Abstract
Lung cancer is the leading cause of cancer-related mortality in men worldwide. Aberrant RARβ promoter methylation has been frequently investigated in non-small-cell lung carcinoma (NSCLC), the most common form of lung cancer. The aim of present study was to carry out a meta-analysis and a systematic review to evaluate clinicopathological significance of RARβ promoter hypermethylation in NSCLC. A systematic literature search was carried out. The data were extracted and assessed by two reviewers independently. The Cochrane software Review Manager 5.2 was used to conduct the review. Odds ratios (ORs) with 95% corresponding confidence intervals (CIs) were calculated. A total of 18 relevant articles were available for meta-analysis which included 1,871 participants. The frequency of RARβ hypermethylation was significantly increased in NSCLC than in nonmalignant lung tissue, and the pooled OR was 5.69 (P<0.00001). RARβ hypermethylation was significantly more frequently observed in adenocarcinoma (AC) than in squamous cell carcinoma (SCC), and the pooled OR was 1.47 (P=0.005). Hypermethylation of RARβ gene in NSCLC was 2.46 times higher in smoking than in nonsmoking individuals, and the pooled OR was 2.46 (P=0.0002). RARβ hypermethylation rate was not significantly correlated with stage of the disease and sex. RARβ gene methylation status was not associated with prognosis of patients with NSCLC. In conclusion, RARβ promoter hypermethylation significantly increased in NSCLC than in non-neoplastic lung tissue and is predominant in AC, suggesting that RARβ methylation contributes to the development of NSCLC, especially AC. RARβ gene is a potential novel target for development of personalized therapy in patients with NSCLC, and is promising in restoration of retinoic acid-target gene induction via demethylation of RARβ1′ promoter.
Introduction
Lung cancer is the leading cause of cancer-related mortality in men worldwide.Citation1 Lung cancers can be classified into two major histological groups: small-cell-lung carcinoma and non-small-cell lung carcinoma (NSCLC). NSCLC consists of adenocarcinoma (AC), squamous cell carcinoma (SCC), large cell carcinoma, and others.Citation2 NSCLC is the most common form, and although the advent of targeted therapies has improved outcomes in a subset of patients, the overall 5-year survival rate remains less than 15%.Citation3 It is critical to identify a molecular predictive marker for monitoring its progression and prognosis. Aberrant DNA methylation is a commonly observed epigenetic modification in human malignancies including NSCLC. Hypermethylation in promoter regions of many tumor suppressor genes inactivates gene transcription and contributes to the development and progression of various cancers by abolishing tumor suppressor gene function.Citation4–Citation6 Currently, many genes were identified as tumor-specifically methylated by using a genome-wide approach to investigate CpG island methylation in a large number of NSCLC patients. Approximately, half of the methylated genes are involved in the regulation of gene expression and cell adhesion, and some of them could be prognostic markers.Citation7,Citation8 Retinoic acid (RA) and its derivatives (retinoids) are required for normal lung development, cell growth, and differentiation, and a deficiency in RA is a risk factor for the development of lung cancer.Citation9–Citation12 RA activity is mediated primarily by retinoic acid receptors (RAR), including RARα, RARβ, and RARγ, which belong to the nuclear receptor superfamily of transcription factors. RARβ gene is located at chromosome 3p24, a region frequently deleted in lung cancer. Loss of expression of RARβ during cancer development is associated with tumorigenesis and retinoid resistance; induction of its expression, on the other hand, can suppress carcinogenesis.Citation13 Previous studies have reported that RARβ hypermethylation was frequently observed in NSCLC tissue, but the reported rates of RARβ hypermethylation in NSCLC were remarkably diverse due to smaller number of patients. In addition, several studies have revealed an association between DNA methylation and tobacco carcinogens in animal models as well as in NSCLC patients, but the results were inconsistent.Citation14–Citation18 In the present study, we systematically reviewed the studies of RARβ promoter hypermethylation in the process of NSCLC onset and progression, and quantified the association between RARβ promoter hypermethylation and the NSCLC by using meta-analysis methods. In addition, we analyzed the relationship of RARβ promoter hypermethylation with clinical features in NSCLC patients and discussed the tumor suppressor function, RA resistance, as well as the clinical significance of RARβ in NSCLC.
