Abstract
The worldwide distributed neogastropod family Muricidae comprises more than 1800 extant species of whelks, rock-shells, murex-shells, drill-shells, and coral-shells. Despite several attempts at a taxonomic revision based on morphological characters, the systematics of this family is still largely debated. Here, we present a molecular revision of the family Muricidae based on the largest dataset analysed so far, which comprises 384 specimens representing 360 species and includes, for the first time, all the currently recognised subfamilies. A molecular dataset of cox1, 12S rRNA, 16S rRNA, and 28S rRNA genetic markers and six fossil-based calibration points were used to produce time-calibrated phylogenetic reconstructions, using a Maximum Likelihood approach. Our results confirmed the monophyly of most of the accepted subfamilies, suggested a revision of the taxonomic composition of Muricopsinae and Muricinae, and highlighted some lineages not immediately comprised in any of the recognised subfamilies. The origin and early diversification of the subfamilies of Muricidae occurred between 32 and 60 million years ago.
Introduction
The neogastropod family Muricidae is one of the most species-rich families of Gastropoda, with an estimate of more than 1800 extant species of whelks, rock-shells, murex-shells, drill-shells, and coral-shells (MolluscaBase eds. Citation2023). The family is distributed worldwide in all oceans, from the lower intertidal down to more than 6000 m (Sysoev Citation1992) and all its members are carnivores, mostly predators with a varying degree of trophic specialisation, from generalists to highly specialised. Muricids have been known to man since ancient times: Mediterranean species were used by the Phoenicians to produce their Tyrian purple dye, and Greeks, Arabians, and Chinese employed muricid species for pharmacological use (Benkendorff et al. Citation2015). Nowadays, some rock shells have economic relevance, either since they are consumed as food (e.g. species of Murex, Concholepas, Hexaplex, Bolinus, and Chicoreus) or being pests of commercial oyster farms (Buhle & Ruesink Citation2009).
The classification of the family was repeatedly revised in the last century based on conchological features of extant and fossil taxa and on radular characters (Cossmann Citation1903; Thiele Citation1929; Keen Citation1971; Radwin & D’Attilio Citation1971; Ponder & Warén Citation1988; Vokes Citation1996; Bouchet & Rocroi Citation2005), while only a single comprehensive attempt at building a molecular phylogenetic framework has been performed to date (Barco et al. Citation2010), along with a few other works at the subfamily level that almost invariably resulted in significant changes (Oliverio & Mariottini Citation2001; Claremont et al. Citation2008, Citation2013; Barco et al. Citation2012, Citation2015, Citation2016). More recently, muricid mitogenomes have been used to address phylogenetic relationships among muricid subfamilies, by Harasewych and Sei (Citation2022) with nine mitogenomes representing six subfamilies, and Yu et al. (Citation2023) with 24 mitogenomes representing five subfamilies.
The classification of muricids is therefore still debated, especially regarding the familial or subfamilial ranking of some groups. A first endeavour by Cossmann (Citation1903) divided the superfamily Muricoidea (as ‘“cénacle” Muricacea) into three families based on shell morphology (Muricidae, Purpuridae, and Coralliophilidae), and recognized five subfamilies of Muricidae based on opercular differences (Muricinae: apical nucleus; Ocenebrinae: lateral nucleus; Trophoninae: sublateral nucleus; Typhinae: apical nucleus, shell with anal tube; Rapaninae: purpurid operculum, muricid-like shell). In recent time, modern recognition of muricid subfamilies (e.g. Bouchet & Rocroi Citation2005) is still largely based on radular differences, identifying a single family, Muricidae, containing 11 subfamilies: Muricinae Rafinesque, 1815, Muricopsinae Radwin & D’Attilio, Citation1971, Ocenebrinae Cossmann, Citation1903, Trophoninae Cossmann, Citation1903, Typhinae Cossmann, Citation1903, Tripterotyphinae D’Attilio & C. M. Hertz, 1988, Ergalataxinae Kuroda, Habe & Oyama, 1971, Rapaninae Gray, 1853, Haustrinae K. S. Tan, Citation2003, Aspellinae Keen, Citation1971, and Coralliophilinae Chenu, 1859. One additional subfamily was more recently recognised: Pagodulinae Barco, Schiaparelli, Houart, Oliverio, Citation2012 (Barco et al. Citation2010, Citation2012). This 12-subfamilies scheme was recently adopted by Houart (Citation2018) and Merle et al. (Citation2022).
In the present study, we investigated the phylogeny of the family Muricidae based on the largest dataset analysed so far, including, for the first time, all the currently recognised subfamilies. The phylogenetic reconstructions were also time-calibrated with several fossil records to produce a robust framework for further studies of the biology and the evolution of this family.
Materials and methods
Dataset
The molecular analysis was based on sequences of four molecular markers: one nuclear (28S rDNA) and three mitochondrial (cox1, 12S rDNA, 16S rDNA), including sequences newly produced for this work as well as sequences retrieved from the GenBank (Supplementary Materials Table S2).
