Abstract
Background
CYP2D6 is one of the most important members of the cytochrome P450 superfamily. Its genetic polymorphism significantly influences the efficacy and safety of some drugs, which might cause adverse effects and therapeutic failure.
Methods and results
The aim of this research was mainly to explore the catalytic activities of 22 newly reported CYP2D6 isoforms (2D6*87, *88, *89, *90, *91, *92, *93, *94, *95, *96,*97, *98, *R25Q, F164L, E215K, F219S, V327M, D336N, V342M, R344Q, R440C, R497C) on dapoxetine in vitro. The research was designed with an appropriate incubation system in test tubes and carried out in the constant temperature water. Through detecting its two metabolites desmethyldapoxetine and dapoxetine-N-oxide, the available data were obtained to explain the influence of CYP2D6 polymorphism on the substrate drug dapoxetine. As a result, the intrinsic clearance (Vmax/Km) values of most variants were significantly altered when compared with the counterpart of CYP2D6*1, with most of these variants exhibiting either reduced Vmax and/or increased Km values. For dapoxetine demethylation pathway (which produces desmethyldapoxetine), 2D6*89 and E215K exhibited no markedly decreased relative clearance of 92.81% and 97.70%, respectively. The relative clearance of rest 20 variants exhibited decrease in different levels, ranging from 20.44% to 90.90%. For the dapoxetine oxidation pathway (which produces dapoxetine-N-oxide), the relative clearance values of three variants, 2D6*90, *94, and V342M, exhibited no markedly increased relative clearance of 106.17%, 107.78%, and 109.98%, respectively; the rest 19 variants exhibited significantly decreased levels ranging from 27.56% to 84.64%. In addition, the kinetic parameters of two CYP2D6 variants (2D6*92 and 2D6*96) could not be detected, due to the defect of the CYP2D6 gene.
Conclusion
As the first report of all aforementioned alleles for dapoxetine metabolism, these data may help in the clinical assessment of the metabolic elimination of dapoxetine and may provide fundamental information for further clinical studies.
Introduction
Polymorphism in the cytochrome P450 (CYP) family may have the most impact on the metabolism of therapeutic drugs. CYP2D6, CYP2C9, CYP2C19, and CYP3A4 polymorphisms account for the most frequent variations in Phase I metabolism of drugs, since almost 80% of drugs are metabolized by these enzymes.Citation1 CYP2D6 is one of the highly polymorphic enzymes and participates in the metabolism of many therapeutic drugs that are commonly used in clinic, including the antidepressants fluoxetine, amitriptyline, and venlafaxine; the antitussive dextromethorphan; the β-adrenergic antagonists bufuralol and metoprolol;Citation2 the opioid analgesics codeine, dihydrocodeine, and tramadol;Citation3 the antipsychotic agent risperidone;Citation4 and the selective serotonin reuptake inhibitor dapoxetine.Citation5 CYP2D6 polymorphisms can lead to no enzyme expression (poor metabolism), very low enzyme activity (intermediate metabolism), or typically associated with overexpression of a metabolic enzyme (ultrarapid metabolism).Citation6 These may cause adverse effects and therapeutic failures, thus more attention should be paid to the CYP2D6 polymorphism.
According to the National Center for Biotechnology Information website report, 370 variable sites of CYP2D6 have been found and more than 100 CYP2D6 alleles have been identified and named by the Human CYP Allele Nomenclature Committee (http://www.cypalleles.ki.se/cyp2d6.htm). In 2013, Qian et alCitation7 analyzed the CYP2D6 gene of 2,129 unrelated healthy Chinese volunteers and detected 165 mutated sites, of which 67 sites were discovered for the first time. Among these, 22 novel mutation sites were nonsynonymous, and of them 12 mutation sites were named as *87–*93, *94A, *94B, and *95–*98 by the Human CYP Allele Nomenclature Committee.Citation7 In later study, 22 newly reported CYP2D6 isoforms were transiently expressed to assess the enzymatic activity of these variants on dextromethorphan and bufuralol.Citation8,Citation9
Premature ejaculation (PE) is a common problem, with a global prevalence estimated to be 20%–40%Citation10 and has significant impact not only on the sufferer, but also on the partner, in terms of self-esteem, interpersonal distress, and sexual satisfaction.Citation11 Dapoxetine, a selective serotonin reuptake inhibitor, is used for the treatment of PE in men aged 18–64 years.Citation12 However, dapoxetine has numerous adverse effects like nausea, diarrhea, dizziness, insomnia, and nasopharyngitis.Citation13–Citation15 In addition, dapoxetine has no pharmacokinetic interactions with food, alcohol, or phosphodiesterase type 5 (PDE5) inhibitors.Citation16,Citation17 Dapoxetine is primarily metabolized by CYP3A4 and CYP2D6.Citation5 Thus, the exploration of CYP3A4 or CYP2D6 gene polymorphisms on dapoxetine metabolism can be meaningful. In this study, we focus on the catalytic activities of 24 CYP2D6 isoforms on dapoxetine metabolism in vitro by detecting its two metabolites generated by CYP2D6 enzyme (shown in ). We hope these findings can provide reference for rational administration of drugs in the clinic and promote the development of personalized medicine.