Materials and methods
Search strategy
We conducted a literature search for articles from the earliest data to August 2015 in PubMed, EMBASE, and Web of Science using the search terms: “lung” and “cancer or tumor or neoplasm or carcinoma”, “methylation”, and “RARβ or retinoic acid receptor-β”. We also screened manually the reference lists of retrieved articles for additional articles. There were 150 articles identified from PubMed, 67 articles from EMBASE, and 340 articles from Web of Science. A total of 557 articles were screened by article titles and abstracts.
Selection criteria
Studies were selected based on the following inclusion criteria: 1) studies that reported the relationship between RARβ hypermethylation and NSCLC clinicopathological parameters and prognosis; 2) RARβ hypermethylation evaluated in the primary NSCLC tissues; 3) RARβ hypermethylation examined by methylation-specific polymerase chain reaction (MSP); and 4) studies provided sufficient information to estimate hazard ratio (HR) about overall survival (OS) and 95% confidence interval (CI). The exclusion criteria included the following: 1) reviews, letters, case reports, expert opinion, conference abstracts, editorials; 2) all publications regarding in vitro/ex vivo studies, cell lines, and human xenografts; 3) studies in which RARβ protein expression was investigated, and RARβ hypermethylation was not reported; and 4) studies in which same population or overlapping data were used.
Although our search did not have language limits initially, for the full-text reading and final evaluation we only performed the review of the studies published in English language. After exclusion of nonrelevant and/or redundant publications from different databases, the 44 remaining papers were evaluated in the full-text version for inclusion and exclusion criteria.
Data extraction and methodological assessment
Two authors (XS, KS) independently reviewed and extracted data from eligible studies. Disagreements were resolved by discussion and consensus. If they could not reach a consensus, a third author (SZ) was consulted. The following information was recorded for each study: the first author name, year of publication, sample source, number of cases, clinicopathological parameters, cancer tumor node metastasis stage, methylation detection method, and methylation rate and site. Data for study characteristics and clinical responses were summarized and organized into a table format. Heterogeneity of investigation was evaluated to determine whether or not the data of the various studies could be analyzed for meta-analysis.
For the methodological evaluation of the studies, three investigators (DPY, ZL, and YH) read through each publication independently, and they assessed and scored them according to the Newcastle Ottawa Quality Assessment Scale (NOQAS). The three readers provided the quality scores and compared them, and then they reached a consensus value for each item. Those scales allocate a maximum of nine points for the quality of selection, comparability, exposure, and outcomes for study participants. The Newcastle–Ottawa scale scores ranged from 0 to 9, and a study with a score of 7 or more indicates a good quality.
Statistical analysis
The meta-analysis was conducted using Review Manager 5.2 (Cochrane Collaboration, Software Update, Oxford, UK). Odds ratios (ORs) with its 95% CIs were calculated. The assessment of statistical heterogeneity was done by using the Cochran’s Q statistic and I2 tests. When the I2 value was below 50%, a fixed effect model was used, and when the I2 value was 50% or greater, a random effect model was used. We also explored reasons for statistical heterogeneity using sensitivity analysis. The pooled frequency of RARβ hypermethylation and 95% CIs were estimated. The frequency of RARβ hypermethylation was compared in different tumor characteristics. The pooled OR was estimated for the association between RARβ hypermethylation and clinicopathological features. P-values tailed less than 0.05 were considered statistically significant. Publication bias was assessed by using a method reported by Egger et al.Citation19
Results
Identification of relevant studies and study quality
Eighteen articles published from 2004 to 2014 were eligible for this meta-analysis, as shown in . The quality of included articles was assessed by using NOQAS. Those scales allocate a maximum of 9 points for the quality of selection, comparability, exposure, and outcomes for study participants. The NOS scores ranged from 0 to 9, and a study with a score of 7 or more indicates a good quality. Among those studies, six scored 9 points, nine scored 8 points, and three scored 7 points. Hence, the studies were of a relatively high quality (data not shown).
Study characteristics
A total of 1,871 NSCLC patients from the People’s Republic of China, Japan, Korea, Finland, Australia, and the USA were enrolled. The variables from 18 relevant studies are listed in .