For the newly produced sequences (170), DNA was extracted from tissue samples at the Service de Systématique Moléculaire (UAR 2AD, MNHN, Paris) using the Epmotion 5075 robot (Eppendorf), following the manufacturer’s recommendations, or at the Department of Biology and Biotechnologies “Charles Darwin” of Sapienza University of Rome, with standard phenol/chloroform (Oliverio & Mariottini Citation2001) or “salting out” protocols (Fassio et al. Citation2022). Polymerase chain reactions (PCR) were performed with 1–3 μL of DNA template in a 25 μL reaction volume, including 2.5–3 μL of 10 × NH4 reaction buffer, 2.5–3 μL of 50 mM MgCl2 solution, 0.15–0.2 μL of BIOTAQ DNA polymerase, 0.4 μL of each 25 pM primer solution, 1 μL of 10% bovine serum albumin solution, and 0.5 μL of 10 mM nucleotide mix solution (Fassio et al. Citation2022). The PCR conditions were as follows: initial denaturation (4 min at 94°C); 35 cycles of denaturation (30 s at 94°C), annealing (40 s at 48–52°C for cox1 and 16S; 40 s at 58–62°C for 28S; 60 s at 58–66°C for 12S) and extension (1 min at 72°C); and final extension (10 min at 72°C). Primers used to amplify the selected markers are reported in Table S1. The PCR products were purified using ExoSAP-IT (USB Corporation) and sequenced at Macrogen, Inc. or by the Eurofins sequencing facility.
Additionally, 3,980 sequences of the barcode markers cox1, 911 of the 16S, 806 of the 12S, and 832 of the 28S, were retrieved from the GenBank.
Sequences were aligned either using the Geneious R7 algorithm (Kearse et al. Citation2012) (cox1) or with the software MAFFT (Kuraku et al. Citation2013; Katoh et al. Citation2019), choosing the Q-INS-I algorithm (12S, 16S, and 28S). The hypervariable regions of the 12S, 16S, and 28S alignments were excluded from the analysis after identification through the software Gblocks (v. 0.91b, Castresana Citation2000 – all options set for the least stringent selection). A concatenated dataset was assembled with SequenceMatrix (Vaidya et al. Citation2011).
Taxonomic identification of every specimen was based on morphological examination of each voucher, where available. The identity of those specimens for which that was not possible was checked – as far as possible – by their position in the cox1 single-gene trees (see below for methods details) preferably in relation to pedigreed sequenced vouchers from publicly available collections included in the dataset. Single gene trees were used to discard contaminations, sequencing artefacts and redundant identical sequences, to have a single or few representatives for each species.
Thereafter, the final dataset was defined through a selection of sequences that maximised the taxonomic coverage of the family and the gene coverage of each species, removing redundancy. Four species of Buccinoidea (Oliverio & Modica Citation2010) were included in the dataset as an outgroup for rooting the trees: Buccinum undatum Linnaeus, 1758, Kelletia lischkei Kuroda, 1938, Penion ormesi (Powell, 1927), Serratifusus lineatus Harasewych, 1991.
Phylogenetic analyses and temporal calibration
The substitution model for each partition (12S, 16S, 28S, and 1st, 2nd, and 3rd codon positions of cox1) was chosen with PartitionFinder 2 (Lanfear et al. Citation2016). Uncalibrated phylogenetic analyses were performed by ML on single gene datasets with IQTree v2.0.3 (Nguyen et al. Citation2014).
The concatenated dataset along with six calibration points were used to produce time-calibrated trees to estimate the node ages of each clade of the family Muricidae using a Maximum Likelihood (ML) approach. We identified the six calibration points for the phylogenetic trees of Muricidae, based on the most reliable fossil data (). The first appearance of the family is witnessed in the Upper Cretaceous of Texas (Merle et al. Citation2011) with the earliest known species attributed to Muricidae (1), Paziella (Flexopteron) cretacea (Garvie, 1991) from the Maastrichtian (c. 70 million years ago, mya); the family was certainly not present before the Albian (Lower Cretaceous, 112 Mya), which was set as the lower bound (Barco et al. Citation2012). The fossil record of Typhinae (2) places the first certain appearance of the subfamily (quite typical morphologically) in the Lower Eocene (Ypresian) (MHNH collection) based on the occurrence of Typhis tubifer (Bruguière, 1792) and on Laevityphis muticus from the lower Ypresian of England and France (Merle et al. Citation2022). Fossils belonging to the subfamily Ocenebrinae (3) are common in the lower Miocene, and the earliest known species is Ocinebrina rarisulcata from the Middle Eocene (Bartonian, 37.8–41.2 mya) (Merle et al. Citation2022). The genus Nucella (Ocenebrinae) (4), has its first documented record in the lower Miocene (Aquitanian, c. 22.5 mya) (Collins et al. Citation1996). For the genus Murex, fossil records of Murex trapa (5) appeared during the Pliocene of Java (Ponder & Vokes Citation1988). The genus Timbellus (6) has the first documented appearance in the Lower Eocene (Danian, 61.6–66 mya) (Merle et al. Citation2022).
Table I. Date intervals (95% confidence range in mya) obtained from least-squares ML analyses (by IQTree) for selected major nodes, along with fossil records used as calibration points. *based on Douglas et al. (Citation2014); samples from Seymour Island suggested to be older (45.8–58.4 mya) based on Montes et al. (Citation2019).
A Bayesian inference approach was attempted using the software BEAST 1.8.0 (Drummond et al. Citation2012) but, despite numerous and long runs, none of the analyses reached convergence. ML time calibrated analyses were performed with the software IQTree v2.0.3 (Nguyen et al. Citation2014) and LSD21 (To et al. Citation2016) implemented in R (version 4.2.1, R Core Team Citation2022). We estimated divergence times with this distance-based calibration method implementing the least-squares dating criterion (–date). We set tips to the present time (–date-tip 0) and calculated a confidence interval based on 100 iterations (–date-ci 100), using the same dataset partition of the not-dated phylogenetic analysis. The default settings employed a birth-death tree prior in combination with a clock.rate = 0 and a ucld.mean value of 0.05. Ultrafast bootstraps (Ufb: 10000 replicates; Hoang et al. Citation2017) were performed, with 25% samples of burn-in, to evaluate the support of tree branches.
