Abstract
Background
The incidence of microorganisms with extended-spectrum beta-lactamase (ESBL) is on the rise, posing a significant public health concern. The current application of machine learning (ML) focuses on predicting bacterial resistance to optimize antibiotic therapy. This study employs ML to forecast the occurrence of bacteria that generate ESBL and demonstrate resistance to multiple antibiotics (MDR).
Methods
Six popular ML algorithms were initially trained on antibiotic resistance test patient reports (n = 489) collected from Al-Hussein/Salt Hospital in Jordan. Trained outcome models predict ESBL and multidrug resistance profiles based on microbiological and patients’ clinical data. The results were utilized to select the optimal ML method to predict ESBL’s most associated features.
Results
Escherichia coli (E. coli, 82%) was the most commonly identified microbe generating ESBL, displaying multidrug resistance. Urinary tract infections (UTIs) constituted the most frequently observed clinical diagnosis (68.7%). Classification and Regression Trees (CART) and Random Forest (RF) classifiers emerged as the most effective algorithms. The relevant features associated with the emergence of ESBL include age and different classes of antibiotics, including cefuroxime, ceftazidime, cefepime, trimethoprim/ sulfamethoxazole, ciprofloxacin, and gentamicin. Fosfomycin nitrofurantoin, piperacillin/tazobactam, along with amikacin, meropenem, and imipenem, had a pronounced inverse relationship with the ESBL class.
Conclusion
CART and RF-based ML algorithms can be employed to predict the most important features of ESBL. The significance of monitoring trends in ESBL infections is emphasized to facilitate the administration of appropriate antibiotic therapy.
Introduction
The emergence of antibiotic-resistant microbes (AMR) has become a significant public health concern globally, impacting various domains including development, animal welfare, and food security. Consequently, there is an approximate annual incidence of 1.27 million sickness cases and 929,000 reported deaths globally.Citation1 According to the World Health Organization (WHO), several Enterobacteriaceae (including Escherichia coli, Klebsiella pneumoniae, Proteus mirabilis, and other species of Enterobacter) have been identified as the most perilous AMR bacteria in relation to their effects on human health and the healthcare system.Citation2 These bacteria are recognized for their ability to cause infections acquired both within hospital (nosocomial) and community settings. β-lactam antibiotics are commonly employed in the management of infections caused by the aforementioned bacterial species.
The continued utilization of β-lactam plays a substantial role in the persistent development of resistance to numerous categories of these medications.Citation3 Consequently, microbial resistance has expanded to encompass newly developed β-lactams, such as cefepime.Citation4 The principal means through which bacteria acquire resistance to β-lactam antibiotics is through the creation of beta-lactamases, particularly the ESBLs.Citation5 The CDC has recognized ESBL-producing Enterobacteriaceae as significant challenges to healthcare systems, given their rising prevalence and consequential effects on clinical and economic aspects.Citation6 Moreover, multidrug resistance (MDR) has increased in ESBL-producing Enterobacteriaceae.Citation7 In fact, increased use of non-β-lactam classes such as carbapenems and fluoroquinolone in response to ESBL and other resistant infections has led to the emergence of MDR.Citation3
ESBLs are commonly acknowledged as acquired β-lactamases, primarily encoded by genes that are typically situated on plasmids. The dissemination of resistance genes carried by ESBL-producing bacteria occurs via horizontal transmission.Citation5 Bacteria with numerous resistance genes exhibit an MDR phenotype.Citation8 Additionally, MDR genes can co-occur on plasmids and chromosomes through a co-selection mechanism.Citation9 MDR may also arise if a single resistance mechanism confers tolerance to multiple antibiotics.Citation8
The literature broadly acknowledges the causal relationship between the creation of ESBLs and the inappropriate usage of antibiotics. However, there remain a multitude of unsolved issues pertaining to this occurrence. Significant knowledge can be obtained from readily available data sources, such as data concerning bacteria that exhibit resistance, including ESBL profiling. The integration of electronic health records with mathematical models enables the development of accurate predictions that elucidate the diverse aspects of AMR. To address the spread of ESBLs, it is possible to utilize insights obtained from prognostications to inform public health initiatives.
