Abstract
A total of 228 lactic acid bacteria colonies were isolated from blue moldy Tulum cheese inoculated with Penicillium roqueforti along with the ripening (4 months). Three different medias (PCA, MRS or M17 agar) were used for isolation, while two API tests were used for identification of LAB. As a result, Enterococcus sp. (53.3%) and Lactobacillus sp. (26.7%) was found as dominant flora at end of ripening. Among them, Enterobacter faecalis (40.0%) and Lactobacillus parabuhneri (13.3%) were determined. The others were also Leuconostoc mesenteroides (10.0%), Pediococcus acetilactici (6.7%) and Lactobacillus bifermentans (6.7%).
INTRODUCTION
Tulum cheese, a kind of local cheese special for Turkey, is not been known worldwide. It is particularly preferred for its characteristic natural moldy taste and flavor. Tulum cheese is a mixture of civil cheese and whey curd (lor) produced in small dairy plants and is also homemade. Civil cheese is manufactured from sour skim milk with a small amount of rennet with heat. The mixture of civil and lor is pressed into a goatskin or a plastic bag and then ripened. During the ripening period, naturally contaminated molds grow and contribute to the ripening process.[Citation1,Citation2] Lactic acid bacteria (LAB) are widespread in nature and predominate the microflora of foods rich in carbohydrates, protein breakdown products, vitamins, and foods with a low oxygen tension. This explains the prevalence of these bacteria in milk and milk products such as cheese. Due to their metabolism (they utilize carbohydrates and produce lactic acid as the major end product), LAB is responsible for the acidification of cheese necessary for coagulation of the milk. In addition, many species of LAB play an important role in the ripening of cheese. This is supported by the number of reports published on the microbiology of varieties of cheese made all around the world.[Citation3] The main goal of this study was to isolate and identify lactic acid bacteria and aerobic mesophilic flora of Tulum cheese inoculated with Penicillium roqueforti. The availability of the mediums used for the isolation of LAB was also discussed.
MATERIAL and METHODS
Sampling
Tulum cheese was prepared as described by Kurt and Öztek.[Citation4] For this purpose, civil cheese made from cow's milk was mixed with lor cheese (1:1, w:w) produced from whey curd. Then, the mixture was inoculated with Penicillium roqueforti isolated from moldy Tulum cheese collected from retail markets,[Citation5] pressed into plastic bags, and ripened at 10-12°C for 4 months. Isolation and identification of lactic acid bacteria from Tulum cheese were carried out monthly during the ripening.
Isolation of Lactic Acid Bacteria
For the propagation of aerobic mesophilic flora, PCA medium was used, while MRS or M17 was used for lactic acid bacteria of blue moldy Tulum cheese. All the mediums were obtained from Merck, Germany; 25-30 colonies from PCA counting plates and 5-15 colonies from MRS or M17 counting plates were randomly selected.[Citation6]
Identification and Evaluation of Lactic acid Bacteria
A total of 237 strains were submitted to Gram staining and the catalase test.[Citation6] Gram-positive and catalase-negative isolates were identified by using the following tests: morphology, colony pigmentation, production of carbon dioxide from glucose (in MRS broth covered with an agar seal trap), growth at 10 and 37°C in BHI broth, salt tolerance (2 and 6.5% NaCl in BHI), starch hydrolysis, and sugar fermentation with imitated the API system (API 50CHL (Bio Merieux, Marcy I'Etoile France) for Leuconostoc sp. and Lactobacillus sp., and API 50 CHE (Bio Merieux, Marcy I'Etoile France) for Lactococcus sp. and Enterococcus sp. according to Lopez-Diaz et al.[Citation3] Identification at species level was carried out following schemes recommended by several authors.[Citation3,Citation7,Citation8,Citation9]
RESULTS
The distribution of LAB isolated from different mediums for Tulum cheese samples was shown in . Aerobic mesophilic LAB was isolated and identified from PCA medium, while for the remaining genera, LAB, MRS, and M17, agar were used. As seen from , LAB isolated from cheese samples generally belong to Enterococcus and Lactobacillus genera. It was also found that bacteria isolated from a PCA medium generally belongs to Enterecoccus sp. (56.7%), as extracted from MRS medium was Lactobacillus sp. (50.0%). Over all, Enterococcus sp., Lactobacillus sp., Leuconostoc sp., Pediococcus sp. and Lactococcus sp. isolated from the cheese were calculated as 35.0, 25.7, 13.9, 13.5, and 8.0%, respectively. The percentage of unidentified microorganisms was also determined as 3.8%.