Phylogenetic trees were drawn using FigTree v. 1.4.4 (Rambaut Citation2018). For all the phylogenetic analyses, nodes with ultrafast Bootstraps (Ufb) ≥95% were considered as statistically supported.
Phylogenetic analyses were run on the Plateforme de Calcul Intensif et Algorithmique PCIA (UAR2700 2AD, MNHN), on the CIPRES Science Gateway (Miller et al. Citation2010), and on Terastat2 (Department of Statistical Science, Sapienza University of Rome; Bompiani et al. Citation2020).
Results
After quality testing all the sequences, checking for consistency and redundancy, and assessing a taxonomic ID to each sequence, we eventually selected the final dataset to maximise the number of represented species. The final dataset was composed of 384 muricid specimens, representative of 360 species, 80 of which type species (Supplementary Material Table S2 and Figure S1), and included 170 newly produced sequences. The final combined alignment after Gblocks was 3179 bp long, of which 455 bp for the 12S, 649 bp for the 16S, 1417 bp for the 28S and 658 bp for the cox1.
The substitution models found by Partition Finder 2 for each partition of our dataset are shown in Table S3.
Phylogenetic reconstruction
The single gene (Supplementary Material Figures S1–S4) and combined dataset ( and ) phylogenetic analyses yielded broadly congruent trees for the major supported nodes.
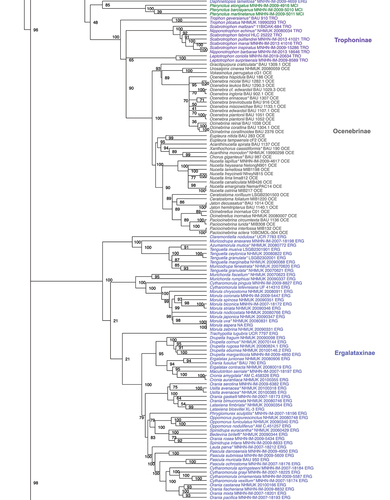
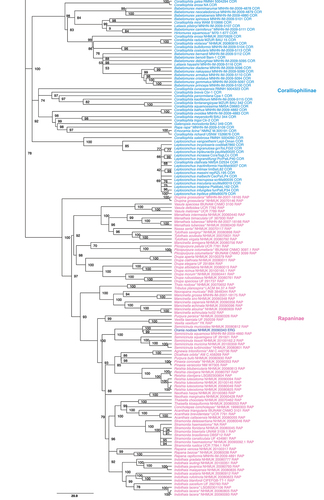
Figure 1. Maximum Likelihood phylogenetic reconstruction of the family Muricidae using IQTree. Values at nodes indicate ultrafast bootstrap support. Three letters at the end of each specimen’s label along with colours (as in ) indicate the current subfamily assignation. Type species are indicated with an * at the end of the name.
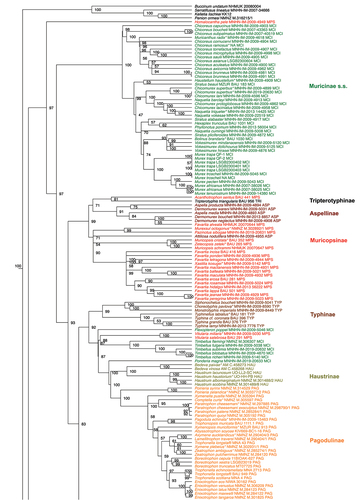
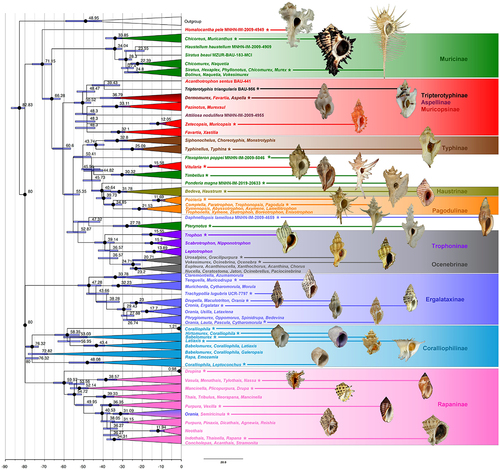
Figure 2. Maximum Likelihood time calibrated phylogenetic reconstruction of the family Muricidae using IQTree. Black dots at nodes indicate ultrafast bootstrap support values ≥ 0.95. Bars at nodes indicate 95% confidence intervals of ages expressed in mya. Branches are largely collapsed (fully uncollapsed tree in Supplementary Material Figure S5). Colours of collapsed clades, taxon names, and figure-connecting lines indicate the traditional subfamilial classification of the taxa represented in the relevant lineage (as also indicated in the subfamily labelling). Shells, not to scale, of representative taxa (indicated by an asterisk), from top/left: Homalocantha pele; Muricanthus radix; Murex pecten; Tripterotyphis fayae; Aspella producta; Muricopsis cristata; typhina coronata; Flexopteron poppei; Vitularia miliaris; Timbellus richeri; Ponderia magna; Haustrum lacunosum; Poirieria zelandica; Pagodula echinata; Daphnellopsis lamellosa; Pterynotus elongatus; Trophon geversianum; Gracilipurpura craticulata; Ocenebra hispidula; Muricodrupa fenestrata; Trachypollia lugubris; Orania fusulus; Ergalatax junionae; Cytharomorula vexillum; Coralliophila galea; Coralliophila violacea; Babelomurex cariniferus; Latiaxis pilsbryi; Leptoconchus peroni; Drupina grossularia; Nassa serta; Drupa ricinus; Vexilla vexillum; Semiricinula muricoides; Rapana bezoar.
