Abstract
Little is known about the RNA polymerase III (Pol III) complex assembly and its transport to the nucleus. We demonstrate that a missense cold-sensitive mutation, rpc128-1007, in the sequence encoding the C-terminal part of the second largest Pol III subunit, C128, affects the assembly and stability of the enzyme. The cellular levels and nuclear concentration of selected Pol III subunits were decreased in rpc128-1007 cells, and the association between Pol III subunits as evaluated by coimmunoprecipitation was also reduced. To identify the proteins involved in Pol III assembly, we performed a genetic screen for suppressors of the rpc128-1007 mutation and selected the Rbs1 gene, whose overexpression enhanced de novo tRNA transcription in rpc128-1007 cells, which correlated with increased stability, nuclear concentration, and interaction of Pol III subunits. The rpc128-1007 rbs1Δ double mutant shows a synthetic growth defect, indicating that rpc128-1007 and rbs1Δ function in parallel ways to negatively regulate Pol III assembly. Rbs1 physically interacts with a subset of Pol III subunits, AC19, AC40, and ABC27/Rpb5. Additionally, Rbs1 interacts with the Crm1 exportin and shuttles between the cytoplasm and nucleus. We postulate that Rbs1 binds to the Pol III complex or subcomplex and facilitates its translocation to the nucleus.
INTRODUCTION
All eukaryotic cells have at least three different RNA polymerases (Pol). Pol I synthesizes the large precursor of rRNA, Pol II produces mainly mRNAs, and Pol III generates tRNAs, 5S rRNA and other small noncoding RNAs.
Pol III, which is the focus of this work, is a heteromultimeric protein complex that contains 17 distinct subunits in Saccharomyces cerevisiae. The two largest subunits, C160 and C128, form opposite sides of the active-center cleft and harbor the catalytic activity, and they are related to the β′ and β components of the α2ββ′ core of bacterial RNA polymerase. Homology to the bacterial α subunit, although less strong, was also observed, with the subunit AC40 common for yeast and Pol III and Pol I. A second α-like subunit, AC19, forms a heterodimer with AC40, which is a functional equivalent to the α2 homodimer in prokaryotes (Citation1). The polymerase core, in addition to the α2ββ′-like structure, contains six small subunits, which are structurally and functionally conserved from yeast to humans. The small core subunits either bind or bridge the two largest catalytic subunits. In addition to the central core, the remaining subunits of Pol III are organized in heterodimeric or trimeric stalks or subdomains. Significantly, the C37 to C53 and C82 to C34 subdomains, which are stably bound to the RNA Pol III core, resemble Pol II general transcription factors TFIIF and TFIIE (Citation2).
Little is known about how polymerase complexes are assembled from the subunits, how they reach their functional destination, and how they dissociate after transcription. In bacteria, the assembly of RNA polymerase starts with the formation of the αα dimer, which then interacts with the β subunit to yield an ααβ intermediate, which finally recruits subunit β′ (Citation3). Although eukaryotic RNA polymerase has never been reconstituted in vitro, the assembly pathway has been examined by early dissociation experiments (Citation4, Citation5). A stepwise dissociation of purified yeast Pol III resulted in a stable subcomplex, C128/AC40/AC19/ABC10β/ABC10α, which was proposed to represent a core subassembly analogous to ααβ from Escherichia coli and β′-like module C160/ABC14.5/ABC27 (Citation5). Assuming similar assembly pathways for all polymerases, it is likely that they together form the Pol III core and that peripheral subcomplexes are subsequently added as preassembled building blocks (Citation6).
Increasing quantities of data suggest that the eukaryotic polymerases are assembled in the cytoplasm and transported to the nucleus as whole complexes (Citation6). Furthermore, RNA polymerases must be delivered to the specific subnuclear locations where they function. Pol I complexes are recruited to multiple copies of rDNAs, which are organized in the nucleous. Pol II must be delivered to specific genes, and the nuclear positioning of a given Pol II-transcribed gene can be important for its expression (Citation7). Finally, Pol III-transcribed tRNA genes occupy distinct subnuclear positions; both the nucleous and nuclear pores are considered in budding yeast (Citation8, Citation9). It is also possible that a nuclear/nucleous structure exists, which functions as a platform to localize these assembly events.
Assembly of polymerases requires proteins which are not components of mature enzymes, and since there is no obvious nuclear localization signal on any of the polymerase subunits, specific carrier proteins participate in the nuclear import of assembled complexes. Only recently has the identification of proteins involved in biogenesis of Pol II been pursued. A cytoplasmic Pol II intermediate in human cells was found to be associated with HSP90 and its prefoldin-like cochaperone RPAP2 (Citation10). A fully assembled enzyme with bound RPAP2 is then imported to the nucleus, followed by CRM1-dependent export of RPAP2 to the cytoplasm (Citation11). Multiple interactions between human Pol II and the small GTPase GPN1 indicated its involvement in Pol II assembly and nuclear import (Citation12). Npa3, the yeast homolog of GPN1, is required for nuclear localization of yeast Pol II and binds it in a GTP-dependent manner (Citation13), which argues that the mechanism involved in the subcellular localization of Pol II requires the catalytic function of GNP proteins and is conserved from yeast to mammals (Citation14). Two other proteins involved in Pol II biogenesis, Iwr1 and Rtp1, were identified in genetic screens for suppressors of the growth defect caused by depletion of NC2, a negative regulator of mRNA transcription (Citation15, Citation16). Iwr1 binds yeast Pol II in the active-center cleft between the two largest subunits, possibly facilitating or sensing complete Pol II assembly in the cytoplasm. Importantly, Iwr1 contains a bipartite nuclear localization signal (NLS) and when associated with Pol II acts as nuclear import factor. Once Pol II engages with the promoter DNA, Iwr1 is released and recycled to the cytoplasm, ready to initiate a new cycle (Citation17). Rtp1 interacts, to different extents, with several Pol II subunits and with members of the R2TP complex. Besides its role in subunit assembly, Rtp1, which is a karyopherin-like protein, is likely involved in nuclear transport of Pol II (Citation16).
Recent studies indicate that mechanisms similar to those identified for Pol II might apply to the other two Pols and that these processes might be interconnected. One common factor is the yeast prefoldin Bud27, which mediates the correct assembly of all three Pol complexes prior to their translocation to the nucleus, in a process dependent on shared subunit, Rpb5 (Citation18). Moreover, the putative yeast GTPase Gpn2 is possibly involved in assembly of Pol II and Pol III (Citation19). The molecular details of the assembly pathway and the assembly factors involved in biogenesis of Pol III remain, however, mostly uncharacterized.
In this study, we have focused on the rpc128-1007 mutation in the gene encoding the second largest subunit of Pol III, C128, which is a homologue of the bacterial β subunit. According to structure prediction, the mutant site is located near the contact points for the association of C128 with the AC40/AC19 subcomplex corresponding to the α2 homodimer. We demonstrate that the rpc128-1007 mutation has severe consequences for the assembly of the active Pol III complex and hence for the capacity of the cell to support transcription activity and growth. These defects are partially suppressed by the overproduction of Rbs1 protein, which might function as an assembly/import factor.
MATERIALS AND METHODS
Strains.
The yeast strains used in these studies are listed in . MJ15-9C, harboring the rpc128-1007 mutation, was generated previously (Citation20) in the genetic background of MB159-4D. MB159-4D and isogenic MB159-2C were considered wild-type controls. MJ15-10A was generated from a backcross. Strain JT2-2D, containing the rpc128-1007 mutation and encoding a hemagglutinin (HA)-tagged C160 subunit of Pol III, was generated by the cross MJ15-9C × MW4415. The rpc31-236 (Citation25), rpc160-112 (Citation26), rpc160-270 (Citation27), rpc160-750 (Citation28), and rpc11-Sp (Citation29) Pol III mutants were from the Service de Biologie Intégrative et Génétique Moléculaire, CEA/Saclay (France).
TABLE 1 Saccharomyces cerevisiae strains used in this study
Rbs1-green fluorescent protein (GFP), C82-GFP, and C53-GFP strains, purchased from Invitrogen, were subsequently crossed with JT2-2D to generate rpc128-1007 mutants encoding the respective Pol III subunits GFP tagged at the 3′ termini. The Rbs1-Myc strain was obtained by transformation of MJ15-10A with the kanMX6 cassette PCR amplified from pFA6-13Myc-kanMX6 plasmid using primers RBS1_F2 (5′-TACTGATTCGGTAGAGATGAAATTTGATAAATTAAACATTCGGATCCCCGGGTTAATTAA-3′) and RBS1_R1 (5′-GTTACGTAATTTTTTCGCTATGTATAGTTCACTCCTCACGGAATTCGAGCTCGTTTAAAC-3′) (Citation30). Transformants were selected on yeast extract-peptone-dextrose (YPD) medium with Geneticin, and the construction of the Rbs1-Myc strain was confirmed by PCR and by Western blotting with anti-Myc antibody (Roche). The Rbs1-Myc strain was crossed with MNY8 (crm1-T359C) and MJ15-9C (rpc128-1007) to generate the respective single and double mutants used in immunofluorescence experiments.
For the deletion of RBS1 in MB159-2C, the Geneticin cassette surrounded on each side by RBS1 flanking regions was synthesized by PCR, using as a template the genomic DNA of BY4741 rbs1Δ and specific primers delRbsF (5′-TTAACAATGATTGCGGCGTA-3′) and delRbsR (5′-ATGGAGGTGGATGAAAACCA-3′). The 2,312-bp product was transformed into MB159-2C cells, and transformants were selected on YPD supplemented with Geneticin. The replacement of RBS1 with the Geneticin cassette was confirmed by PCR and sequencing.
