Abstract
We sequenced and published the chloroplast genome of Commelina caroliniana Walter, which was previously misidentified as C. diffusa owing to their morphological similarities until 1989. The genome of C. caroliniana is 160,857 bp long [large single copy region: 88,064 bp; a small single copy region: 18,549 bp; two inverted repeat regions: 27,122 bp] and has a GC content of 35.7%. The genome comprises 133 genes, including 87 coding sequences (CDSs), 38 tRNAs, and eight rRNAs. The phylogenetic relationship between C. caroliniana and related species was analyzed using the maximum likelihood method based on the 79 CDSs of the chloroplast genome. Phylogenetic analysis revealed that C. caroliniana is closely related to C. communis. Our findings will contribute to studies on species identification and phylogenetic and evolutionary research. These results enhance our understanding of the Commelina genus.
Introduction
Commelina caroliniana Walter, 1788, is a well-known invasive alien species in the United States (Faden Citation1989; Faden Citation1993; Kang et al. Citation2021) (). However, until 1989, it was recognized as the same species as the more commonly encountered C. diffusa Burm.f., with a wider distribution range than C. caroliniana, because of their morphological similarities (Faden Citation1989; Faden Citation1993; Kang et al. Citation2021; Plants of the World Online Citation2023). While C. caroliniana has distinguishable features in the forms of inflorescence, flowers, and seeds from those of C. diffusa, these characteristics are difficult to discern and can be easily overlooked, leading to occasional misidentification.
Figure 1. Photographs of the Commelina caroliniana. (A) The plant has a stem that creeps and spreads diffusely on the ground, rooting at the nodes; (B) Involucral bract is falcate and usually ciliate basally, ciliate on peduncle; (C) Flower has three steminodes with developed antherode; (D) Staminode has yellow antherode, usually with a brown spot in its center; (E) Seed testa is smooth to slightly alveolate. All photographs were taken by Eun Su Kang in Seogwipo-si, Jeju-do, South Korea.

Molecular methods, including the creation and application of DNA barcoding or primer markers, are valuable tools for ascertaining the identity of species that are challenging to be identified morphologically (Nock et al. Citation2011; Li et al. Citation2015). Additionally, molecular taxonomic investigations involving a greater number of species can provide valuable insights for the identification of species within Commelina L. and understanding the dynamic interrelationship with its related taxa (Nock et al. Citation2011; Li et al. Citation2015; Li et al. Citation2016). This can enhance our understanding of the taxonomic status of Commelina.
The incorporation of chloroplast genomes plays a crucial role (Nock et al. Citation2011; Li et al. Citation2015; Li et al. Citation2016; Daniell et al. Citation2016). However, in the genus Commelina, chloroplast genomes have been reported only for two taxa: C. communis L. and C. benghalensis L. (Cui and Liang Citation2019). Given this limitation, the present study aimed to analyze the chloroplast genome sequence of C. caroliniana. The findings of our study aim to provide foundational data for future studies on the identification, evolution, and phylogenetic classification of species within the genus Commelina and its related taxa.
Materials and methods
C. caroliniana was identified and collected by Eun Su Kang in November 2021 from Sagye-ri, Andeok-myeon, Seogwipo-si, Jeju-do, South Korea (33°14'24.8" N, 126°17'41.1" E). The specimen was deposited at the Korea National Arboretum (KH) (http://www.nature.go.kr, Dong Chan Son, e-mail: [email protected], Voucher number: Sagyeri211101-1). DNA was extracted from the dried leaves of the samples using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol provided with the kit. High-purity gDNA was isolated from the extracted DNA on a 2% agarose gel, which was then sequenced on an Illumina MiSeq platform with a 301 bp insert size. A total of 7,438,888 reads were acquired. The reads were merged using NOVOWrap v.1.20 (Wu et al. Citation2021), with reference to C. benghalensis (NC072999) registered in the NCBI database. To confirm the accuracy of the assembly, we utilized Bandage v.0.8.1 to examine the structural characteristics of the combined chloroplast genome (Wick et al. Citation2015) (Figure S1), and the read coverage depth was verified using the Draw_SequencingDepth.py script provided by Ni et al. (Citation2023) (Figure S2). Finally, the genome sequence was annotated using GeSeq (Tillich et al. Citation2017), and the genome map was drawn using CPGView (http://www.1kmpg.cn/cpgview).