Table 1 Main characteristics of included studies
The correlation of RARβ hypermethylation with clinicopathological features
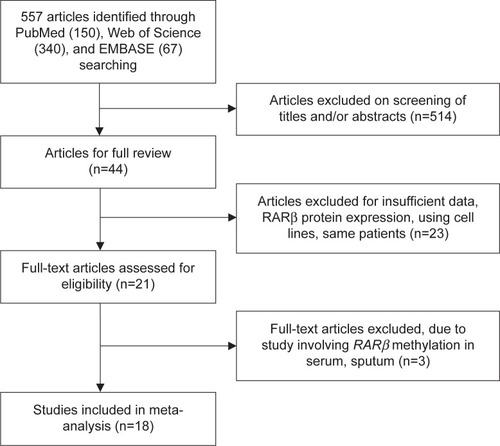
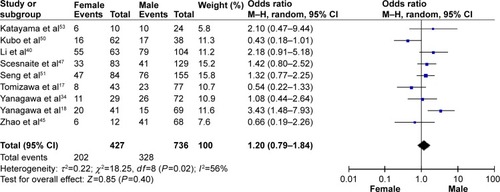
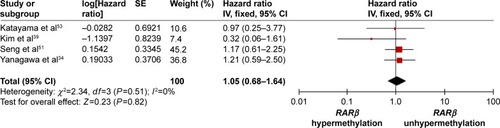
The frequency of RARβ hypermethylation was significantly increased in NSCLC than in nonmalignant lung tissue, and the pooled OR was 5.69 with 95% CI: 3.32–9.76, z=6.32, P<0.00001 (). RARβ hypermethylation was significantly more frequently observed in AC than in SCC, and the pooled OR was 1.47 with 95% CI: 1.12–1.93, z=2.79, P=0.005 (). RARβ hypermethylation rate was not significantly correlated with stage of the disease, and the pooled OR was 0.82 with 95% CI: 0.63–1.08, z=1.4, P=0.16 (). RARβ methylation was 2.46 times higher in smoking than in nonsmoking individuals, and the pooled OR was 2.46 with 95% CI: 1.54–3.93, z=3.76, P=0.0002 (). RARβ gene hypermethylation had no correlation with patient sex, and the pooled OR was 1.20 with 95% CI: 0.79–1.84, z=0.85, P=0.40 (). RARβ gene methylation status was not associated with prognosis of patients with NSCLC, the pooled HR was 1.05 with 95% CI: 0.68–1.64, z=0.23, P=0.82 ().
Figure 2 Forest plot for RARβ hypermethylation in NSCLC and non-neoplastic lung tissue.
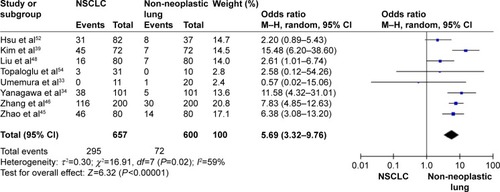
Figure 3 Forest plot for RARβ hypermethylation in AC and SCC.
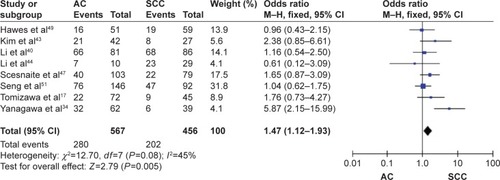
Figure 4 Forest plot for RARβ hypermethylation in advanced and low stage of NSCLC.
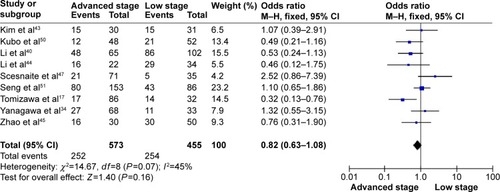
Figure 5 Forest plot for RARβ hypermethylation of NSCLC in smoking and nonsmoking individual.
Sensitivity analyses and publication bias
A sensitivity analysis was conducted by removing one study from the meta-analysis at a time, and the overall results were not significantly affected. The pooled ORs or HRs were not significantly changed, indicating the stability of our analyses. The funnel plots were largely symmetric (), suggesting there were no significant publication biases in the meta-analysis of RARβ hypermethylation and clinicopathological features.
Figure 8 Funnel plot for publication bias.