Plasmids.
pMJ14, selected from a yeast genomic library on centromeric plasmid pRS316 (URA3 CEN6) (constructed by M. Cieśla), contains four genes: RPB10, MGM1, STE4, and SAS5. Analysis of pMJ14 (see Fig. S1 in the supplemental material) involved ClaI/ClaI and XhoI/XhoI deletions and subcloning of BamHI/BamHI. Final deletion of the XhoI/HpaI fragment from the pMJ14 derivative resulted in pMJ24 (called here [RPB10]), which contains RPB10 and the 3′ part of the MGM1 gene. Plasmid pMJ17, cloned from a commercial yeast genomic library on the multicopy YEp13 vector (LEU2 2μ), contains the RBS1 and PPH22 genes. The SacI/SalI fragment of pMJ17 was subcloned in YEp181 (LEU2 2μ), resulting in pMJ22. A genomic fragment with RBS1, PPH22, and UFD2 was independently cloned as plasmid pMJ28 from the genomic library on the multicopy pFL44L vector (URA3 2μ). Both pMJ22 and pMJ28 are here called [RBS1]. Plasmid pLH3, kindly provided by P. Thuriaux, is a pRS316 derivative containing the RPC128 gene on pRS316 and is here called [RPC128]. Plasmid pC160-6, called here [RPC160], contains the RPC160 gene on the pUN75 vector (URA3 CEN4) (Citation26).
Media.
Yeasts were grown in YPD (2% glucose, 2% peptone, 1% yeast extract) or SC (2% glucose, 0.67% yeast nitrogen base without amino acids). SC−ura or SC−Leu contained 20 μg/ml of the amino acids required for growth, except for uracil or leucine, respectively. As required, the YPD medium was supplemented with Geneticin (200 μg/ml) or leptomycin B (LMB) (100 ng/ml). LB medium (1% Bacto tryptone, 0.5% yeast extract, 1% NaCl, pH 7.5) was used for growing E. coli strains. As required, the LB medium was supplemented with ampicillin (60 μg/ml). Solid media contained 2% agar. All reagents used for the media were Difco products.
Cloning of rpc128-1007 suppressors.
The suppressors of the rpc128-1007 mutation were cloned by complementation of the cold-sensitive growth phenotype of the MJ15-9C strain on YPD medium. This strain was transformed with three different yeast genomic libraries: a library based on a single-copy pRS316 vector (constructed by M. Cieśla), a multicopy library on the pFL44L vector provided by F. Lacroute (Citation31), and a multicopy library on the YEp13 vector (Citation32). MJ15-9C cells were transformed with library plasmids by using the lithium acetate method (Citation33) and plated on SC−ura or SC−Leu medium. Single transformants were patched again onto selective medium and replicated on YPD. Plasmids which complemented the rpc128-1007 phenotype were screened during incubation of YPD plates at 16°C for 7 days, with growth monitored every day. To check whether the restored growth on YPD at 16°C correlated with a Leu+ or Ura+ phenotype, plasmids were lost from transformants which passed growth selection, and the correlation of the presence of the plasmid and the complementation phenotype was studied. Then, the plasmids from cells which showed such a correlation were isolated from yeast transformants and introduced into bacteria. Plasmid DNA obtained from single E. coli transformants were used in a second transformation (retransformation) of the initial MJ15-9C strain. Genomic inserts contained in plasmids that complemented growth deficiency on YPD at 16°C were identified by sequencing.
RNA isolation and Northern analysis.
Total RNA was prepared from yeast by the hot-phenol method as described previously (Citation34). The quantity and quality of RNA were analyzed with a NanoDrop 1000 (Thermo ND-1000) and verified by analysis on ethidium bromide (EtBr)-stained agarose gels.
Five micrograms of total RNA isolated from yeast cells was resolved by 10% PAGE with 8 M urea. RNA was hybridized as described previously (Citation35). The probes were 5′-GCGCTCTCCCAACTGAGCT-3′ for tRNA-Phe (GAA), 5′-TATTCCCACAGTTAACTGCGGTCA-3′ for the intron of tRNA-Leu (CCA), 5′-CGAGTCGAACGCCCGAT-3′ for tRNA-Tyr (GUA), and 5′-GCGTTGTTCATCGATGC-3′ for 5.8S rRNA. RNA was quantified using an FLA-7000 PhosphorImager (Fujifilm). Band intensities were quantified using MultiGauge v3.0 software (Fujifilm). The statistical significance was computed using a t test as implemented in OpenOffice.org.
Western analysis.
Western blotting was performed as described previously (Citation35). Protein extracts were separated by 8%, 10%, or 12% SDS-PAGE and hybridized with mouse monoclonal antibodies anti-HA (Covance) at a 1:5,000 dilution for 2 h, anti-GFP (Roche) at a 1:5,000 dilution for 1 h, 9E10 anti-Myc (Roche) at a 1:2,000 dilution for 1 h, and anti-Vma2 (Molecular Probes) at a 1:10,000 dilution for 1 h and with rabbit polyclonal antibodies anti-HA (Sigma) at a 1:2,000 dilution for 1 h, anti-Rpb1 (Santa Cruz Biotechnology) at a 1:1,000 dilution for 1 h, anti-C82 at a 1:5,000 dilution for 1 h, anti-C53 at a 1:20,000 dilution for 1 h, and anti-AC40 at a 1:1,000 dilution for 2 h. Antibodies specific for Pol III subunits were a gift from O. Lefebvre. The nitrocellulose membranes were then incubated with secondary polyclonal goat anti-mouse or anti-rabbit antibodies coupled to horseradish peroxidase (Dako) at a 1:5,000 dilution for 1 h and visualized with a chemiluminescence detection kit (Bio-Rad).
Fluorescence microscopy.
Immunofluorescence was performed as described previously (Citation35). 9E10 anti-Myc mouse antibody (1:500 for 1 h; Roche) and anti-HA mouse antibody (1:500 for 1 h; Covance) were added, followed by secondary Cy3-conjugated anti-mouse antibody (1:250 for 1 h; Jackson ImmunoResearch Laboratories). Immunofluorescence and live imaging of cells expressing GFP-tagged C53 and C82 Pol III subunits were done with a Carl Zeiss Axio ImagerM2 microscope equipped with a 100× oil objective. Images were collected with the AxioVision 4.8 program.
Rbs1-AC40 coimmunoprecipitation.
Cells expressing epitope-tagged Rbs1 or control proteins and strain BY4741 rbs1Δ were grown in YPD medium. Total extracts were prepared from 100 ml of culture that was pelleted and resuspended in 0.5 ml of immunoprecipitation (IP) buffer containing O-Complete protease inhibitor (Roche) (50 mM HEPES-KOH [pH 7.5], 250 mM NaCl, 1 mM EDTA, 0.25% NP-40, 0.5 mM dithiothreitol [DTT], and 20% glycerol, for the IP-Myc experiment and 50 mM HEPES-KOH [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.05% NP-40, 0.5 mM DTT, and 5% glycerol for the IP-GFP experiment). Lysis was performed in the presence of glass beads using a Vibramax disruptor (IKA) at maximum speed for 1 h at 4°C. The cell debris were eliminated by centrifugation (20 min at 4°C and 14,000 rpm). The protein concentration was determined with the Bio-Rad protein assay. Fifty microliters of a suspension of Dynabeads PanMouse IgG magnetic beads (Invitrogen), washed three times with 0.5% bovine serum albumin in phosphate-buffered saline (PBS), was incubated with mouse monoclonal 9E10 anti-Myc antibody (1 μg) or anti-GFP antibody (2 μg) for 3 h at 4°C. After washing in 0.5% bovine serum albumin in PBS (two times) and then in IP buffer (two times), the beads were incubated overnight with 2 mg (IP-Myc) or 3 mg (IP-GFP) of protein extracts in IP buffer with gentle shaking at 4°C. Then beads were washed three times with IP buffer. Immunoprecipitated proteins were released from the beads by boiling them for 5 min. Eluted proteins were separated by SDS-PAGE and analyzed by Western blotting with 9E10 anti-Myc, anti-GFP, and anti-AC40 antibodies.
Coimmunoprecipitation of C160 with Pol III subunits.
Yeast cells expressing HA epitope-tagged C160 (MW4415), the rpc128-1007 mutant (JT2-2D) transformed with empty vector (YEp181) or plasmid with the RBS1 gene (pMJ22), and untagged C160 (YPH499) were preincubated in SC medium and grown in YPD medium to exponential phase. Pellets, corresponding to 100 ml of culture, were resuspended in 0.5 ml IP buffer (50 mM HEPES-KOH [pH 7.5], 100 mM NaCl, 1 mM EDTA, 0.05% NP-40, 0.5 mM DTT, 20% glycerol, O-Complete protease inhibitor [Roche]). Further steps were the same as described above for Rbs1-AC40 coimmunoprecipitation. Beads were incubated with mouse monoclonal anti-HA antibody (3 μg) for 3 h at 4°C and, after washing, were incubated overnight with 2 mg of protein extracts. Eluted proteins were separated by 8% SDS-PAGE and analyzed by Western blotting with the respective antibodies.
GFP affinity purification.