Table 1 Distribution of lactic acid bacteria isolated from different media during the ripening of moldy Tulum cheese
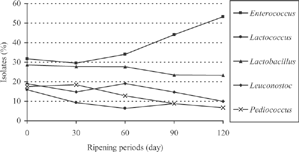
As shown in , all of the lactic acid bacteria strains isolated from moldy Tulum cheese and grown at tested temperatures (10 and 37°C) and salt concentration (2 and 6.5% NaCl) could not hydrolyze starch. It is known that Lb. parabuhneri, P. acidilactici, and P. pentosaceus strains produce CO2 from glucose; and E. faecalis, E. avium, E. durans, and E. faecium strains grow at pH 9.6 in broth medium, while the other strains did not. Therefore, all strains evaluated were gram-positive and catalase-negative.The numbers and percentages of lactic acid bacteria strains isolated and identified from moldy Tulum cheese during ripening were presented in . As shown in , E. faecalis was dominant in all samples during the ripening. Over all, the percentage of these bacteria is 17.5 in fresh cheese. The change in percentages of E. durans and E. faecium strains was very small during ripening. On the other hand, E. avium could not be detected at the end of the period.The percentage of L. lactis ssp lactis is 15.9 in fresh cheese; this percentage decreased to 3.3 at the end of the ripening. On the other hand, the lactis ssp cremoris strain was determined at the end of the period. The percentage of Lb. parabuhneri strains in the facultatively heterofermentative lactobacilli was 15.9 in fresh cheese and increased to 17.0 at 60 days of ripening. This percentage decreased to 13.3 at end of the ripening. While the change in the rates of Lb. paracasei and Lb. bifermentans strains is very small during ripening, Lb. plantarum was not found at the end of the ripening. Leu. mesenteroides ssp cremoris was isolated in significant amounts (19.0%) from fresh cheese, then decreased to 14.8% at 30 days. This rate increased to 19.1% in 60 days. The number of P. acidilactici quickly decreased after storage of 30 days. Nevertheless, the change in the number of P. pentosaceus was low during ripening. However, this strain was not detected at 120 days of ripening. To explain the role of each of these LAB genera found in moldy Tulum cheese, it is convenient to consider how these microorganisms grow with time during cheese ripening. As seen in , Enterococcus sp. and Lactobacillus sp. were dominant; however, Lactococcus sp., Leuconostoc sp. and Pediococcus sp. were present in low proportions.
Table 2 Phenotypic characteristics of the lactic acid bacteria isolated from moldy Tulum cheese
Table 3 Isolation and identification lactic acid bacteria from moldy Tulum cheese during ripening
DISCUSSION
Among the growth mediums (PCA, MRS or M17) used for the isolation of LAB, less isolates were obtained from the MRS medium. Enterococci were found in a higher proportion (61.4%) in PCA, which confirms the usefulness of the other medium for the isolation of these genera.[Citation3,Citation10] Phenotypic characteristics of the lactic acid bacteria isolated in this study corresponded with the results of Durlu-Ozkaya et al.[Citation11] Kalogridou-Vassiliadou et al.[Citation12] isolated E. faecalis, L. lactis, Leu. paramesenteroides from Anthotyro cheese. Lopez-Diaz et al.[Citation3] also isolated E. faecalis (24.7) and Leu. mesenteroides (9.0%) from Valdeon cheese (a moldy cheese variety). In addition, Durlu-Ozkaya[Citation11] expressed that E. faeceum, Lb. paracasei and L. lactis were dominant flora in Beyaz cheese. Gerasi et al.[Citation13] determined that Leu. mesenteroides was 41.6% of surface parts of Manura cheese, and Lb. paracasei was 21.8% of the inner parts of the cheese at the end of ripening. Choisy et al.[Citation14] also stated that lactic acid bacteria was the dominant flora in Roquefort cheese. During ripening, the number of enterococci increased most as the proportion of the other genera decreased. This pattern is common in most cheese varieties. Lactococci develops at the beginning of the process and carries out the acidification of the milk, which leadis to the formation of the curd; subsequently, the lactic acid flora changes, and enterococci and lactobacilli become dominant and participate in the ripening.[Citation3] It was reported that enterocci was dominant in the flora of moldy cheeses that have high pH.[Citation5,Citation11,Citation15,Citation16] The pH undergoes a more moderate decrease on the surface than at the core. This fact can be attributed to the more restricted growth of the LAB on the surface and the same principle applies to the greater evolution of the fungi flora on the cheese surface, whose metabolic activity contributes to neutralizing the acidity.[Citation15] Furthermore, enterococci affected the growth of other LAB, due to their intense proteolytic activity, which favors gas and acid production by Leuconostoc and lactococci.[Citation3,Citation17,Citation18] In addition, Leuconostoc sp. contributes to the development of the inside texture of cheese due to their heterofermentative nature.