Materials and methods
Chemicals and materials
Dapoxetine, desmethyldapoxetine, dapoxetine-N-oxide, and carbamazepine were obtained from Sigma-Aldrich (St Louis, MO, USA). The UPLC® BEH C18 column (2.1 mm ×50 mm, 1.7 μm) was obtained from the Waters (Ireland). Nicotinamide adenine dinucleotide phosphate was obtained from Promega (Madison, WI, USA). Formic acid (analytical reagent grade) was purchased from Sigma-Aldrich. Ultrapure water was obtained from a Milli-Q system (Millipore, Bedford, MA, USA). Liquid chromatography grade methanol and acetonitrile were purchased from Merck Chemicals Co., Ltd. (Darmstadt, Germany). Other chemicals and solvents were of analytical grade from Chemical Industries (Beijing, People’s Republic of China).
Instrumentation
Samples were analyzed by the ultra-performance liquid chromatography tandem mass-spectrometry with ACQUITY UPLC H-Class and XEVO TQD triple-quadrupole mass spectrometer (Waters Corp., Milford, MA, USA) equipped with an electrospray ionization interface. MassLynx 4.1 software (Waters Corp.) was used to control the instrument and process all the data of the samples.
Incubation conditions
Recombinant human CYP2D6 enzymes (the wild-type CYP2D6*1 and 24 CYP2D6 variants) generated in Spodoptera frugiperda 21 insect cells were obtained according to the previously reported method.Citation8 The incubation mixture as the final assay concentration included 176.6 μL 100 mmol/L potassium phosphate buffer (pH 7.4), 5 pmol wild-type CYP2D6*1 or other CYP2D6 recombinant variants, 5 pmol purified cytochrome b5, and 1.71 μL of dapoxetine, which was made to the total volume of 200 μL. Dapoxetine was initially prepared in methanol solution and the concentration range was adjusted from 25 to 3,200 μM. The total concentration of methanol in the mixture was <0.5%. After incubating for 5 minutes, 1 mM nicotinamide adenine dinucleotide phosphate-regenerating system (1.3 mM NADP+, 3.3 mM glucose-6-phosphate, 3.3 mM MgCl2, and 0.4 unit/mL glucose-6-phosphate dehydrogenase) was added. The mixture was incubated at 37°C for 40 minutes. In addition, the experiments were carried out in individual tubes and kept parallel, one for each concentration point. The reaction in tubes was terminated by cooling to −80°C immediately. Then 400 μL acetonitrile and 40 μL internal standard carbamazepine were added. Carbamazepine was dissolved in acetonitrile solution at a concentration of 1 μg/mL. After vortexing for 2 minutes and centrifuging at 13,000 rpm for 10 minutes in 4°C environment, the supernatant of each tube was 1:1 diluted with ultrapure water and 2 μL was injected into the ultra-performance liquid chromatography tandem mass-spectrometry system. The data were analyzed using SPSS Version 13.0 (SPSS Inc., Chicago, IL, USA). All the experimental results were as mean ± standard deviation (SD) of three parallel measurements. All experiments were approved by the Ethics Committee of Beijing Hospital.