For affinity purification of proteins interacting with Rbs1, yeast extracts were prepared from the Rbs1-GFP strain, its derivative harboring the rpc128-1007 mutation, and an isogenic strain with untagged Rbs1, which served as a negative control. Ten liters of cells growing in YPD medium was harvested at an optical density at 600 nm (OD600) of 3, centrifuged, suspended in lysis buffer (250 mM NaCl, 40 mM HEPES-KOH [pH 8.0], 20 mM β-mercaptoethanol), and frozen at −80°C. Yeast cells were broken in a blender with dry ice and lysis buffer (20 ml buffer on 100 g of pellets) with added protease inhibitors (Roche). The extracts were then centrifuged at 20,000 rpm for 25 min at 4°C, and the supernatants were transferred into new tubes and centrifuged again at 35,000 rpm for 1 h 30 min at 4°C. Supernatants were recovered and dialyzed against buffer D (150 mM NaCl, 40 mM HEPES-KOH [pH 8.0], 1 mM DTT, 20% glycerol, protease inhibitors) for 3 h at 4°C. After dialysis, extracts were frozen and kept at −80°C. The epitope-specific resin anti-GFP–Sepharose (prepared as described previously [Citation36]) was incubated with extracts overnight with gentle shaking at 4°C. Resins and captured proteins were transferred into a column (Bio-Rad) and washed three times with 10 ml of IPP150 buffer (150 mM NaCl, 10 mM Tris-HCl [pH 8], 0.1% Triton X-100). Proteins were eluted by addition of 200 μl 0.3 M glycine (pH 2.5), and the supernatant was collected and neutralized by addition of 250 μl of 100 mM Tris (pH 8). The purified proteins were collected into Eppendorf tubes, and 500 μl of H2O was added. The collected samples were divided into 2 equal parts. The bait proteins were precipitated by adding PRM reagent (0.05 mM pyrogallol red, 0.16 mM sodium molybdate, 1 mM sodium oxalate, 50 mM succinic acid, pH 2.5) and incubating for 20 min at room temperature. The precipitated proteins were centrifuged at 14,000 rpm for 15 min at room temperature, and the supernatant was removed. Prior to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis, samples were evaluated by SDS-PAGE and silver staining.
Protein analysis by LC-MS/MS.
LC-MS was performed in the Laboratory of Mass Spectrometry (IBB PAS, Warsaw, Poland) using a nanoAquity ultraperformance liquid chromatography (UPLC) system (Waters) coupled to an LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific) as previously described (Citation37). Raw MS data were analyzed and quantified using Andromeda/MaxQuant (Citation38) software.
RESULTS
A Gly1007Ala substitution in the C128 subunit affects Pol III assembly.
The rpc128-1007 mutant strain was identified in a genetic screen for spontaneous second-site suppressors that bypass the growth defect resulting from the inactivation of the gene encoding a general negative regulator of Pol III, Maf1 (Citation20). This mutant has a single nucleotide change in the RPC128 gene, resulting in a replacement of a Gly with Ala, localized in the C terminus of C128. The rpc128-1007 mutant has 1.6-fold-reduced tRNA levels and therefore suppresses the defect of Maf1 inactivation caused by increased or unbalanced levels of various tRNAs (Citation20). The reason for impaired Pol III activity in rpc128-1007 cells was, however, unknown.
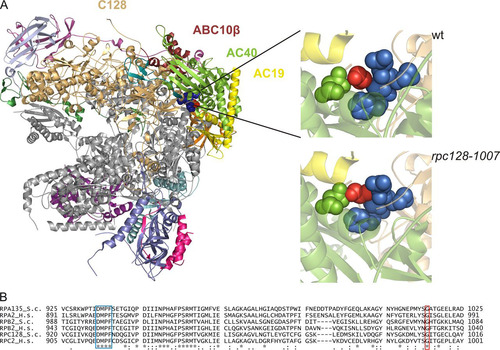
Glycine 1007 in the C128 subunit of Pol III is conserved among the second-largest subunits of Pol I and Pol II and is located within the strictly preserved structural fold (amino acids [aa] 933 to 1052 of C128, corresponding to aa 1001 to 1120 in Rpb2 [Citation39]). To locate this region in the whole Pol III structure, we have generated a comparative model of C128 based on the Pol I crystal structure at 2.8-Å resolution (Citation40) and the available cryo-electron microscopy (cryo-EM) map of Pol III at 9.9-Å resolution (Citation41). As shown in , glycine residue 1007 of the C128 subunit is not exposed on the surface but is located in a loop that contacts two helices: one of subunit AC40 and one of AC19. Gly1007 is also in close proximity to another loop located on the C128 surface adjacent to AC40 and containing a conserved DMPF motif (aa 929 to 932). In the mutant strain an additional methyl group of Ala 1007 (marked in red in , left panel) fills in the space between two loops, which may change flexibility in this region. Close contacts of amino acid 1007 of C128 with subunits AC40 and AC19 make this position very strategic for the assembly and stability of the entire Pol III.
FIG 1 Localization of Gly1007 in the C128 subunit sequence and Pol III structure. (A) The C128 protein structure was modeled based on the respective RNA Pol I subunit A135. In the available 2.8-Å structure of RNA Pol I (PDB ID 4C2M), subunit A135 was replaced by the model of C128. The localization of selected subunits in the complex and position of Gly1007 of C128 are shown. This glycine (marked in red) is located in the C128 structure close to another conserved motif, DMPF (marked in blue), as revealed by magnification of the respective fragment of the polymerase structure in the wild-type (wt) (Gly1007 marked in red) and mutant (rpc128-1007) (Ala1007 marked in red) strains. The structure model was produced using the PyMOL program. (B) the DMPF motif (aa 929 to 932) and glycine 1007 in the C128 subunit of Pol III in S. cerevisiae (RPC128_S.c) are conserved in sequences of the second-largest subunits of yeast Pol I (RPA135_S.c) and Pol II (RPB2_S.c.), as well as in their human homologues (RPC2_H.s, RPA2_H.s, and RPB2_H.s, respectively).
We analyzed the effect of the mutation by growing the rpc128-1007 cells at different temperatures. As shown in and , the rpc128-1007 mutant grew more slowly under standard conditions at 30°C and was sensitive to low temperature.
FIG 2 Inhibited growth and reduced levels of the largest subunit of Pol III in the rpc128-1007 mutant. (A) The rpc128-1007 mutant (MJ15-9C) and the control wt strain (MB159-4D) were inoculated (1/100) into liquid rich glucose medium (YPD) and incubated at 30°C with shaking. The growth was monitored by OD600 measurements. (B and C) rpc128-1007 cells encoding C160-HA were examined by immunofluorescence and Western blotting by using anti-HA antibody and anti-Rpb1 antibody. (D) As indicated, the rpc128-1007 mutant was transformed with empty vector [-], a plasmid harboring RPC128, or a plasmid harboring RPC160. Tenfold serial dilutions of overnight cultures in minimal medium (SC−ura) were plated on SC−ura and incubated at 30°C for 3 days or on rich glucose medium (YPD) and incubated at 16°C for 4 days.
![FIG 2 Inhibited growth and reduced levels of the largest subunit of Pol III in the rpc128-1007 mutant. (A) The rpc128-1007 mutant (MJ15-9C) and the control wt strain (MB159-4D) were inoculated (1/100) into liquid rich glucose medium (YPD) and incubated at 30°C with shaking. The growth was monitored by OD600 measurements. (B and C) rpc128-1007 cells encoding C160-HA were examined by immunofluorescence and Western blotting by using anti-HA antibody and anti-Rpb1 antibody. (D) As indicated, the rpc128-1007 mutant was transformed with empty vector [-], a plasmid harboring RPC128, or a plasmid harboring RPC160. Tenfold serial dilutions of overnight cultures in minimal medium (SC−ura) were plated on SC−ura and incubated at 30°C for 3 days or on rich glucose medium (YPD) and incubated at 16°C for 4 days.](/cms/asset/8b16c0e7-8096-4f66-ac36-e7576e81640e/tmcb_a_12275564_f0002_oc.jpg)
Initial examination of the rpc128-1007 mutant by immunofluorescence suggested a decreased nuclear concentration of C160, the largest subunit of Pol III (). Subsequently, Western blot analysis confirmed that the steady-state levels of the C160 protein in the rpc128-1007 cells were reduced relative to those of the wild-type protein, whereas the level of Rpb1, the largest subunit of Pol II, was not affected (). These observations raise the possibility that instability of the largest Pol III subunit, C160, is a primary reason for the rpc128-1007 growth phenotype. To compensate for conceivable instability, we introduced a C160-encoding plasmid into the rpc128-1007 cells. Increasing the C160 expression in this manner did not, however, overcome the cold sensitivity of the rpc128-1007 mutant (). Thus, we conclude that the defects of Pol III observed in the rpc128-1007 mutant are not caused by the limiting amount of the C160 subunit per se but are instead related to the consequences of inefficient Pol III assembly. We also found it noteworthy that defects in the assembly of multisubunit complexes are most often associated with cold sensitivity, because protein-protein interactions are entropy driven and intrinsically cold sensitive (Citation42).
The cold sensitivity of the Pol III assembly mutant is suppressed by overproduction of the ABC10β subunit or Rbs1, a protein of unknown function.
In an attempt to uncover novel genes that can suppress the cold sensitivity of the rpc128-1007 mutant, we undertook a genetic suppressor screening. We searched for suppressors by transforming the mutant strain MJ15-9C with diverse yeast genomic libraries and selecting colonies that grew at the restrictive temperature of 16°C (). For each transformant, we ascertained that plasmid extraction and retransformation of the original mutant strain suppressed its phenotype and that removing the suppressor plasmid restored the mutant phenotype. Plasmids containing the wild-type RPC128 gene complemented the rpc128-1007 growth defect, as expected. The major class of multicopy suppressors which exhibited a wild-type growth rate at 16°C harbored plasmids with the RBS1 gene, whose function is unknown. The other suppressors restored growth at 16°C only partially. Selection from a single-copy library resulted in isolation of the RPB10 gene, encoding the ABC10β subunit shared by all three RNA polymerases and involved in polymerase assembly (Citation43).
