ABSTRACT
Nitrogen stripping plays a vital role during the sulfate-rich wastewater treatment process by sulfate-reducing bacteria (SRB). However, the interaction between nitrogen stripping, sulfate reduction capacity, S2– concentration, and microbial community structure is rarely studied. Here, two anaerobic systems were conducted to analyze these interactions simultaneously. Results showed that the toxic products (H2S) were stripped by nitrogen at first. Then, the increased relative abundance of fermentative bacteria accelerated the carbon sources conversion, and Desulfovibrio, Dethiosulfovibrio, and Sulfurospirillum facilitated the sulfate reduction after nitrogen stripping. Finally, the produced S2− concentration was increased by 12%, and the sulfate reduction rate was increased by 9.2% compared to that without nitrogen stripping.
1 Introduction
The annual production of mine wastewater in China is more than 1.1 × 1010 m3, and the resource utilization rate is only 20%, with a large amount of mine wastewater is directly discharged [Citation1,Citation2]. Acid mine drainage (AMD) is continuously produced from abandoned mines and contains many high-concentration sulfate ions (SO42–) and various heavy metals with low pH. It brings severe threats to mine equipment, the ecological environment, and human health [Citation3]. There is an urgent need for an economical, practical, and resource-recyclable method to protect the sustainability of ecosystems and reduce resource waste. There are many methods for treating AMD, among which biological technology by SRB has attracted much attention due to their low cost and environmental friendliness [Citation4,Citation5].
S2– plays an essential role in reducing sulfate by SRB [Citation6]. On the one hand, S2– transformed by excessive sulfate can bind with metals to precipitate as metal sulfides under the metabolism of SRB [Citation7]. On the other hand, S2– can be converted to elemental sulfur by sulfur-oxidizing bacteria. Unfortunately, current research predominantly focuses on growth characteristics, utilization of different substrates, and sulfate reduction ability of SRB [Citation8–11]. However, little research has been done on S2– under the metabolic reactions of SRB. Based on previous studies, only a tiny S2– reacts with heavy metals, and a large amount of sulfur is not analyzed [Citation12]. A study by Vasquez et al. reported that 833 mg/L of SO42–-S in the influent and a total of 473 mg L–1 of S2– in the effluent (including sulfate and generated metal sulfides), with approximately 43% of sulfur are not described in the effluent [Citation13]. Therefore, it is necessary to pay attention to S2– when SRB treats sulfate-rich wastewater.
The formation of S2– is affected by many factors. On the one hand, diverse microorganisms have different metabolic profiles because of their inherently different functions. As well, there is a competition between SRB and other bacteria. Additionally, the coexisting hydrolytic bacteria and fermentative bacteria may affect the activity of SRB. The above reason may affect the activity of SRB, which has the most considerable influence on S2– production [Citation14]. Therefore, it is essential to understand the microbial community successions during the sulfate-rich wastewater reduction process. On the other hand, several types of sulfur-containing products (H2S, HS–, and S2–) are found during the SRB metabolism process [Citation15], and the undissociated H2S can penetrate the cell wall and inhibit the activity of SRB, causing the change in S2– production [Citation16]. If toxic products (such as H2S) can be removed, the activity of SRB will not be inhibited, and thus the sulfate reduction reaction will be accelerated. Yao compared the effect of nitrogen stripping on sulfide production when using SRB to treat AMD and found that the sulfide content in the nitrogen stripping group is 2.09 times higher than that in the group without nitrogen stripping [Citation17]. Thusly, nitrogen stripping is a feasible method for removing toxic products in the sulfate-rich wastewater treatment by SRB. Furthermore, the existence of toxic products can influence the microbial community composition. As reported earlier, the added H2S changed the microbial growth environment, which might cause the relative abundance of Hydrogenophaga sp, Clostridium populeti, Bacteroides sp, Pseudomonas sp, and Dysgonomonas wimpennyi to change [Citation18]. Here, the formation of S2– is affected by the microbial community and nitrogen stripping. Meanwhile, the succession of microbial communities is affected by nitrogen stripping in the sulfate-rich wastewater treatment by SRB. However, it is still unclear about the relationship of microbial community succession, nitrogen stripping, and S2– formation during the sulfate-rich wastewater treatment by SRB.