Chromatographic conditions
The liquid chromatographic separations were performed on an UPLC BEH C18 column (2.1 mm ×50 mm, 1.7 μm), with an inline 0.2 mm stainless steel frit filter connected to it. The column temperature was kept at 40°C constantly, while the samples in the autosampler room were maintained at 4°C with an injection volume of 2 μL for detection. The initial mobile phase consisted of acetonitrile and water (containing 0.1% formic acid) at a flow rate of 0.4 mL/min. Elution was in a linear gradient as follow: the acetonitrile content changing from 30% to 85% between 0 and 1 minute, maintained at 85% for 1 minute, then decreased to 30% within 12 seconds. The total run time of the analytes was 3 minutes. After each injection, the sample manager underwent a needle wash process, including a strong wash (methanol-water, 50/50, v/v) and a weak wash (methanol-water, 10/90, v/v). With aforementioned appropriate conditions, the retention times of desmethyldapoxetine, dapoxetine-N-oxide, dapoxetine, and carbamazepine were at 1.40, 1.45, 1.50, and 1.23 minutes, respectively.
Mass spectrometric conditions
A Waters XEVO TQD triple-quadrupole mass spectrometer, equipped with an electrospray ionization source, was set to positive electrospray ionization in multiple reaction-monitoring mode. Nitrogen was used as the desolvation gas (600 L/h) and cone gas (50 L/h). The selected ion monitoring conditions were defined as follows: capillary voltage 2.5 kV; source temperature 150°C; and desolvation temperature 500°C. The multiple reaction-monitoring mode of transitions as quantitative analysis were m/z 306.0→261.1, m/z 292.0→261.1, m/z 322.0→261.1, and m/z 237.0→194.1 for dapoxetine, desmethyldapoxetine, dapoxetine-N-oxide, and IS, respectively. The collision energy was set at 25 V for dapoxetine, desmethyldapoxetine, and dapoxetine-N-oxide, while 20 V for carbamazepine; the cone voltage was set at 50 V for dapoxetine, desmethyldapoxetine, and dapoxetine-N-oxide, while 40 V for carbamazepine, respectively.
Statistical analysis
The kinetic parameters (Km and Vmax) were performed by non-linear regression curve fitting using the computer program Prism version 5 (GraphPad Software Inc., San Diego, CA, USA). One-way analysis of variance was used for intergroup comparison. Dunnett’s test was used to analyze differences in catalytic activity between CYP2D6*1 and other CYP2D6 mutants. Kinetic data for each variant were presented as the mean ± SD of three independent experiments. Statistical analysis was performed using SPSS version 17.0 (SPSS Inc., Chicago, IL, USA), with P<0.05 considered statistically significant.
Results
From the aforementioned study, the catalytic activities of the wild-type and 24 allelic variants of CYP2D6 were assessed, using dapoxetine as the substrate drug. Michaelis–Menten plots of the tested 25 CYP2D6 enzymes are shown in and , and corresponding kinetic parameters are summarized in and . The intrinsic clearance (Vmax/Km) values of dapoxetine were significantly altered in most of the allelic variants, except occasionally a few, according to the analysis of its two metabolites. The relative clearance graph (compared with that of 2D6*1) of dapoxetine demethylation and oxidation is shown in .
Table 1 Kinetic parameters from demethylation by recombinant wild-type and 24 CYP2D6 allelic variants on dapoxetine
Table 2 Kinetic parameters from oxidational activities of recombinant wild-type and 24 CYP2D6 allelic variants toward dapoxetine
Figure 2 Michaelis–Menten curves of the enzymatic activity of the wild-type and 24 variants toward dapoxetine demethylation (each point represents the mean ± standard deviation of three parallel experiments).
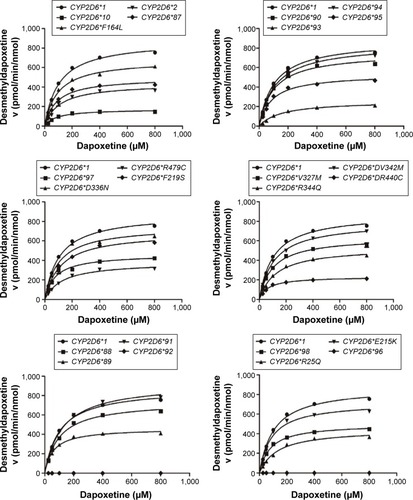
Figure 3 Michaelis–Menten curves of the enzymatic activity of the wild-type and 24 variants toward dapoxetine oxidation (each point represents the mean ± standard deviation of three parallel experiments).