TABLE 2 Overdose suppressors of the rpc128-1007 mutation
Since the growth defect of the rpc128-1007 mutant at 16°C is suppressed by plasmids containing RPB10 and RBS1 (; see Fig. S1 in the supplemental material), we checked whether Pol III activity is restored. Analysis of total RNA on ethidium bromide-stained gels confirmed that the reduced tRNA levels in the rpc128-1007 mutant were restored by overdose of both suppressor genes (). To evaluate the effect of RPB10 and RBS1 on de novo tRNA synthesis, we employed Northern hybridization using specific oligonucleotide probes that identify tRNA precursors. Cells were preincubated in minimal medium and next grown in a glucose medium at 30°C with a shift to the restrictive temperature of 16°C for 2 h. The levels of 5.8S rRNA were comparable in rpc128-1007 and wild-type cells and thus served as an internal control. We monitored the synthesis of three representative intron-containing tRNAs, tRNA-Leu (CAA), tRNA-Phe (GAA), and tRNA-Tyr (GUA) (). The levels of tRNA precursors, both unprocessed initial transcripts and end-matured intron-containing pre-tRNAs, were more than 2-fold decreased in the rpc128-1007 mutant relative to the wild-type control (), confirming the defect in the de novo tRNA synthesis. Although 5S rRNA is also a product of Pol III, its level is not affected in rpc128-1007 cells. This is not surprising, since many other Pol III mutants lead to a decrease of tRNA synthesis but do not alter the transcription of 5S rRNA (Citation44).
FIG 3 ABC10β and Rbs1 as overdose suppressors of the rpc128-1007 mutation. As indicated, the rpc128-1007 mutant (MJ15-9C) and the control wt strain (MB159-4D) were transformed with empty vector [-], a plasmid harboring RBS1, or a plasmid harboring RPB10. (A) Tenfold serial dilutions of overnight cultures were plated on YPD following incubation at 30°C for 3 days or at 16°C for 4 days. For control of plasmid maintenance, the same cells were plated on minimal glucose medium (SC−ura) and incubated at 30°C for 3 days. (B) Cells preincubated in minimal medium were grown in YPD at 30°C, shifted to 16°C for 2 h, and harvested. Total RNA was isolated, separated on a 2.8% agarose gel, and stained with EtBr using equal amounts of RNA per lane (5 μg). (C) RNA was analyzed by Northern blotting using probes specific for pre-tRNA-Leu (CAA), tRNA-Phe (GAA), and tRNA-Tyr (GUA). 5.8S rRNA served as the loading control. Positions of initial transcripts, end-processed intron-containing pre-tRNA (IVS), and mature tRNA are indicated. (D) pre-tRNA forms were quantified. Bars represent levels for primary transcripts and intron-containing precursors normalized to the loading control. Standard deviations were estimated on the basis of three independent experiments. The P values calculated for ratios of pre-tRNAs (rpc128-1007[-]/wt[-], rpc128-1007[-]/rpc128-1007[RPB10], and rpc128-1007[-]/rpc128-1007[RBS1]) showed statistical significance (P < 0.05).
![FIG 3 ABC10β and Rbs1 as overdose suppressors of the rpc128-1007 mutation. As indicated, the rpc128-1007 mutant (MJ15-9C) and the control wt strain (MB159-4D) were transformed with empty vector [-], a plasmid harboring RBS1, or a plasmid harboring RPB10. (A) Tenfold serial dilutions of overnight cultures were plated on YPD following incubation at 30°C for 3 days or at 16°C for 4 days. For control of plasmid maintenance, the same cells were plated on minimal glucose medium (SC−ura) and incubated at 30°C for 3 days. (B) Cells preincubated in minimal medium were grown in YPD at 30°C, shifted to 16°C for 2 h, and harvested. Total RNA was isolated, separated on a 2.8% agarose gel, and stained with EtBr using equal amounts of RNA per lane (5 μg). (C) RNA was analyzed by Northern blotting using probes specific for pre-tRNA-Leu (CAA), tRNA-Phe (GAA), and tRNA-Tyr (GUA). 5.8S rRNA served as the loading control. Positions of initial transcripts, end-processed intron-containing pre-tRNA (IVS), and mature tRNA are indicated. (D) pre-tRNA forms were quantified. Bars represent levels for primary transcripts and intron-containing precursors normalized to the loading control. Standard deviations were estimated on the basis of three independent experiments. The P values calculated for ratios of pre-tRNAs (rpc128-1007[-]/wt[-], rpc128-1007[-]/rpc128-1007[RPB10], and rpc128-1007[-]/rpc128-1007[RBS1]) showed statistical significance (P < 0.05).](/cms/asset/6f17166d-2aba-4520-877e-573e9bf68498/tmcb_a_12275564_f0003_oc.jpg)
In accordance with the suppression of the growth defect, RPB10 and RBS1 overexpression increased the levels of the tRNA precursors in rpc128-1007 cells (the average increases were, respectively, 1.6- ± 0.3-fold and 2- ± 0.5-fold). These results indicate that overexpression of RPB10 or RBS1 overcomes Pol III transcription impairment in the rpc128-1007 mutant.
Next we wanted to ascertain whether the effect of RPB10 or RBS1 overexpression was specific to the rpc128-1007 mutation. To this end, several other available Pol III mutants, defective in initiation (rpc31-236) (Citation25), elongation (rpc160-112 and rpc160-270) (Citation26, Citation27), and termination (rpc160-750 and rpc11-Sp) (Citation28, Citation29) of tRNA gene transcription, were transformed with the plasmids carrying the RPB10 or RBS1 gene. However, we observed no growth of the transformants at restrictive temperatures suitable for the individual mutants, indicating no suppression (). On the basis of these studies and the results of previous genetic screens, rpc128-1007 is the only one polymerase mutation suppressed by RBS1. Suppression by RPB10 overdose was previously reported for some missense mutations in genes encoding AC40 and AC19, indicating mutual interaction of these subunits with ABC10β, which was further confirmed by structure analysis (Citation1, Citation5).
TABLE 3 Specificity of RBS1 and RPB10 suppressorsTable Footnotea
Since C128 is also directly associated with ABC10β in the Pol III complex, it is feasible that suppression by RPB10 overdose enhances this interaction, which is weakened in the rpc128-1007 mutant. Elucidation of RBS1-mediated suppression requires further study.
Synthetic negative growth defect of the double rpc128-1007 rbs1Δ mutant.
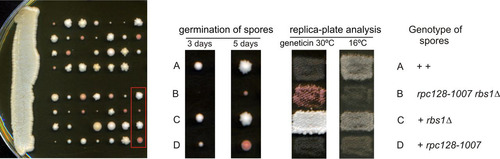
In our studies, the rbs1Δ mutation did not confer a noticeable phenotype on its own. To gain a better understanding of how Rbs1 functions, we assessed the effect of rbs1Δ in the rpc128-1007 mutant showing defect in Pol III assembly. The rbs1Δ deletion was created in a wild-type strain (MB159-2C) and crossed with an isogenic rpc128-1007 mutant (MJ15-9C) of the opposite mating type (). The diploid was sporulated following dissection of 14 tetrads. Three classes of spores were observed: large, small, and very small with delayed germination (). Careful genetic analysis of meiotic progeny of the cross was performed. Both the cold-sensitive phenotype of the rpc128-1007 mutant and Geneticin resistance due to the rbs1Δ deletion marker segregated 2:2 in all 14 tetrads. Since the RPC128 and RBS1 genes are located on different chromosomes, segregation was independent, as expected. Small spores showing the cold-sensitive phenotype and Geneticin sensitivity corresponded to the single rpc128-1007 mutant. Significantly, all very small spores represented rpc128-1007 rbs1Δ double mutants. We thus concluded that the rpc128-1007 rbs1Δ combination results in a strong synthetic growth defect. This suggests a tight functional relationship between Rbs1 and Pol III assembly.
FIG 4 Synthetic negative growth defect of the double rpc128-1007 rbs1Δ mutant. Sporulation of diploids generated by the cross MB159-2C rbs1Δ × MJ15-9C rpc128-1007 was followed by dissection of tetrads. The growth of spores was monitored after 3 days and 5 days (left panel). Genetic analysis of all tetrads was performed by transferring all individual spores a to YPD master plate which was grown for 2 days and replicated to test the genetic phenotypes of Geneticin resistance and cold sensitivity (growth at 16°C). Genetic analysis of one selected tetrad is presented in the right panel.
Rbs1 protein physically interacts with Pol III.
In order to gain insight into the Rbs1 protein function, we used affinity purification followed by quantitative liquid chromatography-tandem mass spectrometry (LC-MS/MS) to identify its binding partners. Thus, we affinity purified Rbs1-GFP from the wild-type and rpc128-1007 mutant strains using anti-GFP resin and estimated the relative amounts of bound proteins using MaxQuant software (Citation38). Five subunits of RNA polymerases which interact with Rbs1 in the wild-type strain were clearly identified in this way: AC40, AC19, A190, B150, and ABC27 (). AC40 and ABC27 were also identified as Rbs1-interacting proteins in extracts from the rpc128-1007 mutant. A control experiment using a non-GFP-tagged strain revealed none of these proteins. Additionally, two Pol I subunits, A135 and A34.5, were enriched in a fraction of proteins interacting with Rbs1-GFP in wild-type cells ().