Therefore, this study conducted two anaerobic systems to simultaneously analyze the interaction of microbial community succession, nitrogen stripping, S2–, and sulfate reduction ability. The SRB inoculum was enriched, and the sulfate reduction ability, the concentration of S2– by controlling nitrogen stripping during sulfate-rich wastewater treatment by SRB were considered. Additionally, the response to microbial communities was also investigated.
2 Materials and methods
2.1 SRB inoculum
The microorganisms were derived from anaerobic sludge at the Nibu Bay Wastewater Treatment Plant (Qingdao, Shandong Province, China). Postgate’S medium C was used for the culture of SRB [Citation19]. This medium was composed of 0.65 g L–1 KH2PO4, 0.06 g L–1 MgCl2 · 6H2O, 1.0 g L–1 Na2SO4, 0.04 g L–1 CaCl2 · 2H2O, 1.0 g L–1 NH4Cl, 1.0 g L–1 yeast extract, and 2.0 mL L–1 sodium lactate. The pH of the media was adjusted to 7 and then sterilized at 121°C for 20 min. After cooling, 1.2 g L–1 of Fe(NH4)2·(SO4)2 · 6H2O and 0.5 g L–1 of L-cysteine sterilized by ultraviolet (UV) radiation for 30 min were added, and the samples were cultured at 28°C and 70 rpm min–1 in a shaker. After two days, when the solution turned ink-colored and the bottle smelled like rotten eggs (detected by using lead acetate test paper), 10% (v/v) of the above culture solution was transferred to fresh Postgate’S medium C, and the same culture conditions were repeated three times to enrich the SRB. Efficient SRBs were obtained and identified by Shanghai Majorbio Bio-pharm Technology Co., Ltd.
Subsequently, the morphology and the growth process of the SRB inoculum were evaluated. The pH and redox potential (ORP) during growth were tested together to investigate the growth characteristics of the SRB inoculum. SRB inoculum (10% [v/v]) were added to the fresh Postgate’S medium C (without the addition of Fe(NH4)2·(SO4)2 · 6H2O, the medium that was not inoculated was used as a blank control, and the OD600, pH, and ORP in the medium were measured every two hours. All experiments were performed three times.
2.2 Design, set up, and operation of anaerobic system SRB reaction device
Considering the influence of initial sulfate concentration and nitrogen stripping on sulfate reduction by SRB, two set anaerobic systems of experimental devices were designed (). Device (a) consisted of bottle A only, filled with sulfate-rich wastewater and SRB. Device (b) was composed of bottles A, B, and nitrogen bottles. Bottle B is filled with CuCl2 solution, absorbing H2S by nitrogen stripped. The practical volumes of bottles A and B were 1 L, and rubber stoppers and polytetrafluoroethylene tapes sealed all the entrances of the bottles to ensure an anaerobic environment.
Figure 1. Experimental reaction device. (a): bottle A was filled with sulfate-rich wastewater and SRB; (b): bottle A, bottle B filled with CuCl2 solution and nitrogen stripping.
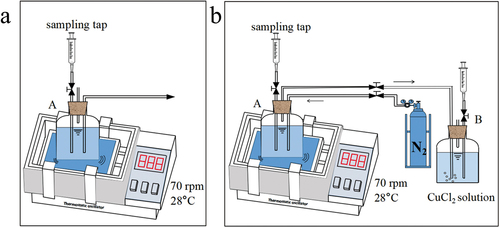
In each group of experiments, the initial sulfate concentrations of wastewater were 600 and 1000 mg L–1, and 30% (v/v) SRB inoculum was added to sulfate-rich wastewater (Stripe nitrogen for 10 min before use to remove oxygen). The concentration of copper ions in bottle B used for absorption was 500 mg L–1. The sulfate concentration, S2– concentration, pH, and ORP during the reaction were measured at 0, 3, 6, 9, 12, and 15 h. All experiments were conducted and analyzed three times.
2.3 Analytical procedures
The morphology of the SRB inoculum was tested through scanning electron microscopy (FEI Quanta 250 FEG), and sample processing steps were based on Shi’s study [Citation20]. A pH meter (PHS-3C) and automatic potentiometric titrator (ZD-2) measured pH and ORP. Sulfate concentration was tested following the ‘Water Quality-Determination of sulfate–barium chromate spectrophotometry’ (HJ/T 342–2007). The S2– concentration was determined using a sulfur ion concentration meter (Banter 931-S). The concentration of Cu2+ remaining in bottle B was measured to account for the H2S produced in bottle A using a flame atomic absorption spectrometer (TAS-986 F).