It was reported that lactobacilli is commonly found in cheeses with long-term ripening and does not extremely affect the increase in pH.[Citation19 Citation,20]Pediococci was found in high amount (17.5%) in fresh cheese; their rates decreased to 6.7% at the end of ripening for 30 days. Reportedly, these bacteria might have contributed to the flavor and accelerated cheese ripening.[Citation13] The results of our work showed that E. faecalis was the dominant strain in blue moldy Tulum cheese during ripening. In addition, Lb. parabuhneri and Leu. mesenteroides were also found in high amounts in the cheese. Therefore, for industrial production of the cheese, these two strains could be used in a starter culture combination. However, further investigations should be carried out for the improvement and standardization of the cheese.
CONCLUSION
The results of this work showed that E. faecalis was the dominant strain in blue moldy Tulum cheese during ripening. In addition, Lb. parabuhneri and Leu. mesenteroides were also found in high amounts in the cheese. Therefore, these two strains could be used in a starter culture combination for the industrial production of this cheese. However, further investigations should be carried out for the improvement and standardization of the cheese.
REFERENCES
- Erdogan , A. , Gurses , M. and Sert , S. 2003 . Isolation of moulds capable of producing mycotoxins from blue mouldy Tulum cheeses produced in Turkey . Int. J. Food Microbiol , 85 : 83 – 85 . [PUBMED] [INFOTRIEVE]
- Gurses , M. , Erdogan , A. and Sert , S. 2004 . Effect on aflatoxin formation of different storage conditions in Tulum cheese ınoculated with Aspergillus parasiticus NRRL 2999 . Turk. J. Vet. Anim. Sci. , 28 : 233 – 238 .
- Lopez-Diaz , T.M. , Alonso , C. , Roman , C. , Garcia-Lopez , M.L. and Moreno , B. 2000 . Lactic acid bacteria isolated from a hand-made blue cheese . Food Microbiol , 17 : 23 – 32 . [CROSSREF]
- Guizani , N , Kasapis , S , Al-Attabi , Z.H and Al-Ruzeiki , M.H. 2002 . Microbiological, physicochemical, and biochemical changes during ripening of Camembert cheese made of pasteurized cow's milk . Int. J. Food Properties , 5 ( 3 ) : 483 – 494 . [CROSSREF]
- Kurt , A. and Öztek , L. 1984 . Savak Tulum peynirinin yap m teknigi üzerine arastrmalar . Atatürk Üniv. Ziraat Fak. Derg. , 15 : 65 – 77 .
- Erdogan , A. and Sert , S. 2004 . The mycotoxin-forming ability of two penicillium Roqueforti strains in blue Tulum cheese ripened at various temperatures . J. Food Prot. , 67 ( 3 ) : 533 – 535 . [PUBMED] [INFOTRIEVE]
- Harrigan , W.F. and MeCance , M.E. 1976 . Laboratory Methods in Food and Dairy Microbiology , 2nd , London : Academic Press .
- Schleifer , K.H. and Kilpper-Balz , R. 1987 . Molecular and chemotaxonomic approaches to the classification of streptococci enterococci and lactococci: a review . Syst. Appl. Microbiol. , 10 : 1 – 19 .