TABLE 4 Components of RNA polymerases copurified with Rbs1Table Footnotea
Significantly, AC40, AC19, and ABC27, which are components of Pol III, were identified as Rbs1-interacting proteins in affinity purification experiments. This argues that Rbs1 physically interacts with the Pol III complex and possibly with other polymerases. Many proteins unrelated to transcription were also copurified with tagged Rbs1 (see Table S1 in the supplemental material). One functional group represents proteins involved in nuclear-cytoplasmic transport. Besides Crm1, a major β-karyopherin for nuclear export of proteins which was previously identified to interact with Rbs1 in a two-hybrid system (Citation45), Npl3, Loc1, Sec13, and Kap104 also were present in the affinity-purified Rbs1 preparation.
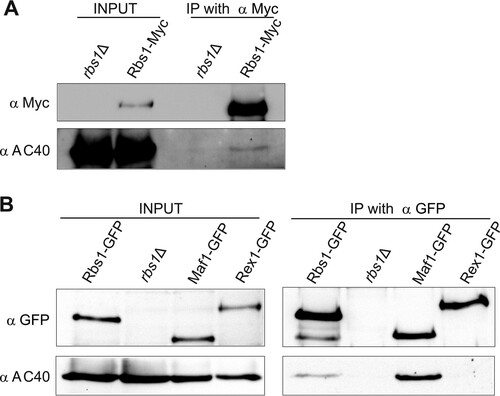
To confirm the interaction between Rbs1 and Pol III, we used a functional Rbs1-Myc fusion protein expressed from a chromosomal locus. Functionality of Rbs1-Myc was confirmed by its ability to suppress the cold sensitivity of the rpc128-1007 mutant when cloned on the multicopy plasmid. Crude extracts were prepared from Rbs1-Myc-tagged cells, and control rbs1Δ cells were grown in glucose medium. Myc-tagged Rbs1 was immunoprecipitated from cell extracts with magnetic beads coated with anti-Myc antibodies, and the immunoprecipitates were examined for the presence of AC40 protein by immunoblotting. As shown in , Rbs1 was immunopurified from crude extracts and AC40 protein was selectively coimmunoprecipitated with Myc-tagged Rbs1. This result indicating that Rbs1 and AC40, a common subunit of Pol I and Pol III, interact with each other was further confirmed by an independent experiment using GFP-tagged Rbs1. Protein immunoprecipitation with magnetic beads coated with anti-GFP antibodies allowed the use of Maf1-GFP- and Rex1-GFP-encoding strains as positive and negative controls, respectively. Clearly, Rbs1 protein interacts with the AC40 subunit (). Stronger interaction of AC40 was observed with the Pol III repressor Maf1, confirming data reported previously (Citation46). As expected, no interaction of AC40 with Rex1 protein, involved in tRNA maturation, was observed.
FIG 5 Rbs1 interacts with AC40. (A) Strain MJ15-10A, expressing Myc-tagged Rbs1, and control strain BY4741 rbs1Δ were grown in YPD. Aliquots of cell extracts (input) and immunoprecipitates with the anti-Myc antibody were analyzed by immunoblotting with anti-Myc and anti-AC40 antibodies. The lane in the middle of the gel is empty and separates input from IP with anti-Myc. (B) Cell extracts prepared from strains encoding GFP-tagged Rbs1, Maf1, and Rex1 proteins and control strain BY4741 rbs1Δ (input) and immunoprecipitates with the anti-GFP antibody were analyzed by immunoblotting with anti-GFP and anti-AC40 antibodies.
Rbs1 shuttles between the cytoplasm and the nucleus.
Our data demonstrate genetic and physical interaction between Rbs1 and Pol III, suggesting a role of Rbs1 in the biogenesis of the Pol III complex. According to the yeast genome database and our observations, Rbs1 is localized in the cytoplasm and excluded from the nucleus. Nevertheless, copurification of Rbs1 with Crm1 encouraged us to study possible nuclear localization of Rbs1 and its Crm1-dependent nuclear export. In order to do so, an Rbs1-Myc crm1-T359C leptomycin (LMB)-sensitive strain was constructed. The effect of LMB was noticeable after 15 min, since Rbs1 became uniformly distributed throughout the cell instead of undergoing nuclear exclusion. After 45 min of incubation with LMB, Rbs1 was localized in the nuclei of all analyzed crm1-T359C cells, whereas in wild-type cells Rbs1 was present in the cytoplasm despite LMB treatment (compare and ). These results indicate that Rbs1 shuttles between the nucleus and cytoplasm and becomes trapped in the nucleus when Crm1 function is inhibited by LMB.
FIG 6 LMB induces nuclear accumulation of Rbs1. Yeast cells were grown to log phase at 30°C (−LMB), and then LMB (100 ng/ml) was added and growth was continued at 30°C for the indicated times before fixation of cells. Localization of Rbs1-Myc was determined by immunofluorescence with anti-Myc antibody in the control wild-type (wt) strain (A), the crm1-T359C mutant (B), and the crm1-T359C rpc128-1007 double mutant (C). Nuclear DNA was stained with DAPI (4′,6′-diamidino-2-phenylindole). (D) The percentage of cells with the given cellular localization of Rbs1 after 1 h of LMB treatment was estimated after inspection of at least 120 single cells.
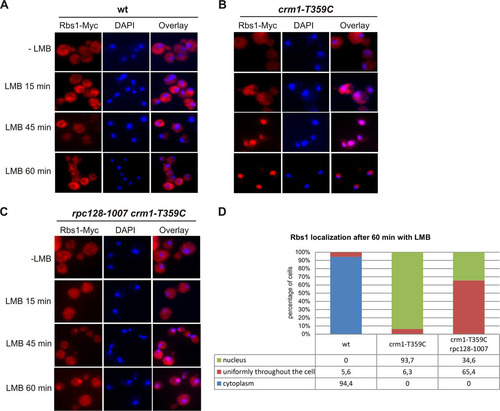
The nuclear-cytoplasmic shuttling of the Rbs1 protein and its interactions with Pol III subunits raise the possibility that Rbs1 is involved in Pol III biogenesis in the cytoplasm, travels to the nucleus together with the Pol III complex, and is exported back to cytoplasm when Pol III is already imported. To explore whether Rbs1 shuttling is related to the biogenesis of the Pol III complex, we examined Rbs1 localization in rpc128-1007 cells. Rbs1-Myc was localized in the cytoplasm in both the single crm1-T359C and double crm1-T359C rpc128-1007 mutants (). Upon LMB treatment of the crm1-T359C rpc128-1007 mutant for 45 min, the majority of cells had Rbs1-Myc uniformly distributed throughout the cytoplasm and nucleus. After 60 min of LMB treatment, we observed nuclear localization of Rbs1-Myc in only about 35% of crm1-T359C rpc128-1007 cells (), whereas the same treatment of the control crm1-T359C mutant harboring wild-type RPC128 resulted in nuclear localization of Rbs1-Myc in about 94% of cells. We thus concluded that cytoplasmic-nuclear shuttling of Rbs1 is compromised when Pol III biogenesis is flawed.
Rbs1 promotes nuclear localization of Pol III subunits when overproduced in rpc128-1007 cells.
We considered the possibility that cytoplasmic assembly of Pol III would be disturbed by the rpc128-1007 mutation, which consequently would affect intracellular localization of the subunits of the complex. Thus, we examined wild-type and rpc128-1007 mutant cells containing C160-HA tagged protein by immunofluorescence using anti-HA antibody. Predominant nuclear fluorescence of C160-HA was observed in a wild-type strain as expected, while in rpc128-1007 cells nuclear staining was more diffused, the overall level of C160 was decreased, and the majority of C160-HA was present in the cytoplasm (). In addition, a high C160-HA concentration in the nucleus was restored when the rpc128-1007 mutation was complemented with the RBS1 gene in a multicopy plasmid (). To validate these results and to monitor the localization of other Pol III subunits, we used strains encoding C53 and C82 genomically tagged with GFP and monitored their localization by live cell imaging (). As in the case of C160, fluorescence for C53 and C82 was detected mainly in the nucleus of the wild-type strain, while the nuclear signal was weak in the rpc128-1007 mutant strain. Again, mutant cells resumed nuclear localization of C53 and C82 when Rbs1 protein was overproduced.
FIG 7 The rpc128-1007 mutation lowers the nuclear concentration of some Pol III subunits, but these effects are suppressed by overproduced Rbs1. Wild-type and rpc128-1007 mutant derivatives transformed with empty vector ([-]) or a multicopy plasmid with the RBS1 gene were grown in rich glucose medium to log phase. (A) Immunofluorescence of cells encoding C160-HA by using anti-HA antibody. Nuclei were stained with DAPI. (B) Live imaging of cells encoding C82-GFP or C53-GFP.
![FIG 7 The rpc128-1007 mutation lowers the nuclear concentration of some Pol III subunits, but these effects are suppressed by overproduced Rbs1. Wild-type and rpc128-1007 mutant derivatives transformed with empty vector ([-]) or a multicopy plasmid with the RBS1 gene were grown in rich glucose medium to log phase. (A) Immunofluorescence of cells encoding C160-HA by using anti-HA antibody. Nuclei were stained with DAPI. (B) Live imaging of cells encoding C82-GFP or C53-GFP.](/cms/asset/030e4640-4684-4edf-a2cc-439ac97522c0/tmcb_a_12275564_f0007_oc.jpg)
These results indicate that the Pol III subunits are partially mislocalized in the rpc128-1007 mutant and that Rbs1 is necessary for their correct nuclear localization.
Rbs1 corrects the defect of Pol III assembly in rpc128-1007 cells.
Next we tested the effect of the rpc128-1007 mutation and Rbs1 protein on the assembly of the Pol III complex. Using an HA-specific antibody, the whole complex was immunopurified from cells expressing C160-HA, and the interactions between different Pol III subunits were examined by coimmunoprecipitation (). It is worth noting that the steady-state level of C160 was lower in rpc128-1007 cells than in the control strain, and amounts of C82 and C53, other Pol III-specific subunits, were also decreased in relation to the loading control (). This is consistent with the overall decrease of the amount of Pol III complex in the mutant strain suggested by microscopy study ().