2.4 Microbial community analysis
The samples (0 h, 15 h-without N2, and 15 h-with N2) at 600 mg L–1 initial sulfate concentration were filtered by a 0.22 µm aqueous filter membrane were collected first for high-throughput sequencing analysis. DNA extraction and polymerase chain reaction (PCR) amplification were performed. According to the manufacturer’s instructions, the samples were using a FastDNA® Spin Kit for Soil (MP Biomedicals, U.S.). The DNA concentration and purity were determined with a NanoDrop 2000 UV–vis spectrophotometer, and DNA extract was checked on 1% agarose gel. PCR amplification was run by two primers of 338 F (ACTCCTACGGGAGGCAGCAG) and 806 R (GGACTACHVGGGTWTCTAAT) for each DNA sample, and the region was V3–V4 hypervariable region. PCR reactions were performed in triplicate. According to the manufacturer’s instructions, the PCR product was extracted from 2% agarose gel, purified using an AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, USA), and quantified using a Quantus™ Fluorometer (Promega, USA). The third step was Illumina MiSeq sequencing. Purified amplicons were pooled in equimolar and paired-end sequenced on an Illumina MiSeq PE300 platform (Illumina, USA) according to the standard protocols by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). The final step was the processing of sequencing data. Operational taxonomic units (OTUs) with 97% similarity cutoff were clustered, and chimeric sequences were identified and removed.
3 Results
3.1 Characteristics of SRB inoculum
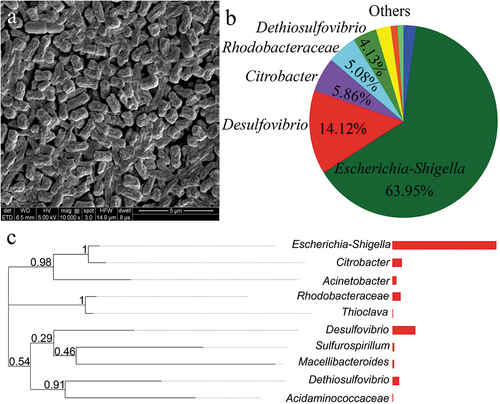
In order to further analyze the succession of microbial community structure during the sulfate reduction process by SRB, we first studied the characteristics of SRB inoculum. The morphology of microorganisms is shown in ). The SRB was arc and rod-shaped, and their length was approximately 0.5 to 2 μm. As shown in , the microorganisms were constituted by Escherichia-Shigella, Desulfovibrio, Citrobacter, Rhodobacteraceae, Dethiosulfovibrio, and others at the genus level. Since the first discovery of SRB by Beijerinck in 1895, more than 40 genera of SRB, such as Desulfovibrio, Desulfotomaculum, Desulfonema, Desulfomonas, Desulfobacter, and Desulfococcus, have been isolated successively [Citation21].
3.2 Effect of different conditions on sulfate reduction ability by microorganisms
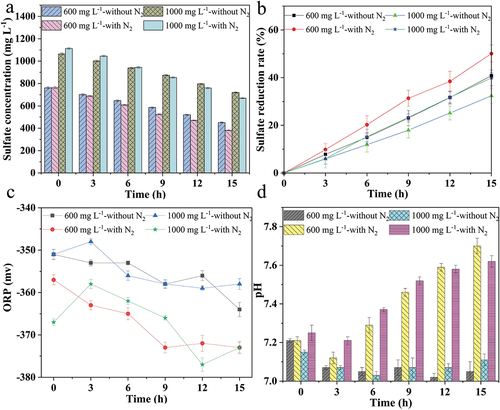
Different initial sulfate concentrations affect microorganisms’ growth and reduction ability due to the different osmotic pressures. As observed in , the sulfate reduction rate was 40.82% when the initial sulfate concentration was 600 mg L–1, and it was 32.40% when the initial sulfate concentration was 1000 mg L–1. Regardless of the initial sulfate concentration, the pH during the reactions was maintained in the range of 7.02–7.11, and the ORP values decreased more obviously at the initial sulfate concentration of 600 mg L–1 than that of the initial sulfate concentration was 1000 mg L–1. With nitrogen stripping, the sulfate reduction rate was improved by 9.25% compared with that without nitrogen stripping at 600 mg L–1 initial sulfate concentration, and there was an approximately 7.48% increase at 1000 mg L–1 initial sulfate concentration. These demonstrated that nitrogen stripping is effective for sulfate-rich wastewater treatment by SRB. In addition, a visible elevation of alkalinity was observed in the SRB treatment of sulfate-rich wastewater after nitrogen stripping.