The phylogenetic relationship between C. caroliniana and related taxa was analyzed using the maximum likelihood (ML) method based on the coding sequences (CDSs) of the complete chloroplast genomes, all of which were registered with the NCBI. Along with C. caroliniana, a total of ten taxa were studied. Hanguana malayana (Jack) Merr. (NC029962), a member of the Hanguaceae family and closely related to Commelinaceae, was used as the outgroup in this study. Using Geneious v.8.0.5. (https://www.geneious.com), 79 CDSs were concatenated and aligned with MAFFT. PhyloSuite v.1.2.2 was used to test the analysis model (Zhang et al. Citation2020), and IQ-tree v.2.1.3 with the setting GTR + F+R3 model and 1,000 bootstrap replicates was used for ML analysis (Minh et al. Citation2020). The ML analysis result was visualized using FigTree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree).
Results
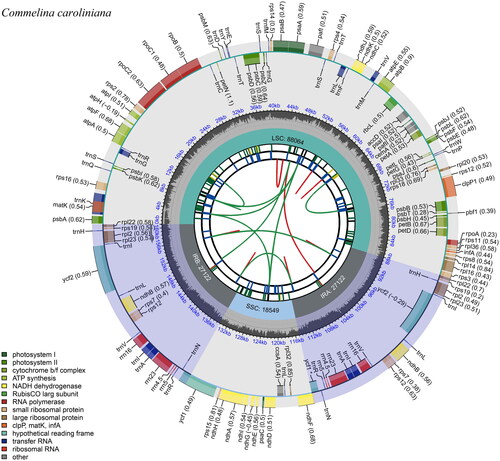
The complete chloroplast genome (GenBank accession no. OR936140) is 160,857 bp long, with a GC content of 35.7% (). The genome structure is divided into four segments, which include the large single copy (LSC) region, a small single copy (SSC) region, and two inverted repeat (IR) regions, commonly observed in angiosperms. The lengths of the LSC, SSC, and IR regions were 88,064 bp, 18,549 bp, and 27,122 bp, respectively. The genome contained a total of 133 genes, including 87 CDS, 38 tRNA, and eight rRNA genes. In the IR regions, there were eight CDS, eight tRNA, and four rRNA genes, totaling 20 genes (CDS: ndhB, rpl2, rpl22, rpl23, rps7, rps12, rps19, and ycf2; tRNA: trnA-UGC, trnH-GUG, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC; and rRNA: rrn4.5, rrn5, rrn16, and rrn23). The cis-splicing genes, atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, and rps16 have one intron, whereas clpP1and pafI have two introns (Figure S3). The trans-spliced gene, rps12, has one intron and spans the LSC and IR regions, with its 5′ end located in the LSC region and its 3′ end in the IR region (Figure S4).
Figure 2. Circular map of Commelina caroliniana. The map consists of six tracks. From the center to the outer part, the first track shows dispersed repeats connected by red and green arcs indicating the direction (forward and reverse, respectively). The second track shows long tandem repeats as blue bands, and the third track shows short tandem repeats or microsatellites as green bands. The fourth track represents the GC content along the plastome. Finally, the sixth track represents the genes as colored boxes, the inner boxes present clockwise transcription, and the outer boxes present counterclockwise transcribed genes.