Fully supported nodes corresponding to subfamilies were as follows: Coralliophilinae (UfB 100%), Rapaninae (UfB 100%), Ocenebrinae (UfB 100%), Haustrinae (UfB 100%), Typhinae (UfB 99%), and Pagodulinae (UfB 100%).
The subfamily Ergalataxinae was recovered as monophyletic (UfB 100%) with two exceptions: Daphnellopsis lamellosa, which was the sister taxon to the clade of Pterynotus (UfB 85%), formerly ascribed to Muricinae; and Orania nodosa that ended up inside the rapanine clade (UfB 100%).
A large clade of core Muricinae (UfB 100%) was defined, which included species of the type genus Murex, as well as of the genera Chicoreus, Muricanthus, Haustellum, Hexaplex, Naquetia, Chicomurex, Phyllonotus, Siratus, Bolinus, Vokesimurex. The genera Timbellus (UfB 100%) and Pterynotus (UfB 100%) and the species Flexopteron poppei and Ponderia magna, traditionally in Muricinae, resulted in four distinct lineages, even if their relationships with other nearby clades were not supported, with the exception of Ponderia magna that was the sister lineage to the clade Haustrinae + Pagodulinae (UfB 100%).
The subfamily Muricopsinae as traditionally conceived, did not form a clade. The clade including the type genus Muricopsis (UfB 99%), as well as the genera Acanthotrophon, Favartia, Murexsul, Pazinotus, Zetecopsis, and Xastilia, included also the genera Attiliosa, Aspella, and Dermomurex (formerly classified in the subfamily Aspellinae), and Tripterotyphis triangularis (formerly in the Tripterotyphinae). Conversely, the genus Vitularia – traditionally included in the Muricopsinae – formed a distinct lineage (UfB 100%) as did also the single specimen of Homalocantha pele.
The subfamily Trophoninae was paraphyletic and divided into three subsequent lineages: Trophon (UfB 100%), Scabrotrophon + Nipponotrophon (UfB 100%), and Leptotrophon (UfB 100%). The subfamily Ocenebrinae was supported as monophyletic (UfB 100%), as also was a clade including ocenebrines+trophonines (UfB 100%).
Overall, two supported major phylogenetic clades can be identified: one including Ergalataxinae, Coralliophilinae, and Rapaninae (Ufb 98%) and another including the rest of the subfamilies and lineages, with Homalocantha pele as the first diverging lineage (UfB 97%). Internal relationships among major lineages were generally not supported except for the clade of Ocenebrinae with the paraphyletic trophonine lineages and the pair Haustrinae + Pagodulinae (UfB 100%).
Several genera as traditionally conceived did not prove monophyletic. For example, within Ergalataxinae, the genus Cytharomorula resulted to be polyphyletic and split into one clade of four species including the type species Cytharomorula vexillum (UfB 100%), and two lineages represented by Cytharomorula pinguis and Cytharomorula lefevreiana, subsequently sisters to the genus Morula (both nodes UfB 100%). The genus Orania was split into seven clades with the type species Orania fusulus sister taxon to Ergalatax junionae, even if without full statistical support (UfB 94%). In addition, Orania nodosa ended up inside the rapanine clade of Semiricinula (UfB 100%) rather than in the ergalataxines. In the subfamily Muricinae, several genera as traditionally conceived appeared to be not monophyletic. In particular, the genus Naquetia resulted to be polyphyletic and split into three independent lineages: (1) Naquetia barclayi, (2) Naquetia triqueter (type species) + Naquetia vokesae (UfB 100%), and (3) Naquetia cumingii.
Dating major lineages
The time-calibrated phylogenies ( and Supplementary material Figure S5; see for 95% confidence interval (CI)) estimated the origin of the family Muricidae at 80 mya (95% CI: 80–80) during the Upper Cretaceous (Campanian). The clade of the Muricinae s.s. was dated at 34.04 mya (95% CI: 30.38–38.75) during the Eocene-Oligocene (Priabonian). The origin of the core Muricopsinae was dated at 50.52 mya (95% CI: 46.22–54.86) during the Eocene (Ypresian). The subfamily Pagodulinae (including the genus Poirieria) is estimated as having originated 34.85 mya (95% CI: 30.37–39.44) during the Eocene-Oligocene (Priabonian). The subfamily Haustrinae is estimated to have arisen 31.78 mya (95% CI: 27.79–36.02) in the Oligocene (Rupelian). The Ergalataxinae were estimated to have arisen in the Eocene (Lutetian), with the node dated at 47.28 mya (95% CI: 41.05–52.93). The Coralliophilinae are suggested to have originated 76.32 mya (95% CI: 73.06–79.60) during the Upper Cretaceous (Campanian). The Ocenebrinae are estimated to have originated during the lower Oligocene (Chattian) 24.71 mya (95% CI: 20.94–30.24). The origin of the Rapaninae was estimated at 59.97 mya (95% CI: 55.69–65.42) during the Paleocene (Selandian). The origin of the Typhinae was dated at 43.74 mya (95% CI: 39.49–48.35) during the Eocene (Lutetian).
Discussion
The family Muricidae is one of the largest groups of marine gastropods, and their phylogenetic systematics has always been controversial (Barco et al. Citation2010). In this work, we have gathered a large dataset based on the molecular information available so far, including for the first time representatives of all the currently recognised subfamilies.
Our phylogenetic hypothesis, based on the ML analyses, confirmed the monophyly of several major clades to be ranked as subfamilies. Ergalataxinae, Rapaninae, Coralliophilinae, Ocenebrinae, Typhinae, Pagodulinae, and Haustrinae were highly supported. The last two, Pagodulinae and Haustrinae, were also recovered as sister clades and share important anatomical features, such as a muricine-like radula (rachidian lacking marginal cusps, and inner denticles independent from lateral cusps), a pallial vas deferens not open to the mantle cavity across its length, the flattened, lensiform egg capsules (Tan Citation2003; Barco et al. Citation2015).