Abbreviations: NSCLC, non-small-cell lung carcinoma; AC, adenocarcinoma; SCC, squamous cell carcinoma; SE, standard error of the mean; OR, odds ratio; df, degree of freedom.
Discussion
The human RARβ gene is located on chromosome 3, locus 3p24, and consists of five distinct isoforms: hRARβ1′, hRARβ1, hRARβ2, hRARβ4, and hRARβ5.Citation20,Citation21 Among those five isoforms, hRARβ1 and hRARβ2 are the functional isoforms that activate distinct cassettes of target genes with different biological effects.Citation22 hRARβ1 is a fetal isoform, which may play an important role during development in human.Citation23 The hRARβ1′ was identified in normal lung tissue and bronchial epithelial cells. The absence of hRARβ1′ is associated with retinoid resistance in human lung carcinogenesis.Citation24,Citation25 The reexpression of hRARβ1′ in lung cancer cells restored RA-target gene induction and growth suppression following retinoid treatment.Citation25 The hRARβ2 is the retinoid-inducible splicing isoform, and multiple hRARβ2 transcript variants are expressed in both normal and neoplastic cells.Citation26,Citation27 Both the hRARβ1′ and hRARβ2 transcripts are obtained from the transcription start sites of P2 promote which contains a CpG-rich region that is susceptible to methylation-induced silencing.Citation28–Citation31
Aberrant methylation of the promoter regions of genes is a major mechanism of gene silencing in tumor.Citation32 A number of studies reported that RARβ promoter hypermethylation was observed in NSCLC, but the rate was remarkably diverse due to small power.Citation33,Citation34 In the present study, we conducted a meta-analysis and pooled 18 studies which included 1,871 NSCLC patients. Our data showed RARβ promoter hypermethylation significantly increased by 5.69 times in NSCLC than in non-neoplastic lung tissue, indicating that RARβ promoter hypermethylation contributed to the initiation and development of NSCLC. On the other hand, the translation of hRARβ1′ initiates at the P2 promoter, and RARβ promoter hypermethylation suppresses hRARβ1′ expression that is required for RA-target gene induction. Therefore, RARβ promoter hypermethylation could lead to the incident of retinoid resistance following retinoid treatment. Confirmation studies need to be carried out in future. RARβ promoter hypermethylation is a reversible event; drug treatment through demethylation may be used not only to delay carcinogenesis and the progression of NSCLC, but also to restore RA-target gene induction. Virmani et al demonstrated that the treatment of lung cancer cell lines with demethylation agent 5-aza-2′-deoxycytidine (5-AZA-CdR) can restore RARβ expression.Citation32 A Phase I/II trial in patients with stage IV NSCLC suggests that 5-AZA-CdR may have some clinical activity against metastatic NSCLC.Citation35 Recently, curcumin, a potent cancer preventive agent,Citation36 was also reported to significantly decrease RARβ promoter methylation and elevate RARβ expression at the messenger RNA and protein levels in lung cancer A549 and H460 cells.Citation37 Taken together, demethylation of RARβ promoter could suppress the development of NSCLC, and potentially restores RA-target gene induction. RARβ is a promising gene target for the development of personalized therapy in patients with NSCLC.
Hua et al conducted a meta-analysis and reported that the frequency of RARβ promoter hypermethylation increased in NSCLC than autologous controls, suggesting the methylation status could be a valuable diagnostic tool for NSCLC.Citation38 However, they only accessed eleven studies. Our analysis included additional seven studies and showed more convincing results. In addition, we also found that the rate of RARβ hypermethylation was increased in AC than SCC and that there was a strong correlation between methylation status of RARβ and smoking in patients with NSCLC.
The rate of RARβ hypermethylation between AC and SCC has been compared in previous studies and the results were contradictory as the studies included limited number of patients.Citation39,Citation40 We pooled eight studies and analyzed the frequency of RARβ promoter hypermethylation in AC and SCC. The rate of RARβ promoter hypermethylation was higher in AC than SCC, suggesting that the molecular mechanisms could be different between AC and SCC. Additionally, RARβ promoter hypermethylation significantly increased in smoking NSCLC patients compared to nonsmoking NSCLC patients, indicating that smoking induced gene methylation which may lead to malignant growth and cancer development.Citation41,Citation42
Several studies reported the correlation between methylation status of RARβ and survival in NSCLC patients, and the results were inconsistent.Citation34,Citation43 In the present meta-analysis, no significant association between RARβ promoter methylation status and survival was found. Additionally, the rate of RARβ promoter hypermethylation was similar between lower and advanced stages of disease. Our study only included the articles published in English or Chinese language, and did not select some relevant papers published in other languages, which may result in certain publication bias. Hence, cautions should be taken when our findings are interpreted among the general populations.