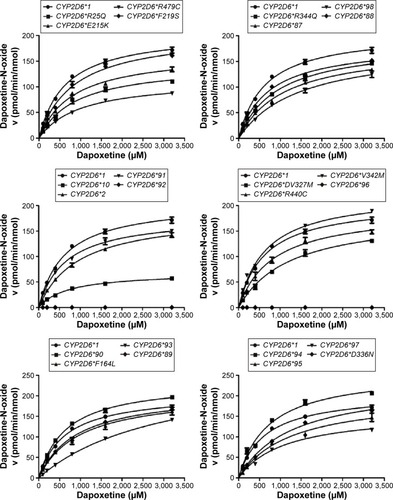
Figure 4 The catalytic activity of expressed CYP2D6 variants toward the two metabolites of dapoxetine, when compared with the counterpart values of the wild-type 2D6*1.
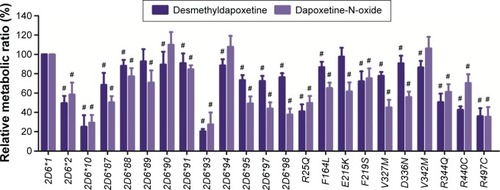
The results of CYP2D6 allelic variants on dapoxetine demethylation are represented in and . Twelve of the allelic variants exhibited obvious reduced Vmax values when compared with 2D6*1 (count as 100%): three mutants (2D6*10, *93, and R440C) obviously reduced to 19.77%–29.41%; nine variants (2D6*2, *87, *89, *95, *97, *98, R25Q, R344Q, and R497C) decreased to 43.64%–61.11%. Moreover, most variants showed no obvious changes on Km values: eight variants (2D6*2, *88, *90, F164L, F219S, V327M, D336N, and V342M) showed no statistical difference; six variants (2D6*10, *87, *95, *97, *98, and E215K) mildly decreased to 78.40%–84.22%; six variants (2D6*91, *93, *94, R25Q, R344Q, and R497C) lightly increased to 108.57%–123.40%. Except two variants (2D6*89 and R440C), their Km values decreased to 56.58% and 62.08%, respectively. As a result, Vmax/Km values for dapoxetine demethylation were altered in majority of these allelic variants (2D6*2, *10, *87, *88, *90, *91, *93, *94, *95, *97, *98, *R25Q, F164L, F219S, V327M, D336N, V342M, R344Q, R440C, and R497C). Definitely, CYP2D6 allelic variants could be classified into categories, according to their intrinsic clearance values when compared with the counterpart of the wild-type (counted as 100%): six variants (2D6*2, *10, *93, R25Q, R440C, and R497C) exhibited obviously decreased intrinsic clearance values (20.44%–49.63% relative clearance); 14 alleles (CYP2D6*87, *88, *90, *91, *94, *95, *97, *98, F164L, F219S, V327M, D336N, V342M, and R344Q) exhibited slight reduction (50.58%–90.90% relative clearance). Another two variants (2D6*89 and E215K) were without distinct difference (92.81% and 97.70% relative clearance), according to the statistical analyses.
The results of the oxidative metabolism of dapoxetine are shown in and . Most of the tested allelic variants exhibited no significant changes on Vmax and/or Km values when compared with the counterparts of 2D6*1 (count as 100%). Two variants (2D6*93 and *94) exhibited obviously increased Vmax values (124.95%–135.58%); five variants (2D6*10, *97, R25Q, E215K, and R497C) decreased to 33.26%–80.63%. Four variants (2D6*93, *97, *98, and D336N) exhibited significantly increased Km values with ~1.78- to 4.98-fold as compared with 2D6*1. Consequently, Vmax/Km values for dapoxetine oxidation were altered in most of the tested allelic variants (2D6*2, *10, *87, *88, *90, *91, *93, *94, *95, *97, *98, *R25Q, F164L, F219S, V327M, D336N, V342M, R344Q, R440C, R497C), except 2D6*90, *94, and V342M without statistical significance. Compared with the Vmax/Km value of 2D6*1 (count as 100%), eight variants (2D6*10, *93, *95, *97, *98, R25Q, V327M, and R497C) significantly declined to 27.56%–49.80%; eleven alleles (CYP2D6*2, *87, *88, *89, *91, F164L, E215K, F219S, D336N, R344Q, and R440C) exhibited mild decreases (50.34%–84.64%). Three variants 2D6*90, 2D6*94, and V342M represented no significant increase (106.17%–109.98% relative clearance).
