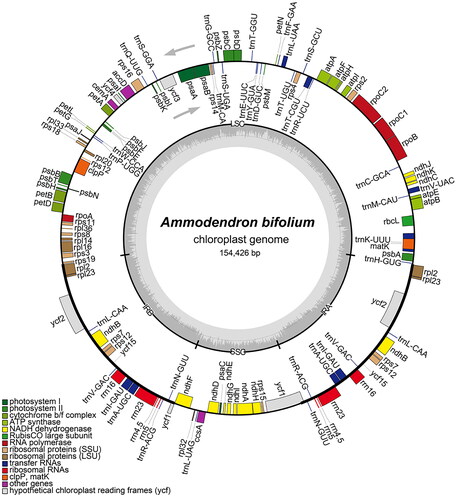
Figure 3. A Phylogenetic tree was reconstructed using the maximum likelihood (ML) method based on the complete chloroplast genome sequences of Ammodendron bifolium (shown in red) along with 18 other related species from the Fabaceae family and one species from the Rosaceae family. The numbers on the nodes represent bootstrap values, derived from 1000 replicates. The following sequences were used: Malus sieversii (Accession number: NC_042192), Ormosia nuda (Accession number: MW450912), Ormosia fordiana (Accession number: MW752862), Ormosia hosiei (Accession number: NC_039418), Ormosia bahiensis (Accession number: MW628939), Salweenia bouffordiana (Accession number: MF449303), Ammopiptanthus nanus (Accession number: MW349015), Ammopiptanthus mongolicus (Accession number: MK704436), Thermopsis lanceolata (Accession number: MN841458), Thermopsis alpina (Accession number: MN841457), Piptanthus nepalensis (Accession number: MN841455), Piptanthus concolor (Accession number: MN841454), Sophora alopecuroides (Accession number: NC_036102), Sophora moorcroftiana (Accession number: NC_056151), Sophora davidii (Accession number: MN841456), Sophora tonkinensis (Accession number: MW376914), Sophora flavescens (Accession number: MH748034), Sophora toromiro (Accession number: NC_057682), Sophora macrocarpa (Accession number: NC_057683).
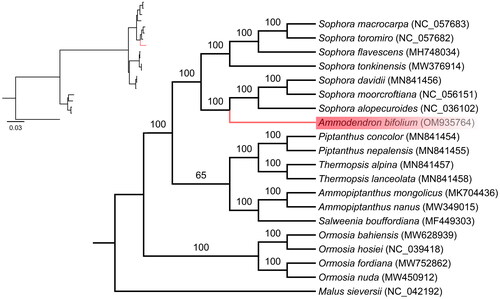
To determine the phylogenetic position of A. bifolium within the Fabaceae family, we obtained 19 complete chloroplast genomes of Fabaceae species from GenBank and used Malus sieversii, a member of the Malus genus in the Rosaceae family as the out-group. We aligned the complete chloroplast genome sequences of A. bifolium and the other 19 species using MAFFT (Katoh and Standley Citation2013) and performed phylogenetic analysis based on maximum likelihood using IQ-TREE 1.6.8 (Nguyen et al. Citation2015) with the TVM + F + I + G4 nucleotide substitution model, which was selected by ModelFinder (Kalyaanamoorthy et al. Citation2017).
Results and discussion
The whole chloroplast genome of A. bifolium (accession number: OM935764) has a length of 154, 426 bp and comprises a large single-copy region (LSC, 84, 486 bp), a small single-copy region (SSC, 17, 946 bp) and two inverted repeats (IRs, 25, 997 bp each) (). The LSC, SSC, and IR regions have GC contents of 33.91%, 30.07%, and 42.66%, respectively, resulting in an overall GC content of 36.41% for the A. bifolium chloroplast genome. The genome contains 128 functional genes, of which 85 are protein-coding genes, 35 are tRNA genes, and 8 are rRNA genes. Each of thirteen protein-coding genes (rpoC1, atpF, rps16, two rps12, petB, petD, rpl16, two rpl2, two ndhB, ndhA) and seven tRNA genes (trnK-UUU, trnV-UAC, trnT-CGU, trnL-UAA, two trnI-GAU, trnA-UGC) have one intron, while two protein-coding genes (ycf3 and clpP) have two introns.
To investigate the phylogenetic relationship between A. bifolium and its related taxa, we obtained the complete chloroplast genomes of 19 other closely related species from the NCBI GenBank database. Subsequently, a maximum likelihood (ML) analysis was performed to construct a phylogenetic tree (). Phylogenetic analysis revealed that A. bifolium was clustered with Sophora alopecuroides, Sophora moorcroftiana and Sophora davidii, with Ammodendron being nested within the Sophora clade. These findings provide support for the non-monophyletic nature of the Sophora genus, which is consistent with the previously research about the phylogenetic relationships among genera in Fabaceae, as determined by using both nrDNA and plastid DNA regions (Duan et al. Citation2019). Further research is required to determine the phylogenetic relationship between Ammodendron and Sophora.
Conclusions
The complete chloroplast genome of Ammodendron bifolium reported for the first time in our study. It had a typical circular structure composed of 154,426 bp and 128 genes. Based on molecular phylogenetic analysis using 20 complete chloroplast genome, we provided evidence that the Sophora genus were the non-monophyletic nature, as A. bifolium was closely related to Sophora in the tribe Sophoreae and nested within the Sophora clade. The published chloroplast genome of A.bifolium will provide the genetic information data for further phylogenetic studies of Ammodendron.
Ethical approval
The data collection was conducted in accordance with the policies of the International Union for Conservation of Nature (IUCN) pertaining to research on species at risk of extinction (refer to the Guidelines for appropriate uses of IUCN Red List data). Additionally, the Convention on Biological Diversity and the Convention on the Trade in Endangered Species of Wild Fauna and Flora were also adhered to.
Authors’ contributions
The study was conceptualized and designed by Xiaojun Shi and Hongxiang Zhang. The plant material was collected and identified by Juan Qiu. Haowen Tian and Xinyu Zhu contributed to the assembly and annotation of the chloroplast genome, conducted the phylogenetic analysis, and participated in manuscript preparation. All authors have approved the final version for publication and have agreed to be accountable for all aspects of the work.
Supplemental Material
Download MS Word (181.8 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number OM935764. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA845065, SRR19537695, and SAMN28854403, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. doi:10.1089/cmb.2012.0021.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- Duan L, Harris AJ, Ye W, Deng SW, Song ZQ, Chen HF, Wen J. 2019. Untangling the taxonomy of the Cladrastis clade (Leguminosae: Papilionoideae) by integrating phylogenetics and ecological evidence. Taxon. 68(6):1189–1203. doi:10.1002/tax.12155.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi:10.1038/nmeth.4285.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Li J, Huang LP, Lv HY, Wang XA. 2013a. Distribution patterns of main populations of Ammodendron argenteum communities in the Takeermohuer Desert. Arid Zone Res. 30:634–639.
- Li J, Wang XA, Lv HY, Wang SX. 2013b. Interspecific relationships of major species in Ammodendron argenteum communities of Takeermohuer Desert. Guihaia. 33:482–487.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13(1):715. doi:10.1186/1471-2164-13-715.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi:10.1111/1755-0998.13729.
- Lohse M, Drechsel O, Bock R. 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52(5–6):267–274. doi:10.1007/s00294-007-0161-y.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi:10.1093/molbev/msu300.
- Yakovlev GP, Sytin AK, Roskov YR. 1996. Legumes of Northern Eurasia. A checklist: 1–724. Royal Botanic Gardens, Kew.