FIG 8 The defect of Pol III assembly in the rpc128-1007 mutant is partially rescued by overproduced Rbs1. Extracts were prepared from control wild-type cells and cells encoding C160-HA (wild-type and rpc128-1007 mutant) transformed with empty vector ([-]) or a multicopy plasmid with the RBS1 gene (input). Pol III was immunoprecipitated from yeast extracts by HA-specific antibody (IP with α HA). (A) Primary extracts (lanes 1 to 4) and IP fractions (lanes 6 to 9) were examined by Western blotting with antibody specific for HA, C82, C53, AC40, or Vma2 (used as a loading control). An additional higher band which was repeatedly recognized by anti-C82 antibody in mutant cells appeared to be a result of an unspecific cross-reaction. Lane 5 is empty and separates input from IP with anti-HA. (B) Quantification of the levels of Pol III subunits in total extracts from the rpc128-1007 mutant transformed with empty vector or a multicopy plasmid with the RBS1 gene. Band intensities from Western blot images were quantified by MultiGauge v3.0 software (Fujifilm). Bars represent the means with standard deviations from three independent experiments. Levels from the wild-type strain were assumed to be 1. (C) Quantification of the amount of Pol III subunits coimmunopurified with C160 from extracts of the rpc128-1007 mutant transformed with empty vector or a multicopy plasmid with the RBS1 gene. Bands intensities from Western blot images were calculated as for panel B. The subunit ratios in the wild-type, rpc128-1007 [-], and rpc128-1007 [RBS1] strains were calculated and referred to levels of the respective subunits in the wild-type strain, which were assumed to be 1.
![FIG 8 The defect of Pol III assembly in the rpc128-1007 mutant is partially rescued by overproduced Rbs1. Extracts were prepared from control wild-type cells and cells encoding C160-HA (wild-type and rpc128-1007 mutant) transformed with empty vector ([-]) or a multicopy plasmid with the RBS1 gene (input). Pol III was immunoprecipitated from yeast extracts by HA-specific antibody (IP with α HA). (A) Primary extracts (lanes 1 to 4) and IP fractions (lanes 6 to 9) were examined by Western blotting with antibody specific for HA, C82, C53, AC40, or Vma2 (used as a loading control). An additional higher band which was repeatedly recognized by anti-C82 antibody in mutant cells appeared to be a result of an unspecific cross-reaction. Lane 5 is empty and separates input from IP with anti-HA. (B) Quantification of the levels of Pol III subunits in total extracts from the rpc128-1007 mutant transformed with empty vector or a multicopy plasmid with the RBS1 gene. Band intensities from Western blot images were quantified by MultiGauge v3.0 software (Fujifilm). Bars represent the means with standard deviations from three independent experiments. Levels from the wild-type strain were assumed to be 1. (C) Quantification of the amount of Pol III subunits coimmunopurified with C160 from extracts of the rpc128-1007 mutant transformed with empty vector or a multicopy plasmid with the RBS1 gene. Bands intensities from Western blot images were calculated as for panel B. The subunit ratios in the wild-type, rpc128-1007 [-], and rpc128-1007 [RBS1] strains were calculated and referred to levels of the respective subunits in the wild-type strain, which were assumed to be 1.](/cms/asset/8eee961f-000f-4bb2-9f9e-14f1ee6d1f5d/tmcb_a_12275564_f0008_oc.jpg)
The total amount of AC40 subunit common to Pol I and Pol III was relatively less decreased. Presumably, the rpc128-1007 mutation has no effect on the stability of Pol I-derived AC40, which contributes to the overall steady-state level of this subunit.
The effect of the rpc128-1007 mutation on the interaction between C160 and other subunits was estimated as the efficiency of coimmunoprecipitation in relation to that observed in the wild-type strain ( and ). The association of C160 with C82 was not significantly altered, whereas C160-C53 and C160-AC40 associations were clearly decreased in the mutant cells, around 2- and 6-fold, respectively (). This result indicates that the rpc128-1007 mutation negatively affects Pol III assembly. A marked decrease in the association of C160 and AC40 subunits suggests a defect in interaction between β- and α-like Pol III modules.
To investigate whether Rbs1 participates in the assembly of Pol III, we also immunoprecipitated C160-HA from a mutant strain containing a multicopy plasmid containing the RBS1 gene. The overproduction of Rbs1 corrected the expression of the C160, C82, and C53 subunits in the rpc128-1007 mutant and increased C160-C53 interaction, whereas the C160-C82 interaction appeared to be less affected. Moreover, Rbs1 had a minor effect on the expression of AC40 but significantly enhanced C160-AC40 association.
All these data, taken together, demonstrate that the point mutation in the C128 subunit affects the assembly and stability of Pol III, decreasing the amount of C160 and additionally lowering its interaction with other subunits. Importantly, RBS1 overexpression suffices to correct the assembly and stability defects caused by the rpc128-1007 mutation in the C128 subunit of Pol III.
DISCUSSION
In this work we present evidence that the rpc128-1007 mutation in the second-largest subunit of Pol III causes a defect in the assembly of the polymerase complex. This defect is corrected by overexpression of Rbs1 protein, which interacts with Pol III and shuttles between the cytoplasm and nucleus in a Crm1-dependent manner.
The experimental data presented in this paper, together with the position of the rpc128-1007 mutation in the Pol III structure, strongly support the hypothesis that the rpc128-1007 mutation prevents the correct assembly of the RNA Pol III complex. First, Gly1007 in the C128 subunit is highly conserved and is located in region which closely contacts two other Pol III subunits, AC19 and AC40. Second, decreased interaction between the largest subunit C160 and the AC40 subunit is observed by coimmunoprecipitation. Therefore, it can be concluded that Gly1007 in the second-largest subunit of Pol III is important for the proper assembly of the entire enzyme.
Correct assembly is essential for maintaining the integrity or stability of the polymerase complex. In the rpc128-1007 mutant, we observed decreased levels of Pol III subunits C160, C82, and C53. Meanwhile, transcription of the C160-encoding gene is unaffected in this mutant (data not shown), so the largest Pol III subunit is probably degraded when unassembled to the complex. Nuclear degradation of the largest Pol II subunit by an Asr1-independent mechanism was detected in mutants defective in the assembly of the Pol II complex (Citation47). Neither the mechanism of C160 degradation nor the location where this process occurs in rpc128-1007 cells is known, and this requires further study. Altogether, inefficient interaction of C160 with the rest of the complex and its degradation indicate a decrease in the total amount of Pol III in the rpc128-1007 mutant.
We also considered the possibility that incorrect assembly would lead to specific modifications of some Pol III subunits. An additional band observed for the C82 subunit in the rpc128-1007 mutant suggested its sumoylation. Sumoylation of C82 and other subunits of Pol III (C160, C128, AC40, C53, C37, and ABC23) has been previously reported (Citation48Citation–Citation50), but its role is unknown. However, we have experimentally excluded a link of the additional band with sumoylation of C82 (data not shown).
Assembly of the RNA polymerases in both yeast and humans is proposed to occur in the cytoplasm as a requirement for their nuclear import (Citation6). Only one earlier study focused on Pol III biogenesis has indicated the coordinated nuclear import of at least a subset of Pol III subunits (Citation51). A potential NLS was described within 16 amino acids near the N terminus of C128 subunit, and its deletion led to cytoplasmic accumulation of C128, which caused cytoplasmic accumulation of a subset of Pol III subunits, such as C160, C53, and C11, whereas other Pol III subunits, i.e., C82, AC40, and C25, remained nuclear. Here we show that the point mutation in the C-terminal part of the C128 subunit leads to decreased nuclear concentration of the C160 and C53 subunits, as well as that of C82. Presumably, low nuclear concentrations of these proteins correspond to a small amount of Pol III dictated by the assembly defect and subsequent instability of subunits.
Pol III is functional only as a full complex of 17 subunits, in contrast to Pol II, in which not all components are essential. Therefore, every Pol III subcomplex, if it exists, is nonfunctional and would probably be unstable. Since the enzymatic activity of Pol III is restricted to the assembled and properly localized enzyme, the tRNA transcription in the rpc128-1007 mutant is very low.
Conditional mutations in a given Pol I, Pol II, or Pol III subunit are often suppressed by increasing the gene dosage of another subunit of the same assembly (Citation1, Citation52, Citation53). Here we identified suppression of the rpc128-1007 mutation by overdose of the ABC10β subunit, which is common to all three RNA polymerases. It is known that ABC10β binds to the second-largest subunit of polymerases and plays a role in their assembly (Citation43, Citation54). The role in assembly was documented also for other small subunits common to Pol I, Pol II, and Pol III: Rpb5 (ABC27) facilitates the assembly step mediated by prefoldin Bud27 (Citation18), whereas ABC14.5 selectively affects Pol III assembly by its specific interaction with C160 (Citation55). Significantly, suppression by a high dosage of ABC10β was previously identified for temperature sensitive alleles of genes encoding the α-like subunits AC19 and AC40, which are common to Pol I and Pol III (Citation1). It would be interesting to investigate whether Pol III assembly is compromised in these mutants. Thus, suppression of rpc128-1007 by overdose of ABC10β could be interpreted as a correction of the assembly defect. Also noteworthy is that the overexpression of RPB10 compensated the low steady-state levels and nuclear concentrations of the C160, C82, and C53 subunits of Pol III in the rpc128-1007 mutant (data not shown).