3.3 Effect of different conditions on S2– production during sulfate reduction process by microorganisms
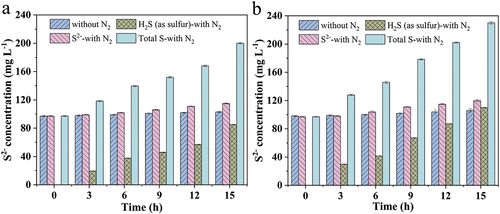
S2- as the target product of SRB metabolism and can form stable metal sulfide precipitation with heavy metal ions in wastewater, the S2- concentration with different nitrogen stripping conditions was studied, and the results were shown in . The S2– concentration increased from 97 mg L–1 to 103 mg L–1 with 600 mg L–1 initial sulfate concentration, and it grew from 98 mg L–1 to 106 mg L–1 with 1000 mg L–1 initial sulfate concentration. After nitrogen stripping, the concentration of S2–increased from 97 mg L–1 to 115 mg L–1 and 120 mg L–1 when the initial sulfate concentration increased from 600 mg L–1 to 1000 mg L–1, which was approximately 12% higher than the final S2– concentrations in the system without nitrogen stripping. After 15 h bioreaction, the concentration of H2S (as sulfur) blown off was 85.14 and 109.99 mg L–1 with an initial sulfate concentration of 600 and 1000 mg L–1, respectively.
3.4 Microbial community differences on sulfate-rich wastewater treatment by controlling nitrogen stripping
3.4.1 Alpha diversity analysis of different samples
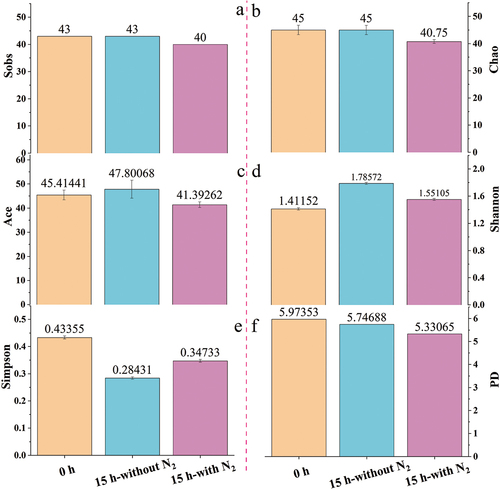
16S rRNA genes were amplified and sequenced through high-throughput sequencing, and all OTUs were clustered at a setting cut off 97% similarity. A total of 32,048 sequences per sample were collected after subsampling to determine microbial diversity. Community richness and diversity in each sample are estimated, and the results are shown in . As shown in , the microbial community richness has a slight downward trend after nitrogen stripping. As for microbial community diversity, an increasing tendency was found under the sulfate-rich wastewater treatment process of SRB (). Moreover, the results indicated that nitrogen stripping negatively influenced microbial community diversity.
3.4.2 Microbial community composition analysis of different samples
The distribution of microorganisms was examined to reveal the functional microbial community among the three samples. The phylum with the highest representation was Proteobacteria representing 77.86%, 68.91%, and 59.96% among the samples of 0 h, 15 h-without N2, and 15 h-with N2, respectively, followed by Desulfobacterota (14.12%, 15.58%, and 16.63%), Bacteroidota, Synergistota, and Firmicutes ()).
Figure 6. Species composition analysis at different levels among three samples. (a) the sample-species relationship circle diagram at the phylum level; (b) the ternary analysis at the class level; (c) the heatmap at the genus level; (d) the distribution of specific functional genera among three samples.