The monophyly of Trophoninae, at variance with results by Barco et al. (Citation2015), was not recovered. The available developmental data on the trophonine and ocenebrine radulae, with adult “ocenebrine” features observed also during trophonine ontogeny (Pio et al. Citation2014), are fully compatible with this pattern and suggest trophonines as representing a primitive grade in an ocenebrine clade, rather than a distinct subfamily. We take this position conservatively, pending further testing with the analysis of a wider, genome-scale molecular dataset.
Relationships among the major lineages were not resolved unequivocally across the tree. A close relationship of Rapaninae, Ergalataxinae, and Coralliophilinae was supported; this was recently questioned by the results of the analysis of mitogenomes in Harasewych and Sei (Citation2022) who recovered the coralliophiline as the sister taxon to the remaining muricids (unfortunately, Yu et al. (Citation2023) did not include the coralliophiline mitogenome published by Harasewych and Sei (Citation2022) in their larger dataset). However, a close relationship between Rapaninae and Ergalataxinae is also supported by morphological data (see, e.g. Herbert et al. Citation2007 for ontogenetic data on the radula), whereas the strong anatomical differences of Coralliophilinae may be related to their parasitic way of life (Richter & Luque Citation2002, Citation2003).
There seem to be good reasons to restrict the concept of the subfamily Muricinae s.s. to the clade including the type genus (the core Muricinae), thus excluding from the subfamily the genera Timbellus, Flexopteron, Ponderia, and Pterynotus as in part already suggested by Barco et al. (Citation2010) and Merle et al. (Citation2011).
Similarly, a revision of the scope of the subfamily Muricopsinae is urged. We detected a clade that can be proposed as Muricopsinae s.s., that – along with the type species of the genus Muricopsis – includes the former aspellines Aspella, Dermomurex, and Attiliosa, and also Tripterotyphis triangularis (formerly in the Tripterotyphinae, that thus, quite probably represent a mere muricopsine lineage). Conversely, the genera Homalocantha and Vitularia, traditionally considered as muricopsines, should be excluded from Muricopsinae.
Several taxa were placed either independently or rather unstably in the trees and may represent independent lineages: this is the case of Homalocantha, Flexopteron, Timbellus, Vitularia, Daphnellopsis, and Pterynotus. Their actual position should be tested by a phylogenomic approach and may either end in one of the recognised subfamilies or be recognised as worthy of suprageneric taxonomic recognition. Analyses of mitogenomic datasets were not conclusive, especially with different relationships supported by either ML or BI analyses (Harasewych & Sei Citation2022; Yu et al. Citation2023). It seems reasonable to expect more resolution by phylogenomic approaches on a wider nuclear scale.
In their recent critical analysis of the fossil records, Merle et al. (Citation2022) pointed out as the first known unquestionable muricid record is that of “Poirieria (Paziella) cretacea” Garvie, 1991, from the Late Cretaceous of Texas (Maastrichtian, 66–72 mya), for which they convincingly highlighted the morphological similarity with modern Flexopteron spp. In the Early Paleocene, other species with a shell morphology considered as “typical muricine” are known, classified in Flexopteron, Poirieria, and Timbellus. We agree with Merle et al. (Citation2022) that the shell morphology of Flexopteron is closer to Pagodulinae than to Muricinae.
There seems to be robust evidence that the origin and early diversification of the subfamilies of Muricidae occurred between 32 and 60 mya (95% intervals ranging from 28 to 65 mya) which largely agrees with the available knowledge from the fossil records. This is broadly the same period estimated for the origin of the lineages not assigned to a subfamily (Flexopteron, Timbellus, Daphnellopsis, Pterynotus) with the exception of Vitularia, which is estimated to be younger (15.58 mya, 95% CI: 11.48–21.56), and Homalocantha that is estimated to have diverged rather earlier (71.15 mya, 95% CI: 66.44–76.27). A remarkable exception is represented by the Coralliophilinae, for which an origin at 76 mya was estimated (95% CI: 73–79.6 mya), i.e. very close to the estimated origin of the family Muricidae; this is extremely older than any reliable fossil record for the subfamily (the oldest being from the Middle Eocene, c. 40 mya). As already suggested by Barco et al. (Citation2010), this estimate is probably biased by the very long branches of the coralliophiline lineages, which are very likely to be produced by the “Davison-effect”, i.e. the accelerated accumulation of mutations in mitochondrial (and, to a lesser extent, nuclear) genes in protandrous hermaphrodites (Davison Citation2006).
Taking into consideration the results from the phylogenetic analysis and the estimates of the node ages, rather than proposing a new classification of the family, we present the following annotated scheme (derived with modifications after Merle et al. Citation2022), with comments intended as hypotheses to be tested by further studies based on integrative approaches on anatomical and genomic data (taxa included according to results on samples assayed herein). In this scheme, the number of recognizable subfamilies is reduced to nine, but at the same time we highlighted seven lineages incertae sedis, which may prove worthy of subfamilial recognition.
Family Muricidae Rafinesque, 1815
Subfamily Muricinae Rafinesque, 1815 – To be restricted to the type genus (Murex) and related taxa (Bolinus, Chicomurex, Chicoreus, Haustellum, Hexaplex, Muricanthus, Naquetia, Phyllonotus, Siratus, Vokesimurex).
Subfamily Muricopsinae Radwin & D’Attilio, Citation1971 – To include also former Aspellinae Keen Citation1971 and Tripterotyphinae D’Attilio & Hertz, 1988 (Muricopsis, Acanthotrophon, Aspella, Attiliosa, Dermomurex, Favartia, Murexsul, Pazinotus, Tripterotyphis, Zetecopsis, Xastilia).