Interestingly, another strong overdose suppressor of rpc128-1007 is the Rbs1 protein, a function which was previously unknown. Several lines of evidence support the hypothesis that Rbs1 participates in the assembly and/or nuclear transport of the Pol III complex. First, genetic data show a strong synthetic negative effect of rbs1Δ and Pol III assembly mutant rpc128-1007 and allele-specific suppression of rpc128-1007 by RBS1 overdose. Second, several Pol III subunits have been copurified with Rbs1, and Rbs1 interacts with Pol III by coimmunoprecipitation. Third, Rbs1 shuttles between the nucleus and cytoplasm in a Crm1-dependent fashion. Finally, the overexpression of Rbs1 improves the interaction between Pol III subunits in the rpc128-1007 mutant, which possesses a defect in Pol III assembly. Taken together, these data are consistent with a model previously presented (Citation6) in which Rbs1 functions similarly to other factors by facilitating polymerase biogenesis. This model assumes that Rbs1 binds and facilitates Pol III assembly in the cytoplasm, travels with the Pol III complex to the nucleus, and then dissociates and is exported by Crm1 back to the cytoplasm.
It is unknown whether the Rbs1 function is specific for Pol III and the given subassembly. According to affinity purification data, Rbs1 interacts only with a subset of polymerase subunits: AC19 and AC40, common to Pol I and Pol III, and ABC27, a component of all three polymerases. Whereas AC19 and AC40, together with ABC10β, form an ααβ-like subassembly, ABC27 is considered to be a part of a β′-like subassembly (Citation6). Moreover, ABC27 participates in the assembly of all three polymerases mediated by Bud27 (Citation18).
Among proteins which copurified with Rbs1, we also detected rare Pol I- and Pol II-specific subunits, although in smaller amounts. Taking these data together, we cannot exclude the possibility that Rbs1 is involved in the biogenesis of all three RNA polymerases. Rbs1 is not an essential protein, and thus its role in the assembly/nuclear import of RNA polymerase(s) could be substituted for by other, so-far-unknown factors. No defect in Pol III assembly in the rbs1Δ strain was observed (data not shown). The relationship between Bud27, Iwr1, and other factors which participate in the process is unknown and requires further study.
Although we have shown that Rbs1 shuttles between the cytoplasm and the nucleus, NLS motifs are not obvious in its sequence. In case of nuclear export signals, NetNES1.1 Server revealed that Rbs1 contains two sequences: KPLLQELQL (aa 95 to 103) and LEQERIEKERL (aa 173 to 183). These motifs only partially match the consensus NES recognized by the exportin Crm1p (F1-[X]2–3-F2-[X]2–3-F3-X-F4, where F is hydrophobic and X is any amino acid) (Citation56). The significance of these sequences has not been studied yet. Another interesting motif in the Rbs1 sequence is the R3H domain (aa 57-RLLSH-61), which potentially binds single-stranded nucleic acids (Citation57). The R3H domain is conserved in Rbs1 sequences from other yeast species and is also present in other yeast and mammalian proteins, including those from humans (Citation57). Inactivation of this domain by mutagenesis prevents Rbs1-mediated suppression of the rpc128-1007 mutation (M. Cieśla, unpublished data).
In our affinity purification experiments, we identified several interesting proteins which interact with Rbs1, namely, mitochondrial proteins (Cor1, Cor2, Mas1, Mas2, and Mas5), and prion Rnq1. Rbs1 copurifies with other Rbs1 molecules, which may indicate its aggregation. This is consistent with the predicted prion-like properties of Rbs1 (Citation58). The participation of prions in polymerase biogenesis is a fascinating issue which we would like to investigate in the future.
tmcb_a_12275564_sm0001.pdf
Download PDF (176.7 KB)tmcb_a_12275564_sm0002.pdf
Download PDF (156.2 KB)tmcb_a_12275564_sm0003.xls
Download MS Excel (113.5 KB)ACKNOWLEDGMENTS
We thank Olivier Lefebvre for providing antibodies specific for Pol III subunits and yeast strains. We acknowledge Dominik Cysewski for his help with processing of mass spectrometry data.
This work was supported by the Foundation for Polish Science (Parent-Bridge Programme/2010-2/2) for M.C. and by the National Science Centre (UMO-2012/04/A/NZ1/00052) for M.B.
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01230-14.
REFERENCES
- Lalo D, Carles C, Sentenac A, Thuriaux P. 1993. Interactions between three common subunits of yeast RNA polymerases I and III. Proc Natl Acad Sci U S A 90:5524–5528. http://dx.doi.org/10.1073/pnas.90.12.5524.
- Carter R, Drouin G. 2010. The increase in the number of subunits in eukaryotic RNA polymerase III relative to RNA polymerase II is due to the permanent recruitment of general transcription factors. Mol Biol Evol 27:1035–1043. http://dx.doi.org/10.1093/molbev/msp316.
- Ishihama A. 1981. Subunit of assembly of Escherichia coli RNA polymerase. Adv Biophys 14:1–35.
- Kimura M, Ishiguro A, Ishihama A. 1997. RNA polymerase II subunits 2, 3, and 11 form a core subassembly with DNA binding activity. J Biol Chem 272:25851–25855. http://dx.doi.org/10.1074/jbc.272.41.25851.
- Lane LA, Fernández-Tornero C, Zhou M, Morgner N, Ptchelkine D, Steuerwald U, Politis A, Lindner D, Gvozdenovic J, Gavin A-C, Müller CW, Robinson CV. 2011. Mass spectrometry reveals stable modules in holo and apo RNA polymerases I and III. Structure 19:90–100. http://dx.doi.org/10.1016/j.str.2010.11.009.
- Wild T, Cramer P. 2012. Biogenesis of multisubunit RNA polymerases. Trends Biochem Sci 37:99–105. http://dx.doi.org/10.1016/j.tibs.2011.12.001.
- Schoenfelder S, Clay I, Fraser P. 2010. The transcriptional interactome: gene expression in 3D. Curr Opin Genet Dev 20:127–133. http://dx.doi.org/10.1016/j.gde.2010.02.002.
- Bertrand E, Houser-Scott F, Kendall A, Singer RH, Engelke DR. 1998. Nucleolar localization of early tRNA processing. Genes Dev 12:2463–2468. http://dx.doi.org/10.1101/gad.12.16.2463.
- Chen M, Gartenberg MR. 2014. Coordination of tRNA transcription with export at nuclear pore complexes in budding yeast. Genes Dev 28:959–970. http://dx.doi.org/10.1101/gad.236729.113.
- Boulon S, Pradet-Balade B, Verheggen C, Molle D, Boireau S, Georgieva M, Azzag K, Robert M-C, Ahmad Y, Neel H, Lamond AI, Bertrand E. 2010. HSP90 and its R2TP/prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Mol Cell 39:912–924. http://dx.doi.org/10.1016/j.molcel.2010.08.023.
- Forget D, Lacombe A-A, Cloutier P, Lavallée-Adam M, Blanchette M, Coulombe B. 2013. Nuclear import of RNA polymerase II is coupled with nucleocytoplasmic shuttling of the RNA polymerase II-associated protein 2. Nucleic Acids Res 41:6881–6891. http://dx.doi.org/10.1093/nar/gkt455.
- Forget D, Lacombe A-A, Cloutier P, Al-Khoury R, Bouchard A, Lavallée-Adam M, Faubert D, Jeronimo C, Blanchette M, Coulombe B. 2010. The protein interaction network of the human transcription machinery reveals a role for the conserved GTPase RPAP4/GPN1 and microtubule assembly in nuclear import and biogenesis of RNA polymerase II. Mol Cell Proteomics 9:2827–2839. http://dx.doi.org/10.1074/mcp.M110.003616.
- Staresincic L, Walker J, Dirac-Svejstrup AB, Mitter R, Svejstrup JQ. 2011. GTP-dependent binding and nuclear transport of RNA polymerase II by Npa3 protein. J Biol Chem 286:35553–35561. http://dx.doi.org/10.1074/jbc.M111.286161.
- Di Croce L. 2011. Regulating the shuttling of eukaryotic RNA polymerase II. Mol Cell Biol 31:3918–3920. http://dx.doi.org/10.1128/MCB.06093-11.
- Peiró-Chova L, Estruch F. 2007. Specific defects in different transcription complexes compensate for the requirement of the negative cofactor 2 repressor in Saccharomyces cerevisiae. Genetics 176:125–138. http://dx.doi.org/10.1534/genetics.106.066829.
- Gómez-Navarro N, Peiró-Chova L, Rodriguez-Navarro S, Polaina J, Estruch F. 2013. Rtp1p is a karyopherin-like protein required for RNA polymerase II biogenesis. Mol Cell Biol 33:1756–1767. http://dx.doi.org/10.1128/MCB.01449-12.
- Czeko E, Seizl M, Augsberger C, Mielke T, Cramer P. 2011. Iwr1 directs RNA polymerase II nuclear import. Mol Cell 42:261–266. http://dx.doi.org/10.1016/j.molcel.2011.02.033.
- Mirón-García MC, Garrido-Godino AI, García-Molinero V, Hernández-Torres F, Rodríguez-Navarro S, Navarro F. 2013. The prefoldin bud27 mediates the assembly of the eukaryotic RNA polymerases in an rpb5-dependent manner. PLoS Genet 9:e1003297. http://dx.doi.org/10.1371/journal.pgen.1003297.
- Minaker SW, Filiatrault MC, Ben-Aroya S, Hieter P, Stirling PC. 2013. Biogenesis of RNA polymerases II and III requires the conserved GPN small GTPases in Saccharomyces cerevisiae. Genetics 193:853–864. http://dx.doi.org/10.1534/genetics.112.148726.