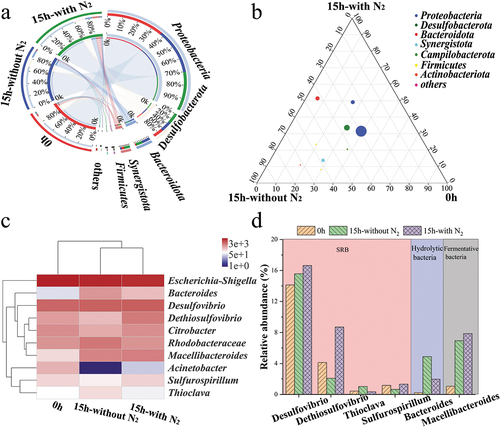
At the class level, the relative abundance of nine bacterial phyla was identified with over 1%. The most abundant phyla were Gammaproteobacteria, as the main part of Proteobacteria representing 72.32%, 58.36%, and 54.61% among the samples of 0 h, 15 h-without N2, and 15 h-with N2, respectively, followed by Desulfovibrionia, Baceroidia, Alphaproteobacteria, Synergistia, Campy-lobacteria, Clostridia, Actinobacteria, and Negativicutes. Alphaproteobacteria were classified as Proteobacteria, accounting for 5.54%, 10.55%, and 5.37% among the samples ()).
As shown in ), the bacterial community of samples altered after sulfate reduction (only the top 10 genera). The distribution of some specific functional genera among the three samples is illustrated in . The principal genera responsible for sulfate reduction were Desulfovibrio, accounting for 14.12%, 15.58%, and 16.63% among the samples of 0 h, 15 h-without N2, and 15 h-with N2, respectively, which performed an increasing tendency after the bioreaction. At the same time, Dethiosulfovibrio accounted for 4.13%, 2.06%, and 8.70%, respectively. Thioclava accounted for 0.44%, 1.02%, and 0.35%, which played an essential role in the biogeochemical sulfur cycle and sulfide and sodium thiosulfate oxidization [Citation22]. Here, the relative abundance of Desulfovibrio, Macellibacteroides, Dethiosulfovibrio, Citrobacter, and Sulfurospirillum increased, while the relative abundance of Escherichia-Shigella, Rhodobacteraceae, and Bacteroides decreased after nitrogen stripping.
4 Discussion
This study comprehensively investigated the sulfate reduction rate, S2– concentration, and microbial community by controlling nitrogen stripping during sulfate-rich wastewater treatment by SRB. In the sulfate reduction process, SRB generated several reduction products, including H2S, HS–, and S2–, especially when H2S caused the most toxicity to the growth and performance of SRB [Citation23]. Consequently, nitrogen gas can strip the H2S generated during the sulfate reduction by SRB to decrease the toxicity. Other studies suggested that when the concentration of H2S reached 40–50 mg L–1, SRB was inhibited, and SRB activity was irreversibly destroyed when the concentration of H2S exceeded the toxicity level for 3–6 h [Citation24]. Different SRB possessed different tolerances to H2S, and the toxicity of H2S produced during sulfate reduction to SRB may be direct and reversible [Citation25,Citation26].
In this paper, the concentration of H2S (as sulfur) blown off by nitrogen was 85.14 mg L–1 after 15 h bioreaction with an initial sulfate concentration of 1000 mg L–1, respectively. At the same time, the S2− production concentration was increased from 103 to 120 mg L–1 compared to that without nitrogen stripping. Moreover, the sulfate reduction rate was increased by 9.2% to 50.08%. In other words, toxicity products were blown by nitrogen, and SRB activity was restored, thereby enhancing the sulfate reduction ability.
Furthermore, the H2S was blown off by nitrogen stripping, with the evolution of microbial community structure. Specifically, the relative abundance of Desulfovibrio, Macellibacteroides, Dethiosulfovibrio, and Sulfurospirillum increased. Desulfovibrio has an essential role in anaerobic systems [Citation27], and it was a typical hydrogenotrophic SRB, indicating that these SRB can use hydrogen to reduce sulfate [Citation28]. Moreover, Dethiosulfovibrio reduced thiosulfate and elemental sulfur but not sulfate to hydrogen sulfide [Citation29]. Macellibacteroides, a type of fermentative bacteria, produced hydrogen for hydrogenotrophic and enhanced the activity of Desulfovibrio [Citation30]. Sulfurospirillum, a type of sulfite-reducing bacteria [Citation31] and Nogueira et al listed Sulfurospirillum as SRB genera [Citation5]. In addition, the species of Bacteroides has a relatively high abundance in 15 h-with N2 sample. Bacteroides was a hydrolytic bacterium that could participate in protein degradation and convert amino acids to acetate and had the function of being tolerant to high concentrations of metals and metalloids [Citation32]. It is inferred from these results that the increased fermentative bacteria (Macellibacteroides) accelerated the conversion of carbon sources. The sulfate was transformed to sulfite by Desulfovibrio, and the bacteria of Dethiosulfovibrio and Sulfurospirillum converted sulfite to S2–. Thus, the produced S2− concentration increased, and sulfate reduction ability improved.