- Schleifer , K.H. , Kraus , J. , Dvorak , C. , Kilpper-Balz , R. , Collins , M.D. and Fischer , W. 1985 . Transfer of streptococcus lactis and related streptococci to the genus lactococcus gen Nov. Syst . Appl. Microbiol. , 6 : 183 – 195 .
- Facklam , R.R. and Collins , M.D. 1989 . Identification of enterococcus species isolated from human infections by a conventional test scheme . J Clinical Microbiol. , 27 : 731 – 734 .
- Reuter , G. 1985 . Elective and selective media for lactic acid bacteria . Int. J. Food Microbiol. , 2 : 55 – 68 . [CROSSREF]
- Durlu-Ozkaya , F. , Xanthopoulos , V. , Tunail , N. and Litopoulou-Tzanetaki , E. 2001 . Technologically important properties of lactic acid bacteria isolated from Beyaz cheese made from raw ewes milk . J. App. Microbiol. , 91 : 861 – 870 . [CROSSREF]
- Kalogridou-Vassiliadou , D. , Tzanetakis , N. and Litopoulou-Tzanetaki , E. 1994 . Microbiological and physicochemical characteristics of Anthotyro, a Greek traditional whey cheese . Food Microbiol. , 11 : 15 – 19 . [CROSSREF]
- Gerasi , E. , Litopoulou-Tzanetaki , E. and Tzanetakis , N. 2003 . Microbiological study of Manura, a hard cheese made from raw ovine milk in the Greek island Sifnos . Int. J. Dairy Tech. , 56 : 117 – 122 . [CROSSREF]
- Choisy , G. , Gueguen , M. , Lenoir , J. , Schmidt , J.L. and Tourneur , C. 1987 . “ Les phenomenes microbiens ” . In Le fromage Edited by: Eck , A. 259 – 290 . Paris Lavoisier (Tech. and Doc.)
- Erdogan , A. Erzurum yöresinden toplanan küflü Tulum Peynirlerinden izole edilen . 2000 . Penicillium roqueforti suslarinin Tulum Peynirindeki aktivitesi ile olusan toksikolojik ve kimyasal degismeler (Doktora tezi). Atatürk Üniv. Fen Bilimleri Enstitüsü Erzurum.
- García Fontan , M.C. , Franco , I. , Prieto , B. , Tornadijo , M.E. and Carballo , J. 2001 . Microbiological changes in San Simon cheese throughout ripening and its relationship with physico-chemical parameters . Food Microbiol. , 18 : 25 – 33 . [CROSSREF]
- Caridi , A. 2003 . Identification and first characterization of lactic acid bacteria isolated from the artisanal ovine cheese Pecorino del Poro . Int. J. Dairy Technol. , 56 : 105 – 110 . [CROSSREF]
- Rehman , S. , Banks , J.M. , Brechany , E.Y. , Muir , D.D. , McSweeney , P.L.H. and Fox , P.F. 2000 . Influence of ripening temperature on the volatiles profile and flavour of cheddar cheese made from raw or pasteurised milk . Int Dairy J. , 10 : 55 – 65 . [CROSSREF]
- Rehman , S. , Banks , J.M. , McSweeney , P.L.H. and Fox , P.F. 2000 . Effect of ripening temperature on the growth and significance of non-starter lactic acid bacteria in cheddar cheese made from raw or pasteurised milk . Int Dairy J. , 10 : 45 – 53 . [CROSSREF]
- Fortina , M.G. , Ricci , G. , Acquati , A. , Zeppa , G. , Gandini , A. and Manachini , P.L. 2003 . Genetic characterization of some lactic acid bacteria occurring in an artisanal protected denomination origin (PDO) Italian cheese,the Toma piemontese . Food Microbiol. , 20 : 397 – 404 . [CROSSREF]
- Adamberg , K. , Kask , S. , Laht , T. and Paalme , T. 2003 . The effect of temperature and pH on the growth of lactic acid bacteria: pH-auxostat study . Int. J. Food Microbiol. , 85 : 171 – 183 . [PUBMED] [INFOTRIEVE] [CROSSREF]