Furthermore, two defective alleles (CYP2D6*92 and CYP2D6*96) showed extremely low activity or no activity. Thus, the concentrations of both metabolites were below the detection limit and the kinetic parameters could not be determined.
Discussion
CYP2D6 is a highly polymorphic enzyme and is involved in the metabolism of many drugs. Nowadays, it participates in the metabolism of ~30% of drugs in clinic.Citation18 The polymorphism of CYP2D6 significantly affects the pharmacokinetics of its substrate drugs in clinic.Citation19 The study of CYP2D6 polymorphism, which mainly cause drug metabolism differences and result in side effects, can provide references to the clinical research. There are significant differences among various racial and ethnic population on the frequency of CYP2D6 polymorphism.Citation20,Citation21 In our previous study, we performed a large-scale genetic investigation by sequencing the CYP2D6 gene in 2,129 healthy Chinese volunteers and 22 novel nonsynonymous variants were discovered and detected.Citation7 This finding may greatly contribute to the development of personalized medicine for the Chinese Han population.
Dapoxetine has been recently evaluated for the treatment of PE by several countries.Citation22,Citation23 It is the only drug that has been approved for the on-demand treatment of PE.Citation24 But in the process of treatment, a variety of adverse reactions occurred, so a considerable number of patients chose to spontaneously discontinue treatment with it.Citation14 Thus, the effects of CYP2D6 polymorphism on dapoxetine metabolism in vitro have a great significance for the basic research and personalized treatment.
Two typical variants CYP2D6*2 and CYP2D6*10 were served as the positive controls for the functional analysis to ensure the reliability of this study. Recently, a study demonstrated that CYP2D6*2 exhibited a significantly decreased Vmax/Km values for both bufuralol and dextromethorphan (~40% of CYP2D6*1) with the baculovirus expression system.Citation9 For CYP2D6*10, the most common allelic variant in oriental populations, several functional analyses have been conducted in various expression systemsCitation25 and it has higher Km, lower Vmax, and lower Vmax/Km values for bufuralol, dextromethorphan, debrisoquine, atomoxetine, and nortriptyline in vitro.Citation18 Our investigation revealed that both variants presented significantly decreased Vmax/Km value on dapoxetine (approximately decreased by 50% for CYP2D6*2 and 75% for CYP2D6*10); these findings were consistent with the aforementioned studies.
To better understand the effects of CYP2D6 allelic variants on the metabolism of dapoxetine, we analyzed the 22 novel CYP2D6 variants in detail. In particular, three isoforms, 2D6*93, R25Q, and R497C, exhibited significant changes on Vmax/Km values, indicating that the amino acids in these sites have a vital impact on the metabolism of dapoxetine. For allelic isoform CYP2D6*93, the Vmax/Km valve decreased by 70%. For the other two variants, the Vmax/Km valve of R497C decreased bŷ65%, while R25Q decreased by more than 50%. Furthermore, two allelic isoforms (CYP2D6*92 and CYP2D6*96) exhibited absent metabolic activity. CYP2D6*93 (Thr249Pro) contains one nucleotide substitution (A>C) at position 745 in the complementary DNA (cDNA), which causes an amino acid change from Thr to Pro at position 249.Citation8 The Thr-249 residue is adjacent to the residues Phe-247 and Leu-248, which are located on the border of CYP2D6 active site cavity,Citation26 because Leu-248, Leu-110, and Phe-112 constitute one access channel for substrate entrance. We speculate that the replacement of the hydrophilic Thr with the hydrophobic Pro at position 249 might influence the spatial structure of the adjacent egress channel and thus blocks the normal entrance of the CYP2D6 substrate.