5 Conclusions
In this study, the contributions of nitrogen stripping to sulfate reduction by SRB were comprehensively investigated. Stripped H2S, favorable S2– production, and considerable sulfate reduction ability implied a vital contribution of nitrogen stripping in the sulfate-rich wastewater. The hydrolytic and fermentative microorganisms coexistent with the SRB, and the increased fermentative bacteria accelerated the conversion of carbon sources. The relative abundance of Desulfovibrio, Dethiosulfovibrio, and Sulfurospirillum increased after nitrogen stripping, thereby facilitating the sulfate reduction. These findings provide advanced incisive insights into the effect of nitrogen stripping on sulfate-rich wastewater treatment by SRB from S2–, sulfate reduction ability, and microbial community successions.
Acknowledgments
This work was supported by the scientific research fund project of the National Natural Science Foundation of China (grant numbers 52070123), the Natural Science Foundation of Shandong Province (grant numbers ZR2020ME224); Project of Shandong Province Higher Educational Young Innovative Talent Introduction and Cultivation Team [Wastewater treatment and resource innovation team], and high-quality curriculum construction project of graduate education of Shandong University of science and technology.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Liu CS, Zhao DF, Yan LH, et al. Elemental sulfur formation and nitrogen removal from wastewaters by autotrophic denitrifiers and anammox bacteria. Bioresour Technol. 2015;191:332–336.
- Sun MY, Cui GW, Wang JF, et al. The Sulfur Distribution Regularities of Huaheng High Sulfur Coal. Journal of Shandong University of Science and Technology (Natural Science). 2013;32:36–41.
- Zhang WQ, Li B, Gao B. Research Status and Development Trend of Mine Roof Water Inrush Prediction. Journal of Shandong University of Science and Technology (Natural Science). 2017;36:15–23.
- Chen J, Zhang G, Ma C, et al. Antimony removal from wastewater by sulfate-reducing bacteria in a bench-scale upflow anaerobic packed-bed reactor. Acta Geochimica. 2020;39(2):203–215.
- Nogueira EW, Godoi L, Yabuki L, et al. Sulfate and metal removal from acid mine drainage using sugarcane vinasse as electron donor: performance and microbial community of the down-flow structured-bed bioreactor. Bioresour Technol. 2021;330:124968.
- Sanchez-Andrea I, Sanz JL, Bijmans MFM, et al. Sulfate reduction at low pH to remediate acid mine drainage. J Hazard Mater. 2014;269:98–109.
- Mao ZG, Meng LQ, Liu XT, et al. Geochemical Characteristics and Ore Genesis of Nanxiaoyao Gold Deposit in Central Yishu Fault Zone, Shandong Province. Journal of Shandong University of Science and Technology(Natural Science). 2019;38:1–13.
- Pruden A, Messner N, Pereyra L, et al. The effect of inoculum on the performance of sulfate-reducing columns treating heavy metal contaminated water. Water Res. 2007;41(4):904–914.
- Lindsay MBJ, Wakeman KD, Rowe OF, et al. Microbiology and Geochemistry of Mine Tailings Amended with Organic Carbon for Passive Treatment of Pore Water. Geomicrobiol J. 2011;28(3):229–241.
- Hiibel SR, Pereyra LP, Breazeal MVR, et al. Effect of Organic Substrate on the Microbial Community Structure in Pilot-Scale Sulfate-Reducing Biochemical Reactors Treating Mine Drainage. Environ Eng Sci. 2011;28(8):563–572.
- Hiibel SR, Pereyra LP, Inman LY, et al. Microbial community analysis of two field-scale sulfate-reducing bioreactors treating mine drainage. Environ Microbiol. 2008;10(8):2087–2097.
- Martins M, Santos ES, Pires C, et al. Production of irrigation water from bioremediation of acid mine drainage: comparing the performance of two representative systems. J Clean Prod. 2010;18(3):248–253.