Subfamily Typhinae Cossmann, Citation1903 – Type genus based on a fossil (Typhis tubifer Bruguière, 1792). Morphologically very distinctive (Choreotyphis, Monstrotyphis, Siphonochelus, Typhina, Typhinellus).
Subfamily Haustrinae Tan, Citation2003 – Haustrum, Bedeva. Relative ranking with Pagodulinae to be tested.
Subfamily Pagodulinae Barco, Schiaparelli, Houart & Oliverio, Citation2012 – Poirieria definitely represents a plesiomorphic lineage of this subfamily (Pagodula, Abyssotrophon, Axymene, Boreotrophon, Comptella, Enixotrophon, Lamellitrophon, Paratrophon, Trophonella, Trophonopsis, Xymene, Xymenopsis, Zeatrophon).
Subfamily Ocenebrinae Cossmann, Citation1903 – Molecular evidence to include typical ocenebrines (Ocenebra, Acanthina, Acanthinucella, Ceratostoma, Chorus, Eupleura, Gracilipurpura, Jaton, Nucella, Ocinebrellus, Ocinebrina, Paciocinebrina, Urosalpinx, Vokesinotus, Xanthochorus) and also former Trophoninae (Trophon, Leptotrophon, Nipponotrophon, Scabrotrophon).
Subfamily Ergalataxinae Kuroda & Habe, 1971 – Monophyly of some genera as traditionally conceived to be tested (Ergalatax, Azumamorula, Bedevina, Claremontiella, Cronia, Cytharomorula, Drupella, Lataxiena, Lauta, Maculotriton, Morula, Murichorda, Muricodrupa, Oppomorus, Orania, Pascula, Phrygiomurex, Spinidrupa, Tenguella, Trachypollia, Usilla).
Subfamily Coralliophilinae Chenu, 1859 – Monophyly of several genera as traditionally conceived (including the type genus, Coralliophila) to be tested (Babelomurex, Emozamia, Hirtomurex, Galeropsis, Latiaxis, Leptoconchus, Rapa).
Subfamily Rapaninae Gray, 1853 – Monophyly of a few genera as traditionally conceived to be tested (Rapana, Acanthais, Stramonita, Agnewia, Concholepas, Dicathais, Drupa, Drupina, Indothais, Mancinella, Menathais, Nassa, Neorapana, Neothais, Pinaxia, Plicopurpura, Purpura, Reishia, Semiricinula, Thais, Thaisella, Tribulus, Tylothais, Vasula, Vexilla).
Incertae sedis Homalocantha – An odd position for a morphologically odd taxon.
Incertae sedis Flexopteron – If extant species are actually related to the upper Cretaceous and Early Paleogene lineages, it may be worthy of recognition as a separate subfamily (potentially useful to define the plesiomorphic Bauplan of Muricidae).
Incertae sedis Vitularia – Another long branch which may be biased by the Davison effect (Barco et al., Citation2010).
Incertae sedis Timbellus – Likely to represent an independent lineage, worthy of subfamilial rank.
Incertae sedis Ponderia – It may represent the plesiomorphic lineage at the base of the pagoduline-haustrine diversification.
Incertae sedis Daphnellopsis – Not an ergalataxine, relationships with Pterynotus (unsupported) to be tested.
Incertae sedis Pterynotus – Likely to represent an independent lineage (maybe with Daphnellopsis) worthy of subfamilial ranking.
Supplemental Material
Download MS Word (1.2 MB)Acknowledgments
MNHN specimens were obtained during research cruises and expeditions organized by the MNHN and ProNatura International as part of the Our Planet Reviewed program, and by the MNHN and the Institut de Recherche pour le Développement as part of the Tropical Deep-Sea Benthos program (PANGLAO 2004, PANGLAO 2005 and AURORA 2007 in the Philippines; BIOPAPUA (dx.doi.org/10.17600/10100040) and PAPUA NIUGINI (dx.doi.org/10.17600/18000841) in Papua New Guinea; CONCALIS (dx.doi.org/10.17600/8100010), EBISCO (dx.doi.org/10.17600/5100080), NORFOLK 1 (http://dx.doi.org/10.17600/1100050), NORFOLK 2 (dx.doi.org/10.17600/3100030) and TERRASSES (dx.doi.org/10.17600/8100100) in New Caledonia; SANTO 2006 in Vanuatu, SALOMON 2 (dx.doi.org/10.17600/4100090) in the Solomons, ATIMO VATAE (dx.doi.org/10.17600/10110040) and MIRIKY in Madagascar; MAINBAZA in Mozambique, KARUBENTHOS 2012 in Guadeloupe, and GUYANE 2014 in French Guiana – more information can be found at expeditions.mnhn.fr). These expeditions operated under the regulations then in force in the countries in question and satisfy the conditions set by the Nagoya Protocol for access to genetic resources. We are grateful to Barbara Buge (MNHN), Dario Zuccon (MNHN), Virginie Héros (MNHN), Philippe Maestrati (MNHN), and Philippe Bouchet (MNHN) for their help with MNHN materials and Thomas Lemarcis (MNHN) for his help with the MNHN cluster. Lab work and bioinformatic analyses were performed partly in the Service de Systématique Moléculaire and on the Plateforme de Calcul Intensif et Algorithmique PCIA (UAR2700 2AD, MNHN), partly in the Molecular Systematics lab at BBCD and HPC TeraStat2 (Sapienza University of Rome). We are thankful to Thu Hien To (Norwegian University of Life Sciences) and to Edoardo Bompiani (Sapienza University of Rome) for their help with bioinformatic analyses.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/24750263.2023.2283517
Additional information
Funding
References
- Barco A, Claremont M, Reid DG, Houart R, Bouchet P, Williams ST, Cruaud C, Couloux A, Oliverio M. 2010. A molecular phylogenetic framework for the Muricidae, a diverse family of carnivorous gastropods. Molecular Phylogenetics and Evolution 56(3):1025–1039. DOI: 10.1016/j.ympev.2010.03.008.