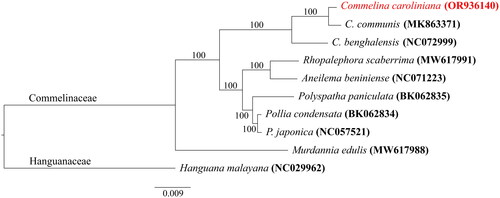
The phylogenetic relationship with closely related species of C. caroliniana was investigated using ML analysis (). The results showed that the Commelinaceae species formed a monophyletic clade, and Murdannia edulis (Stokes) Faden (MW617988) diverged first from the other Commelinaceae species within this clade. Subsequently, the three Commelina species formed a single cluster, diverging from the other Commelinaceae species [Aneilema beniniense (P.Beauv.) Kunth, Pollia condensate C.B.Clarke, P. japonica Thunb., Polyspatha paniculate Benth., and Rhopalephora scaberrima (Blume) Faden]. Within the Commelina group, C. benghalensis diverged first and C. caroliniana was closely related to C. communis.
Figure 3. Maximum likelihood phylogenetic tree of nine Commelinaceae species and the outgroup, one Hanguanaceae species based on concatenated CDSs of chloroplast genomes. Numbers on the nodes indicated the bootstrap proportion. The following sequences were used: OR936140, BK062834 (Jung et al. unpublished), BK062835 (Jung et al. in press), NC072999 (unpublished), NC071223 (unpublished), NC057521 (unpublished), MW617991 (Jung et al. Citation2021), MW617988 (Jung et al. Citation2021), MK863371 (Cui and Liang Citation2019), NC029962 (Barrett et al. Citation2016).
Discussion and conclusion
Commelina is the largest genus in the Commelinaceae family, with approximately 170 species worldwide (Gajurel and Shrestha Citation2010; Kaul and Koul Citation2012; Nandikar and Gurav Citation2018; Hassemer Citation2019). Commelina is well known for its persistent nomenclature and taxonomy issues, with several taxa presenting challenges that remain unresolved to date (Faden Citation1989; Hassemer Citation2019). Even with unresolved taxonomic issues, new species of Commelina have been reported, exacerbating confusion for the identification of Commelina species (Hassemer Citation2019; Kang et al. Citation2021). To address these issues, various taxonomic studies, including nomenclatural, morphological, and phylogenetic analyses, are essentially required. This multidisciplinary approach is crucial for enhancing our understanding of this genus.
The present study elucidates the nucleotide sequence of the chloroplast genome of C. caroliniana and ML analysis based on CDSs revealed that C. caroliniana is closely related to C. communis. These findings can provide foundational data and contribute to the resolution of taxonomic issues within the Commelina genus. Furthermore, considering that this species was previously misidentified as C. diffusa, and given the current limitations in available data, the scarcity of information further emphasizes the considerable significance of this study.
Authors’ contributions
E. S. Kang collected photographs, specimens, and samples of C. caroliniana and wrote the paper; S.C. Kim conducted the experiments and analyzed the data. S. R. Lee and B. K. Park created the figures presented in the paper. D. C. Son reviewed the paper, provided critical feedback to revise the content, and was responsible for the final approval of the version to be published. All authors contributed to this study and have read and agreed to the published version of the manuscript.
Ethical approval
The materials used in this study were not included in the IUCN Red List, and the collection area was not protected. This study was conducted in compliance with the Act on the Creation and Furtherance of Arboretums and Gardens.
Supplemental Material
Download MS Word (603.4 KB)Supplemental Material
Download JPEG Image (1.2 MB)Supplemental Material
Download JPEG Image (1.1 MB)Supplemental Material
Download JPEG Image (1.4 MB)Supplemental Material
Download JPEG Image (1.1 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The complete chloroplast genome sequence data for C. caroliniana can be found in GenBank [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/), and the accession numbers are No. OR936140. The associated BioProject, Bio-Sample, and SRA numbers were PRJNA1050870, SAMN38756118, and SRR27270114, respectively.
Additional information
Funding
References
- Barrett CF, Baker WJ, Comer JR, Conran JG, Lahmeyer SC, Leebens‐Mack JH, Li J, Lim GS, Mayfield-Jones DR, Perez L, et al. 2016. Plastid genomes reveal support for deep phylogenetic relationships and extensive rate variation among palms and other commelinid monocots. New Phytol. 209(2):855–870. doi:10.1111/nph.13617.