- Vasquez Y, Escobar MC, Saenz JS, et al. Effect of hydraulic retention time on microbial community in biochemical passive reactors during treatment of acid mine drainage. Bioresour Technol. 2018;247:624–632.
- Weber KP, Werker A, Gehder M, et al. Influence of the Microbial Community in the Treatment of Acidic Iron-Rich Water in Aerobic Wetland Mesocosms. Bioremed J. 2010;14(1):28–37.
- Icgen B, Harrison S. Exposure to sulfide causes populations shifts in sulfate-reducing consortia, research in microbiology. Research in Microbiology. 2006;157(8):784–791.
- Moosa S, Harrison S. Product inhibition by sulphide species on biological sulphate reduction for the treatment of acid mine drainage. Hydrometallurgy. 2006;83(1–4):214–222.
- Yao Q, Huang J, Yang L, et al. Characteristic of metabolism for sulfur-containing components during sulfate bioreduction process. Chin J Environ Eng. 2018;12:2783–2790.
- Gao M, Li L, Liu JX. Simultaneous removal of hydrogen sulfide and toluene in a bioreactor: performance and characteristics of microbial community. J Environ Sci. 2011;23(3):353–359.
- Postgate JR. The Sulphate-Reducing Bacteria. Cambridge: Cambridge University Press; 1984.
- Shi K, Xue JL, Xiao XF, et al. Mechanism of Degrading Petroleum Hydrocarbons by Compound Marine Petroleum-Degrading Bacteria: surface Adsorption, Cell Uptake, and Biodegradation. Energy Fuels. 2019;33(11):11373–11379.
- Castro W. Ogram, Phylogeny of sulfate-reducing bacteria. FEMS Microbiol Ecol. 2000;31(1):1–9.
- Sorokin DY, Tourova TP, Spiridonova EM, et al. Thioclava pacifica gen. nov., sp. nov., a novel facultatively autotrophic, marine, sulfur-oxidizing bacterium from a near-shore sulfidic hydrothermal area. Int J Syst Evol Microbiol. 2005;55(3):1069–1075.
- Li X, Fan M, Liu L, et al. Treatment of high-concentration chromium-containing wastewater by sulfate-reducing bacteria acclimated with ethanol. Water Sci Technol. 2020;80. 10.2166/wst.2020.057.
- Stucki G, Hanselmann KW, Hurzeler RA. Biological sulfuric acid transformation: reactor design and process optimization. Biotechnol Bioeng. 1993;41(3):303–315.
- Yuan H, Zhu N. Progress in inhibition mechanisms and process control of intermediates and by-products in sewage sludge anaerobic digestion. Renew Sust Energ Rev. 2016;58:429–438.
- Chen JL, Ortiz R, Steele TWJ, et al. Toxicants inhibiting anaerobic digestion: a review. Biotechnol Adv. 2014;32(8):1523–1534.
- Ferraz RM, Costa JM, Verola LM, et al. Effects of the inoculum source, electron donor, and immobilization on the microbial community of sulfidogenic bioreactors. Chem Eng J. 2021;404:126549.
- Angélica Marcia Dos S, Martins Costa J, Kawanishi Braga J, et al. Lactate as an effective electron donor in the sulfate reduction: impacts on the microbial diversity. Environ Technol. 2021;1–12. 10.1080/09593330.2021.1916092
- Whitman WB. Dethiosulfovibrio. New York: John Wiley & Sons, Inc;2015.
- Jabari L, Gannoun H, Cayol J-L, et al. Macellibacteroides fermentans gen. nov., sp. nov., a member of the family Porphyromonadaceae isolated from an upflow anaerobic filter treating abattoir wastewaters. Int J Syst Evol Microbiol. 2012;62(Pt_10):2522–2527.
- Simon J, Kroneck PMH. Chapter Two-Microbial Sulfite Respiration. Poole RK, Ed. Adv Microb Physiol. 62 2013. 10.1016/B978-0-12-410515-7.00002-0.
- Kindaichi T, Ito T, Okabe S. Ecophysiological interaction between nitrifying bacteria and heterotrophic bacteria in autotrophic nitrifying biofilms as determined by microautoradiography-fluorescence in situ hybridization. Appl Environ Microbiol. 2004;70(3):1641–1650.