For the isoform R25Q, which exhibited significantly decreased enzymatic activity on dapoxetine, it has been deduced that Arg25 is located within the transmembrane domain and acts as a halt transfer signal; thus, changes in this site might significantly decrease the enzymatic activity in vitro.Citation27 For R497C, previous functional predictions revealed that this allelic isoform might deleteriously affect the CYP2D6 protein using bioinformation tools.Citation7
Similar to the previously reported variants CYP2D6*20 (211frameshift), CYP2D6*8 (Gly169STOP), CYP2D6*92 (218frameshift), and CYP2D6*96 (Gln424STOP) exhibited no enzymatic activity.Citation1 CYP2D6*92 variant has a one-nucleotide deletion (nucleotide C) at site 1995 in exon 4 and causes a frameshift effect: a disrupted reading frame and the premature termination of protein synthesis during its translation. CYP2D6*96 has a single-nucleotide mutation in exon 8 C>T at position 3895 in the DNA (site 1270 in cDNA) and causes the codon 424 changing from CAG to one stop codon TAG. For CYP2D6*92 and CYP2D6*96, immunoblotting results revealed that both variants were expressed as truncated proteins, which caused the functional loss of enzymatic activity.Citation8 We speculate that responsibility for the deficiency of enzymatic function might be the frameshift and resulting premature termination.
Particularly, the Vmax/Km values of CYP2D6*89 and E215K exhibited no difference compared with 2D6*1 for demethylation, with 92.81% and 97.70%, respectively. This phenomenon is not consistent with the previous research results.
For CYP2D6*89 (L142S), it contains one T to C substitution in site 1678 of the DNA sequence (425T>C in the cDNA) and results in one amino acid change from Leu to Ser at position 142.Citation7 On the other hand, for E215K, Rowland et alCitation26 found that many important amino acid residues were essential to the active site cavity. The Glu216 residue, which is located in the F helix, plays an important role in substrate recognition and binding.Citation28 In the tested variants, E215K and F219S were located in the F helix.Citation7 Dai et alCitation8 indicated that CYP2D6*89 and E215K exhibited >90% decrease in catalytic activity on bufuralol and dextromethorphan compared with the wild-type CYP2D6*1.Citation8 Cai et alCitation9 reported that CYP2D6*89 decreased the metabolism of bufuralol, but had no effect on the metabolism of dextromethorphan in vitro; E215K exhibited >97% decrease in catalytic activity on bufuralol and dextromethorphan.
As per our speculation, the different specificities of the substrate drug may be the main reason for this inconsistency. In another point of view, probing drugs cannot rule out some special factors in the process of metabolism of other drugs by one or two allelic variants.
In summary, we screened the enzymatic activity of the 24 variants of CYP2D6 on the metabolism of dapoxetine in vitro, especially the 22 novel isoforms. As the first report of all aforementioned alleles on dapoxetine metabolism, the research provided new information about CYP2D6 genetic polymorphism and related impact on its substrate drug dapoxetine; on the other hand, this study could help clinical assessment of the metabolic effects on dapoxetine, provide fundamental information to guide rational drug usage, and promote personalized medicine.
Acknowledgments
This work was supported by the Ministry of Health of the People’s Republic of China (201302008), the First Affiliated Hospital of Wenzhou Medical University (FHY2015010), and a grant from the National Natural Science Foundation of China (No 31371280).
Disclosure
The authors report no conflicts of interest in this work.