Conclusion
In summary, RARβ promoter hypermethylation significantly increased in NSCLC tumor than in non-neoplastic lung tissue, suggesting that RARβ methylation contributes to the development of NSCLC. RARβ gene is a potential novel target for demethylation treatment in patients with NSCLC, and is promising in restoration of RA-target gene induction via demethylation of RARβ1′ promoter. RARβ promoter hypermethylation is associated with smoking behavior as well as histologic subtypes of NSCLC in patients with NSCLC. The rate of RARβ promoter hypermethylation is higher in AC than SCC. RARβ methylation status is not correlated to survival of patients with NSCLC.
Authors contributions
All authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work.
Acknowledgments
This work was not supported by any fund. This work did not get contribution from other people except authors.
Disclosure
The authors report no conflicts of interest in this work.
References
- JemalASiegelRXuJWardECancer statistics, 2010CA Cancer J Clin20106027730010.3322/caac.20073caac.20073 [pii]20610543
- TravisWDPathology of lung cancerClin Chest Med20113266969210.1016/j.ccm.2011.08.005 S0272-5231(11)00080-3 [pii]22054879
- TsaiMFWangCCChenJJTumour suppressor HLJ1: a potential diagnostic, preventive and therapeutic target in non-small cell lung cancerWorld J Clin Oncol2014586587310.5306/wjco.v5.i5.86525493224
- BaylinSBHermanJGGraffJRVertinoPMIssaJPAlterations in DNA methylation: a fundamental aspect of neoplasiaAdv Cancer Res1998721411969338076
- BaylinSBEstellerMRountreeMRBachmanKESchuebelKHermanJGAberrant patterns of DNA methylation, chromatin formation and gene expression in cancerHum Mol Genet20011068769211257100
- MerloAHermanJGMaoL5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancersNat Med199516866927585152
- HellerGBabinskyVNZieglerBGenome-wide CpG island methylation analyses in non-small cell lung cancer patientsCarcinogenesis20133451352110.1093/carcin/bgs36323172663
- SandovalJMendez-GonzalezJNadalEA prognostic DNA methylation signature for stage I non-small-cell lung cancerJ Clin Oncol2013314140414710.1200/JCO.2012.48.551624081945
- GrummerMAThetLAZachmanRDExpression of retinoic acid receptor genes in fetal and newborn rat lungPediatr Pulmonol1994172342388208594
- MendelsohnCLohnesDDécimoDFunction of the retinoic acid receptors (RARs) during development (II). Multiple abnormalities at various stages of organogenesis in RAR double mutantsDevelopment1994120274927717607068
- ChytilFRetinoids in lung developmentFASEB J1996109869928801181
- MinnaJDMangelsdorfDJRetinoic acid receptor expression abnormalities in lung cancer: important clues or major obstacles?J Natl Cancer Inst1997896026049150178
- BogosKRenyi-VamosFKovacsGTovariJDomeBRole of retinoic receptors in lung carcinogenesisJ Exp Clin Cancer Res2008271810.1186/1756-9966-27-18 1756-9966-27-18 [pii]18625040
- LeeYWKleinCBKargacinBCarcinogenic nickel silences gene expression by chromatin condensation and DNA methylation: a new model for epigenetic carcinogensMol Cell Biol199515254725577537850
- SwaffordDSMiddletonSKPalmisanoWAFrequent aberrant methylation of p16INK4a in primary rat lung tumorsMol Cell Biol199717136613749032263
- RomWNHayJGLeeTCJiangYTchou-WongKMMolecular and genetic aspects of lung cancerAm J Respir Crit Care Med20001611355136710.1164/ajrccm.161.4.990801210764334
- TomizawaYIijimaHNomotoTClinicopathological significance of aberrant methylation of RARbeta2 at 3p24, RASSF1A at 3p21.3, and FHIT at 3p14.2 in patients with non-small cell lung cancerLung Cancer200446305312 S0169500204002181 [pii]10.1016/j.lungcan.2004.05.00315541815