- Barco A, Herbert G, Houart R, Fassio G, Oliverio M. 2016. A molecular phylogenetic framework for the subfamily Ocenebrinae (Gastropoda, Muricidae). Zoologica Scripta 46(3):322–335. DOI: 10.1111/zsc.12219.
- Barco A, Marshall B, Houart R, Oliverio M. 2015. Molecular phylogenetics of Haustrinae and Pagodulinae (Neogastropoda: Muricidae) with a focus on New Zealand species. Journal of Molluscan Studies 81(4):476–488. DOI: 10.1093/mollus/eyv020.
- Barco A, Schiaparelli S, Houart R, Oliverio M. 2012. Cenozoic evolution of Muricidae (Mollusca, Neogastropoda) in the Southern Ocean, with the description of a new subfamily. Zoologica Scripta 41(6):596–616. DOI: 10.1111/j.1463-6409.2012.00554.x.
- Benkendorff K, Rudd D, Nongmaithem BD, Liu L, Young F, Edwards V, Avila C, Abbott CA. 2015. Are the traditional medical uses of Muricidae molluscs substantiated by their pharmacological properties and bioactive compounds? Marine Drugs 13(8):5237–5275. DOI: 10.3390/md13085237.
- Bompiani E, Petrillo UF, Lasinio GJ, Palini F 2020. High-performance computing with terastat. 2020 IEEE Intl Conf on Dependable, Autonomic and Secure Computing, Intl Conf on Pervasive Intelligence and Computing, Intl Conf on Cloud and Big Data Computing, Intl Conf on Cyber Science and Technology Congress (DASC/PiCom/CBDCom/CyberSciTech) IEEE, Calgary, Alberta, Canada. pp. 499–506.
- Bouchet P, Rocroi JP. 2005. Classification and nomenclator of gastropod families. Malacologia 47:1–397.
- Buhle ER, Ruesink JL. 2009. Impacts of invasive oyster drills on Olympia oyster (Ostrea lurida Carpenter 1864) recovery in Willapa Bay, Washington, United States. Journal of Shellfish Research 28(1):87–96. DOI: 10.2983/035.028.0115.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution 17(4):540–552. DOI: 10.1093/oxfordjournals.molbev.a026334.
- Claremont M, Houart R, Williams ST, Reid DG. 2013. A molecular phylogenetic framework for the Ergalataxinae (Neogastropoda: Muricidae). Journal of Molluscan Studies 79(1):19–29. DOI: 10.1093/mollus/eys028.
- Claremont M, Reid DG, Williams ST. 2008. A molecular phylogeny of the Rapaninae and Ergalataxinae (Neogastropoda: Muricidae). Journal of Molluscan Studies 74(3):215–221. DOI: 10.1093/mollus/eyn005.
- Collins TM, Frazer K, Palmer AR, Vermeij GJ, Brown WM. 1996. Evolutionary history of Northern Hemisphere Nucella (Gastropoda, Muricidae): Molecular, morphological, ecological, and paleontological evidence. Evolution 50(6):2287–2304. DOI: 10.2307/2410698.
- Cossmann M. 1903. Essais de Paleoconchologie Comparée. Paris: The Author & F.R de Rudeval.
- Davison A. 2006. The ovotestis: An underdeveloped organ of evolution. BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology 28(6):642–650. DOI: 10.1002/bies.20424.
- Douglas PMJ, Affek HP, Ivany LC, Houben AJP, Sijp WP, Sluijs A, Schouten S, Pagani M. 2014. Pronounced zonal heterogeneity in Eocene southern high-latitude sea surface temperatures. Proceedings of the National Academy of Sciences 111:6582–6587. DOI: 10.1073/pnas.1321441111.
- Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution 29(8):1969–1973. DOI: 10.1093/molbev/mss075.
- Fassio G, Russo P, Bonomolo G, Fedosov AE, Modica MV, Nocella E, Oliverio M. 2022. A molecular framework for the systematics of the Mediterranean spindle-shells (Gastropoda, Neogastropoda, Fasciolariidae, Fusininae). Mediterranean Marine Science 23(3):623–636. DOI: 10.12681/mms.29935.
- Fassio G, Stefani M, Russini V, Buge B, Bouchet P, Treneman N, Malaquias MAE, Schiaparelli S, Modica MV, Oliverio M. 2022. Neither slugs nor snails: A molecular reappraisal of the gastropod family Velutinidae. Zoological Journal of the Linnean Society 197(4):924–964. DOI: 10.1093/zoolinnean/zlac091.
- Griffin M, Pastorino G. 2005. The genus Trophon Monfort, 1810 (Gastropoda: Muricidae) in the tertiary of Patagonia. Journal of Paleontology 79(2):296–311. DOI: 10.1666/0022-3360(2005)079<0296:TGTMGM>2.0.CO;2.
- Harasewych MG, Sei M. 2022. The complete mitochondrial genome of Coralliophila richardi (P. Fischer, 1882)(Neogastropoda: Muricidae: Coralliophilinae). The Nautilus 136:1–11.
- Herbert GS, Merle D, Gallardo CS. 2007. A developmental perspective on evolutionary innovation in the radula of the predatory neogastropod family Muricidae. American Malacological Bulletin 23(1):17–32. DOI: 10.4003/0740-2783-23.1.17.