References
- ZhouSFLiuJPChowbayBPolymorphism of human cytochrome P450 enzymes and its clinical impactDrug Metab Rev20094128929519514967
- HaufroidVHantsonPCYP2D6 genetic polymorphisms and their relevance for poisoning due to amphetamines, opioid analgesics and antidepressantsClin Toxicol (Phila)201553650151025998998
- SusceMTMurray-CarmichaelEde LeonJResponse to hydrocodone, codeine and oxycodone in a CYP2D6 poor metabolizerProg Neuropsychopharmacol Biol Psychiatry20063071356135816631290
- OkuboMMurayamaNMiuraJChibaYYamazakiHEffects of cytochrome P450 2D6 and 3A5 genotypes and possible coadministered medicines on the metabolic clearance of antidepressant mirtazapine in Japanese patientsBiochem Pharmacol201593110410925475885
- DapoxetinePremature ejaculation: not worth the riskPrescrire Int201019105121420455331
- CaiWMNikoloffDMPanRMCYP2D6 genetic variation in healthy adults and psychiatric African-American subjects: implications for clinical practice and genetic testingPharmacogenomics J20066534335016550211
- QianJCXuXMHuGXGenetic variations of human CYP2D6 in the Chinese Han populationPharmacogenomics201314141731174324192122
- DaiDPGengPWWangSHIn vitro functional assessment of 22 newly identified CYP2D6 allelic variants in the Chinese populationBasic Clin Pharmacol Toxicol20151171394325469868
- CaiJDaiDPGengPWEffects of 22 novel CYP2D6 variants found in the Chinese population on the bufuralol and dextromethorphan metabolisms in vitroBasic Clin Pharmacol Toxicol Epub2015827
- McCartyEDinsmoreWDapoxetine: an evidence-based review of its effectiveness in treatment of premature ejaculationCore Evid2012711422315582
- OwenRTDapoxetine: a novel treatment for premature ejaculationDrugs Today (Barc)200945966967819956808
- JanniniEAIsidoriAMAversaALenziAAlthofSEWhich is first? The controversial issue of precedence in the treatment of male sexual dysfunctionsJ Sex Med201310102359236924112352
- YueFGDongLHuTTQuXYEfficacy of dapoxetine for the treatment of premature ejaculation: a meta-analysis of randomized clinical trials on intravaginal ejaculatory latency time, patient-reported outcomes, and adverse eventsUrology201585485686125817107
- JernPJohanssonAPihaJWestbergLSanttilaPAntidepressant treatment of premature ejaculation: discontinuation rates and prevalence of side effects for dapoxetine and paroxetine in a naturalistic settingInt J Impot Res2015272758025410962
- AndrewsPWThomsonJAJrAmstadterANealeMCPrimum non nocere: an evolutionary analysis of whether antidepressants do more harm than goodFront Psychol2012311722536191
- KendirciMSalemEHellstromWJDapoxetine, a novel selective serotonin transport inhibitor for the treatment of premature ejaculationTher Clin Risk Manag20073227728918360636
- ModiNBDresserMDesaiDEdgarCWesnesKDapoxetine has no pharmacokinetic or cognitive interactions with ethanol in healthy male volunteersJ Clin Pharmacol200747331532217322143
- ShenHHeMMLiuHComparative metabolic capabilities and inhibitory profiles of CYP2D6.1, CYP2D6.10, and CYP2D6.17Drug Metab Dispos20073581292130017470523
- Ingelman-SundbergMGenetic polymorphisms of cytochrome P450 2D6 (CYP2D6): clinical consequences, evolutionary aspects and functional diversityPharmacogenomics J20055161315492763
- JajaCBurkeWThummelKEdwardsKVeenstraDLCytochrome p450 enzyme polymorphism frequency in indigenous and native American populations: a systematic reviewCommunity Genet200811314114918376110
- WeberWWPopulations and genetic polymorphismsMol Diagn19994429930710671640
- GurSSikkaSCThe characterization, current medications, and promising therapeutics targets for premature ejaculationAndrology20153342444225951512
- GiulianoFClementPPharmacology for the treatment of premature ejaculationPharmacol Rev201264362164422679220
- CormioLMassenioPLa RoccaRVerzePMironeVCarrieriGThe Combination of dapoxetine and behavioral treatment provides better results than dapoxetine alone in the management of patients with lifelong premature ejaculationJ Sex Med20151271609161526077706
- HiratsukaMIn vitro assessment of the allelic variants of cytochrome P450Drug Metab Pharmacokinet2012271688422041138
- RowlandPBlaneyFESmythMGCrystal structure of human cytochrome P450 2D6J Biol Chem2006281117614762216352597
- SakuyamaKSasakiTUjiieSFunctional characterization of 17 CYP2D6 allelic variants (CYP2D6.2, 10, 14A–B, 18, 27, 36, 39, 47–51, 53–55, and 57)Drug Metab Dispos200836122460246718784265
- GuengerichFPHannaIHMartinMVGillamEMRole of glutamic acid 216 in cytochrome P450 2D6 substrate binding and catalysisBiochemistry20034251245125312564927