- YanagawaNTamuraGOizumiHEndohMSadahiroMMotoyamaTInverse correlation between EGFR mutation and FHIT, RASSF1A and RUNX3 methylation in lung adenocarcinoma: relation with smoking statusAnticancer Res20113112111214 31/4/1211 [pii]21508367
- EggerMDavey SmithGSchneiderMMinderCBias in meta-analysis detected by a simple, graphical testBMJ19973156296349310563
- BenbrookDLernhardtEPfahlMA new retinoic acid receptor identified from a hepatocellular carcinomaNature198833366967210.1038/333669a02836738
- BrandNPetkovichMKrustAIdentification of a second human retinoic acid receptorNature199833285085310.1038/332850a02833708
- SwiftCBHaysJLPettyWJDistinct functions of retinoic acid receptor beta isoforms: implications for targeted therapyEndocr Metab Immune Disord Drug Targets20088475018393922
- ToulouseAMorinJPelletierMBradleyWEStructure of the human retinoic acid receptor beta 1 geneBiochim Biophys Acta1996130914 S0167-4781(96)00126-1 [pii]8950166
- MaYKoza-TaylorPHDiMattiaDAMicroarray analysis uncovers retinoid targets in human bronchial epithelial cellsOncogene2003224924493210.1038/sj.onc.12067281206728 pii12894236
- PettyWJLiNBiddleABoundsRA novel retinoic acid receptor beta isoform and retinoid resistance in lung carcinogenesisJ Natl Cancer Inst20059716451651 97/22/1645 [pii]10.1093/jnci/dji37116288117
- LotanRXuXCLippmanSMSuppression of retinoic acid receptor-beta in premalignant oral lesions and its up-regulation by isotretinoinN Engl J Med19953321405141010.1056/NEJM1995052533221037723796
- PengXMehtaRGTonettiDAChristovKIdentification of novel RARbeta2 transcript variants with short 5′-UTRs in normal and cancerous breast epithelial cellsOncogene20052412961301 1208284[pii]10.1038/sj.onc.120828415558014
- Di CroceLRakerVACorsaroMMethyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factorScience20022951079108210.1126/science.1065173 295/5557/1079 [pii]11834837
- FaziFZardoGGelmettiVHeterochromatic gene repression of the retinoic acid pathway in acute myeloid leukemiaBlood200710944324440 doi:blood-2006-09-045781[pii]10.1182/blood-2006-09-04578117244680
- ToulouseAMorinJDionPAHouleBBradleyWERARbeta2 specificity in mediating RA inhibition of growth of lung cancer-derived cellsLung Cancer200028127137 S0169-5002(99)00122-1 [pii]10717330
- SunSYWanHYuePHongWKLotanREvidence that retinoic acid receptor beta induction by retinoids is important for tumor cell growth inhibitionJ Biol Chem2000275171491715310.1074/jbc.M000527200 M000527200 [pii]10747926
- VirmaniAKRathiAZöchbauer-MüllerSPromoter methylation and silencing of the retinoic acid receptor-beta gene in lung carcinomasJ Natl Cancer Inst2000921303130710944551
- UmemuraSFujimotoNHirakiAAberrant promoter hypermethylation in serum DNA from patients with silicosisCarcinogenesis2008291845184910.1093/carcin/bgn169 bgn169 [pii]18632757
- YanagawaNTamuraGOizumiHPromoter hypermethylation of RASSF1A and RUNX3 genes as an independent prognostic prediction marker in surgically resected non-small cell lung cancersLung Cancer200758131138 S0169-5002(07)00315-7 [pii]10.1016/j.lungcan.2007.05.01117606310
- MomparlerRLBouffardDYMomparlerLFDionneJBelangerKAyoubJPilot phase I–II study on 5-aza-2′-deoxycytidine (Decitabine) in patients with metastatic lung cancerAnticancer Drugs199783583689180389
- TuorkeyMJCurcumin a potent cancer preventive agent: mechanisms of cancer cell killingInterv Med Appl Sci2014613914610.1556/IMAS.6.2014.4.1 []IMAS_6(2014)4/1 [pii]25598986