- Cui Y, Liang R. 2019. The chloroplast genome sequence of Commelina communis (Commelinaceae). Mitochondrial DNA Part B Resour. 4(2):2631–2632. doi:10.1080/23802359.2019.1642153.
- Daniell H, Lin CS, Yu M, Chang WJ. 2016. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17(1):134. doi:10.1186/s13059-016-1004-2.
- Faden RB. 1989. Commelina caroliniana (Commelinaceae): a misunderstood species in the United States is an old introduction from Asia. Taxon. 38(1):43–53. doi:10.2307/1220885.
- Faden RB. 1993. The misconstrued and rare species of Commelina (Commelinaceae) in the eastern United States. Ann Missouri Bot Gard. 80(1):208–218. doi:10.2307/2399824.
- Gajurel JP, Shrestha KK. 2010. Taxonomy of the genus Commelina Plum. ex L. (Commelinaceae) in Nepal. Botanica Orientalis. 6:25–31. doi:10.3126/botor.v6i0.2907.
- Hassemer G. 2019. Further advances to the nomenclatural, taxonomic and geographic knowledge of the New World Commelina (Commelinaceae): toward a continental treatment. Phytotaxa. 400(3):89–122. doi:10.11646/phytotaxa.400.3.1.
- Jung J, Kim C, Kim JH. 2021. Insights into phylogenetic relationships and genome evolution of subfamily Commelinoideae (Commelinaceae Mirb.) inferred from complete chloroplast genomes. BMC Genomics. 22(1):231. doi:10.1186/s12864-021-07541-1.
- Kang ES, Lee KH, Son DC. 2021. An overlooked invasive alien plant of Jejudo Island: Commelina caroliniana (Commelinaceae). Korean J Pl Taxon. 51(1):10–17. doi:10.11110/kjpt.2021.51.1.10.
- Kaul V, Koul AK. 2012. Staminal variation and its possible significance in Commelina benghalensis L. and Commelina caroliniana Walter. Curr Sci. 103(4):419–426. https://www.jstor.org/stable/24085091.
- Li B, Cantino PD, Olmstead RG, Bramley GL, Xiang CL, Ma ZH, Tan YH, Zhang DX. 2016. A large-scale chloroplast phylogeny of the Lamiaceae sheds new light on its subfamilial classification. Sci Rep. 6(1):34343. doi:10.1038/srep34343.
- Li X, Yang Y, Henry RJ, Rossetto M, Wang Y, Chen S. 2015. Plant DNA barcoding: from gene to genome. Biol Rev Camb Philos Soc. 90(1):157–166. doi:10.1111/brv.12104.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534. doi:10.1093/molbev/msaa015.
- Nandikar MD, Gurav RV. 2018. A new species of Commelina (Commelinaceae) from India. Webbia. 73(2):233–237. doi: 10.1080/00837792.2018.1540743.
- Ni Y, Li J, Zhang C, Liu C. 2023. Generating sequencing depth and coverage map for organelle genomes. doi:10.17504/protocols.io.4r3l27jkxg1y/v1.
- Nock CJ, Waters DL, Edwards MA, Bowen SG, Rice N, Cordeiro GM, Henry RJ. 2011. Chloroplast genome sequences from total DNA for plant identification. Plant Biotechnol J. 9(3):328–333. doi:10.1111/j.1467-7652.2010.00558.x.
- Plants of the World Online. 2023. Website: Plants of the World Online (accessed November 21, 2023. Available at http://www.plantsoftheworldonline.org.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. doi:10.1093/nar/gkx391.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352. doi:10.1093/bioinformatics/btv383.
- Wu P, Xu C, Chen H, Yang J, Zhang XC, Zhou SL. 2021. NOVOWrap: an automated solution for plastid genome assembly and structure standardization. Mol Ecol Resour. 21(6):2177–2186. doi:10.1111/1755-0998.13410.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355. doi:10.1111/1755-0998.13096.