- Cieśla M, Towpik J, Graczyk D, Oficjalska-Pham D, Harismendy O, Suleau A, Balicki K, Conesa C, Lefebvre O, Boguta M. 2007. Maf1 is involved in coupling carbon metabolism to RNA polymerase III transcription. Mol Cell Biol 27:7693–7702. http://dx.doi.org/10.1128/MCB.01051-07.
- Kwapisz M, Smagowicz WJ, Oficjalska D, Hatin I, Rousset J-P, Zoładek T, Boguta M. 2002. Up-regulation of tRNA biosynthesis affects translational readthrough in maf1-delta mutant of Saccharomyces cerevisiae. Curr Genet 42:147–152. http://dx.doi.org/10.1007/s00294-002-0342-7.
- Soutourina J, Bordas-Le Floch V, Gendrel G, Flores A, Ducrot C, Dumay-Odelot H, Soularue P, Navarro F, Cairns BR, Lefebvre O, Werner M. 2006. Rsc4 connects the chromatin remodeler RSC to RNA polymerases. Mol Cell Biol 26:4920–4933. http://dx.doi.org/10.1128/MCB.00415-06.
- Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27.
- Neville M, Rosbash M. 1999. The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J 18:3746–3756. http://dx.doi.org/10.1093/emboj/18.13.3746.
- Thuillier V, Stettler S, Sentenac A, Thuriaux P, Werner M. 1995. A mutation in the C31 subunit of Saccharomyces cerevisiae RNA polymerase III affects transcription initiation. EMBO J 14:351–359.
- Dieci G, Hermann-Le Denmat S, Lukhtanov E, Thuriaux P, Werner M, Sentenac A. 1995. A universally conserved region of the largest subunit participates in the active site of RNA polymerase III. EMBO J 14:3766–3776.
- Thuillier V, Brun I, Sentenac A, Werner M. 1996. Mutations in the alpha-amanitin conserved domain of the largest subunit of yeast RNA polymerase III affect pausing, RNA cleavage and transcriptional transitions. EMBO J 15:618–629.
- Rozenfeld S, Thuriaux P. 2001. A genetic look at the active site of RNA polymerase III. EMBO Rep 2:598–603. http://dx.doi.org/10.1093/embo-reports/kve136.
- Chédin S, Riva M, Schultz P, Sentenac A, Carles C. 1998. The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination. Genes Dev 12:3857–3871. http://dx.doi.org/10.1101/gad.12.24.3857.
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953–961.
- Stettler S, Chiannilkulchai N, Hermann-Le Denmat S, Lalo D, Lacroute F, Sentenac A, Thuriaux P. 1993. A general suppressor of RNA polymerase I, II and III mutations in Saccharomyces cerevisiae. Mol Gen Genet 239:169–176.
- Nasmyth KA, Reed SI. 1980. Isolation of genes by complementation in yeast: molecular cloning of a cell-cycle gene. Proc Natl Acad Sci U S A 77:2119–2123. http://dx.doi.org/10.1073/pnas.77.4.2119.
- Chen DC, Yang BC, Kuo TT. 1992. One-step transformation of yeast in stationary phase. Curr Genet 21:83–84. http://dx.doi.org/10.1007/BF00318659.
- Schmitt ME, Brown TA, Trumpower BL. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res 18:3091–3092. http://dx.doi.org/10.1093/nar/18.10.3091.
- Cieśla M, Mierzejewska J, Adamczyk M, Farrants A-KÖ, Boguta M. 2014. Fructose bisphosphate aldolase is involved in the control of RNA polymerase III-directed transcription. Biochim Biophys Acta 1843:1103–1110. http://dx.doi.org/10.1016/j.bbamcr.2014.02.007.
- Płociński P, Laubitz D, Cysewski D, Stoduœ K, Kowalska K, Dziembowski A. 2014. Identification of protein partners in mycobacteria using a single-step affinity purification method. PLoS One 9:e91380. http://dx.doi.org/10.1371/journal.pone.0091380.
- Orlowska KP, Klosowska K, Szczesny RJ, Cysewski D, Krawczyk PS, Dziembowski A. 2013. A new strategy for gene targeting and functional proteomics using the DT40 cell line. Nucleic Acids Res 41:e167. http://dx.doi.org/10.1093/nar/gkt650.
- Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372. http://dx.doi.org/10.1038/nbt.1511.
- Jasiak AJ, Armache K-J, Martens B, Jansen R-P, Cramer P. 2006. Structural biology of RNA polymerase III: subcomplex C17/25 X-ray structure and 11 subunit enzyme model. Mol Cell 23:71–81. http://dx.doi.org/10.1016/j.molcel.2006.05.013.
- Engel C, Sainsbury S, Cheung AC, Kostrewa D, Cramer P. 2013. RNA polymerase I structure and transcription regulation. Nature 502:650–655. http://dx.doi.org/10.1038/nature12712.
- Fernández-Tornero C, Böttcher B, Rashid UJ, Steuerwald U, Flörchinger B, Devos DP, Lindner D, Müller CW. 2010. Conformational flexibility of RNA polymerase III during transcriptional elongation. EMBO J 29:3762–3772. http://dx.doi.org/10.1038/emboj.2010.266.
- Hampsey M. 1997. A review of phenotypes in Saccharomyces cerevisiae. Yeast 13:1099–1133.
- Gadal O, Shpakovski GV, Thuriaux P. 1999. Mutants in ABC10beta, a conserved subunit shared by all three yeast RNA polymerases, specifically affect RNA polymerase I assembly. J Biol Chem 274:8421–8427. http://dx.doi.org/10.1074/jbc.274.13.8421.
- Archambault J, Friesen JD. 1993. Genetics of eukaryotic RNA polymerases I, II, and III. Microbiol Rev 57:703–724.
- Jensen TH, Neville M, Rain JC, McCarthy T, Legrain P, Rosbash M. 2000. Identification of novel Saccharomyces cerevisiae proteins with nuclear export activity: cell cycle-regulated transcription factor ace2p shows cell cycle-independent nucleocytoplasmic shuttling. Mol Cell Biol 20:8047–8058. http://dx.doi.org/10.1128/MCB.20.21.8047-8058.2000.
- Pluta K, Lefebvre O, Martin NC, Smagowicz WJ, Stanford DR, Ellis SR, Hopper AK, Sentenac A, Boguta M. 2001. Maf1p, a negative effector of RNA polymerase III in Saccharomyces cerevisiae. Mol Cell Biol 21:5031–5040. http://dx.doi.org/10.1128/MCB.21.15.5031-5040.2001.
- Garrido-Godino AI, García-López MC, Navarro F. 2013. Correct assembly of RNA polymerase II depends on the foot domain and is required for multiple steps of transcription in Saccharomyces cerevisiae. Mol Cell Biol 33:3611–3626. http://dx.doi.org/10.1128/MCB.00262-13.
- Panse VG, Hardeland U, Werner T, Kuster B, Hurt E. 2004. A proteome-wide approach identifies sumoylated substrate proteins in yeast. J Biol Chem 279:41346–41351. http://dx.doi.org/10.1074/jbc.M407950200.
- Wohlschlegel JA, Johnson ES, Reed SI, Yates JR. 2004. Global analysis of protein sumoylation in Saccharomyces cerevisiae. J Biol Chem 279:45662–45668. http://dx.doi.org/10.1074/jbc.M409203200.
- Albuquerque CP, Wang G, Lee NS, Kolodner RD, Putnam CD, Zhou H. 2013. Distinct SUMO ligases cooperate with Esc2 and Slx5 to suppress duplication-mediated genome rearrangements. PLoS Genet 9:e1003670. http://dx.doi.org/10.1371/journal.pgen.1003670.
- Hardeland U, Hurt E. 2006. Coordinated nuclear import of RNA polymerase III subunits. Traffic 7:465–473. http://dx.doi.org/10.1111/j.1600-0854.2006.00399.x.
- Archambault J, Schappert KT, Friesen JD. 1990. A suppressor of an RNA polymerase II mutation of Saccharomyces cerevisiae encodes a subunit common to RNA polymerases I, II, and III. Mol Cell Biol 10:6123–6131.
- Thuriaux P, Mariotte S, Buhler JM, Sentenac A, Vu L, Lee BS, Nomura M. 1995. Gene RPA43 in Saccharomyces cerevisiae encodes an essential subunit of RNA polymerase I. J Biol Chem 270:24252–24257. http://dx.doi.org/10.1074/jbc.270.41.24252.
- Flores A, Briand JF, Gadal O, Andrau JC, Rubbi L, Van Mullem V, Boschiero C, Goussot M, Marck C, Carles C, Thuriaux P, Sentenac A, Werner M. 1999. A protein-protein interaction map of yeast RNA polymerase III. Proc Natl Acad Sci U S A 96:7815–7820. http://dx.doi.org/10.1073/pnas.96.14.7815.
- Voutsina A, Riva M, Carles C, Alexandraki D. 1999. Sequence divergence of the RNA polymerase shared subunit ABC14.5 (Rpb8) selectively affects RNA polymerase III assembly in Saccharomyces cerevisiae. Nucleic Acids Res 27:1047–1055. http://dx.doi.org/10.1093/nar/27.4.1047.
- Kutay U, Güttinger S. 2005. Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol 15:121–124. http://dx.doi.org/10.1016/j.tcb.2005.01.005.
- Grishin NV. 1998. The R3H motif: a domain that binds single-stranded nucleic acids. Trends Biochem Sci 23:329–330. http://dx.doi.org/10.1016/S0968-0004(98)01258-4.
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. 2009. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137:146–158. http://dx.doi.org/10.1016/j.cell.2009.02.044.