- JiangAWangXShanXCurcumin reactivates silenced tumor suppressor gene RARbeta by reducing DNA methylationPhytother Res2015291237124510.1002/ptr.537325981383
- HuaFFangNLiXZhuSZhangWGuJA meta-analysis of the relationship between RARbeta gene promoter methylation and non-small cell lung cancerPLoS One20149e9616310.1371/journal.pone.009616324796328
- KimYTParkSJLeeSHPrognostic implication of aberrant promoter hypermethylation of CpG islands in adenocarcinoma of the lungJ Thorac Cardiovasc Surg20051301378 doi:S0022-5223-(05)01052-4 [pii]10.1016/j.jtcvs.2005.06.01516256792
- LiWDengJTangJXCombined effects methylation of FHIT, RASSF1A and RARbeta genes on non-small cell lung cancer in the Chinese populationAsian Pac J Cancer Prev2014155233523725040980
- International Agency for Research on CancerTobacco smoke and involuntary smokingIARC Monographs on the Evaluation of the Carcinogenic Risk of Chemicals to HumansLyon, FranceInternational Agency for Research on Cancer832004
- Husgafvel-PursiainenKGenotoxicity of environmental tobacco smoke: a reviewMutat Res2004567427445 doi:S1383-5742(04)00063-8 [pii]10.1016/j.mrrev.2004.06.00415572289
- KimYTLeeSHSungSWKimJHCan aberrant promoter hypermethylation of CpG islands predict the clinical outcome of non-small cell lung cancer after curative resection?Ann Thorac Surg20057911801188 discussion. 1180–1188. doi:S0003-4975(04)02007-7 [pii]10.1016/j.athoracsur.2004.09.06015797047
- LiWDengJWangSSAssociation of methylation of the RAR-beta gene with cigarette smoking in non-small cell lung cancer with Southern-Central Chinese populationAsian Pac J Cancer Prev201415109371094125605205
- ZhaoXWangNZhangMXueSShiKChenZDetection of methylation of the RAR-beta gene in patients with non-small cell lung cancerOncol Lett2012365465810.3892/ol.2011.527 ol-03-03-0654 [pii]22740970
- ZhangCYJinYTXuHYRelationship between promoter methylation of p16, DAPK and RAR beta genes and the clinical data of non-small cell lung cancerZhonghua Yi Xue Yi Chuan Xue Za Zhi201128232810.3760/cma.j.issn.1003-9406.2011.01.006 940628006 [pii]21287504
- ScesnaiteAJarmalaiteSMutanenPSimilar DNA methylation pattern in lung tumours from smokers and never-smokers with second-hand tobacco smoke exposureMutagenesis20122742342910.1093/mutage/ger092 ger092 [pii]22217548
- LiuZLiWLeiZCpG island methylator phenotype involving chromosome 3p confers an increased risk of non-small cell lung cancerJ Thorac Oncol2010579079720521346
- HawesSESternJEFengQDNA hypermethylation of tumors from non-small cell lung cancer (NSCLC) patients is associated with gender and histologic typeLung Cancer20106917217910.1016/j.lungcan.2009.11.002 S0169-5002(09)00579-0 [pii]19945765
- KuboTYamamotoHIchimuraKDNA methylation in small lung adenocarcinoma with bronchioloalveolar carcinoma componentsLung Cancer20096532833210.1016/j.lungcan.2008.12.001 S0169-5002(08)00663-6 [pii]19144441
- SengTJCurreyNCooperWADLEC1 and MLH1 promoter methylation are associated with poor prognosis in non-small cell lung carcinomaBr J Cancer20089937538210.1038/sj.bjc.66044526604452 pii18594535
- HsuHSChenTPWenCKMultiple genetic and epigenetic biomarkers for lung cancer detection in cytologically negative sputum and a nested case-control study for risk assessmentJ Pathol200721341241910.1002/path.224617973238
- KatayamaHHirakiAAoeKAberrant promoter methylation in pleural fluid DNA for diagnosis of malignant pleural effusionInt J Cancer20071202191219510.1002/ijc.2257617285579
- TopalogluOHoqueMOTokumaruYDetection of promoter hypermethylation of multiple genes in the tumor and bronchoalveolar lavage of patients with lung cancerClin Cancer Res2004102284228815073103