- Hoang DT, Chernomor O, Von Haeseler A, Minh BQ, Vinh LS. 2017. UFBoot2: Improving the ultrafast bootstrap approximation. Molecular Biology and Evolution 35:518–522. DOI: 10.1093/molbev/msx281.
- Houart R. 2018. Historique et classification des espèces actuelles de Muricidae (Neogastropoda, Muricoidea). Novapex 19(2):37–66.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20(4):1160–1166. DOI: 10.1093/bib/bbx108.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649. DOI: 10.1093/bioinformatics/bts199.
- Keen AM. 1971. A review of the Muricacea. The Echo 4:35–36.
- Kuraku S, Zmasek CM, Nishimura O, Katoh K. 2013. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Research 41:22–28. DOI: 10.1093/nar/gkt389.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2016. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution 34:772–773. DOI: 10.1093/molbev/msw260.
- Merle D, Garrigues B, Pointier JP. 2011. Fossil and recent Muricidae of the world. Part Muricinae. Harxheim: ConchBooks.
- Merle D, Garrigues B, Pointier JP. 2022. Fossil and recent Muricidae of the world: Part Muricopsinae. Harxheim: ConchBooks.
- Miller MA, Pfeiffer W, Schwartz T 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans. pp. 1–8.
- MolluscaBase eds. 2023. MolluscaBase. Muricidae Rafinesque, 1815. Accessed through: World Register of Marine Species at: https://www.marinespecies.org/aphia.php?p=taxdetails&id=148.
- Montes M, Beamud E, Nozal F, Santillana S. 2019. Late Maastrichtian-Paleocene chronostratigraphy from Seymour (Marambio) island (James Ross Basin, Antarctic Peninsula). Eustatic controls of sedimentation. Advances in Polar Science 30(3):303–327. DOI: 10.13679/j.advps.2019.2.00.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2014. IQ-TREE: A fast and effective stochastic algorithm for estimating Maximum-Likelihood Phylogenies. Molecular Biology and Evolution 32:268–274. DOI: 10.1093/molbev/msu300.
- Oliverio M, Mariottini P. 2001. A molecular framework for the phylogeny of Coralliophila and related muricoids. Journal of Molluscan Studies 67:215–224. DOI: 10.1093/mollus/67.2.215.
- Oliverio M, Modica MV. 2010. Relationships of the haematophagous marine snail Colubraria (Rachiglossa: Colubrariidae), within the neogastropod phylogenetic framework. Zoological Journal of the Linnean Society 158(4):779–800. DOI: 10.1111/j.1096-3642.2009.00568.x.
- Pio MJ, Herbert GS, Pastorino G. 2014. Developmental origins of complex radular characters in the Muricidae: The bifid rachidian edge. Invertebrate Biology 133(1):64–73. DOI: 10.1111/ivb.12040.
- Ponder W, Vokes E. 1988. A revision of the indo-west pacific fossil and recent species of Murex ss and Haustellum (Mollusca: Gastropoda: Muricidae). Records of the Australian Museum Supplement 8:1–157. DOI: 10.3853/j.0812-7387.8.1988.96.
- Ponder WF, Warén A. 1988. Classification of the Caenogastropoda and Heterostropha – a list of the family-group names and higher taxa. Malacological Review 4:288–326.
- Radwin GE, D’Attilio A. 1971. Muricacean supraspecific taxonomy based on the shell and radula. The Echo 4:55–67.
- Rambaut A. 2018. FigTree, a graphical viewer of phylogenetic trees (version 1.4. 4). Edinburgh: Institute of evolutionary biology, University of Edinburgh.
- R Core Team. 2022. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. Available: https://www.R-project.org/.
- Richter A, Luque ÁA. 2002. Current knowledge on Coralliophilidae (Gastropoda) and phylogenetic implication of anatomical and reproductive characters. Bollettino Malacologico 38(4):5–18.
- Richter A, Luque ÁA. 2003. Reproductive anatomy of three Mediterranean species of Coralliophilidae (Mollusca: Gastropoda: Neogastropoda). Journal of the Marine Biological Association of the United Kingdom 83(5):1029–1045. DOI: 10.1017/S0025315403008245h.
- Sysoev AV. 1992. A new hadal species of Boreotrophon (Gastropoda, Muricidae) from Northwestern Pacific. Ruthenica 2:166–167.
- Tan KS. 2003. Phylogenetic analysis and taxonomy of some southern Australian and New Zealand Muricidae (Mollusca: Neogastropoda). Journal of Natural History 37(8):911–1028. DOI: 10.1080/00222930110120610.
- Thiele J. 1929. Handbuch der systematischen Weichtierkunde. Jena: Gustav Fischer.
- To TH, Jung M, Lycett S, Gascuel O. 2016. Fast dating using least-squares criteria and algorithms. Systematic Biology 65(1):82–97. DOI: 10.1093/sysbio/syv068.
- Vaidya G, Lohman DJ, Meier R. 2011. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27(2):171–180. DOI: 10.1111/j.1096-0031.2010.00329.x.
- Vermeij GJ, Carlson SJ. 2000. The muricid gastropod subfamily Rapaninae: Phylogeny and ecological history. Paleobiology 26(1):19–46. DOI: 10.1666/0094-8373(2000)026<0019:TMGSRP>2.0.CO;2.
- Vokes EH. 1996. One last look at the Muricidae. American Conchologist 24:4–6.
- Yu Y, Kong L, Li Q. 2023. Mitogenomic phylogeny of Muricidae (Gastropoda: Neogastropoda). Zoologica Scripta 52(4):413–425. DOI: 10.1111/zsc.12598.