The application of Machine Learning (ML) algorithms in the context of antibiotic resistance shows potential for generating precise predictions that shed light on various facets of the issue.Citation10 For instance, Chen et al used Random Forest (RF) and cross-correlation algorithms to predict MDR microorganisms at the hospital level.Citation11 Cánovas-Segura and Moran et al employed ML to create a clinical decision and antibiotic selection system for antibiotic management in a hospital.Citation12,Citation13 Moreover, incorporating ML algorithms into systems frequently used for microbiology screening and detection of pathogenic bacteria, such as flow cytometry and mass spectrometry technologies, has the potential to expedite antimicrobial resistance diagnosis.Citation14–16 Despite their limitations, previous studies demonstrate the robust capacity of ML algorithms to enhance the effective use of medical data for evidence-based decision-making by identifying relevant rules and generating predictions with greater precision than those made by humans in real-life contexts.
Enhancing comprehension of the primary determinants influencing the dissemination of ESBL can be achieved by incorporating multiple parameters into the forecasting process. These parameters encompass the socio-demographic characteristics of patients and the various sources of medical isolates. These parameters contribute to the predictive accuracy of the model and exhibit diverse epidemiological and etiological impacts that are comparable to the selection pressure imposed by antibiotics on prevalent clinical bacteria. This is primarily due to the fact that bacterial pathogens engage in interactions with hosts that exhibit various levels of resistance to infection. One example of this phenomenon is the decline in natural immunity to infection as individuals age.
This study employed ML analysis to forecast the occurrence of bacteria producing ESBL and multidrug resistance against frequently prescribed antibiotics in the Jordanian Al-Salt hospital. The primary goal is to design a computerized system to predict ESBL based on analyzing a dataset comprising ESBL laboratory profiling. The proposed pipeline includes six popular ML algorithms trained on patient records and antibiotic resistance measures. The trained outcome model predicts ESBL and resistance profiles, guiding the selection of the optimal ML method for predicting ESBL distribution.
Methodology
The Process of Gathering and Validating Data
We conducted a retrospective analysis of ESBL data related to patients treated at Al-Hussein/Salt Hospital, the sole healthcare facility in Al-Salt, Jordan, involving 180,090 individuals. Within this population, 51% are male, and 49% are female.
The antibiotic susceptibility data were acquired by cultivating bacteria for identification purposes. Subsequently, these bacteria underwent testing with routinely employed antibiotics using the VITEK 2 system. The duration of this operation may extend to a maximum of 72 hours or even longer. The acquisition of this information is crucial for the effective management of bacterial infections. The minimum inhibitory concentration (MIC) of an antibiotic, necessary to effectively impede the growth or eradicate a pathogenic organism within a controlled laboratory setting, is typically communicated to medical practitioners, along with predetermined thresholds for medicine susceptibility, categorizing the pathogen as either resistant or susceptible.
The allocation of data is presently being carried out within the Jordanian electronic health record initiative, known as Hakeem, which serves as a platform for managing and reporting health-related information in Jordan.
Throughout the period from January 20, 2021, to December 31, 2022, a single sample was collected from each patient exhibiting symptoms related to infection, in addition to the samples obtained for screening tests. Hence, the collected data exhibit representativeness across all patients in a consecutive manner, devoid of any specific group selection. The original dataset comprises a total of 2893 patient records of microbiology tests. However, this study included only those records with ESBL profiling data (n=489). The clinical and demographic features included in the analysis are bacterial species, sex (female or male), age (in years), organism quantity (categorized in an ordered manner), diagnosis, Gram staining (positive or negative), antimicrobial substances, date of sample collection, and source of the clinical sample.
In the original dataset, more than 14 different types of bacterial infections have been diagnosed over the study period; however, this study includes only the species being routinely tested for ESBL phenotype, comprising Escherichia coli, Klebsiella pneumoniae, Proteus mirabilis, and other species of Enterobacter. Additionally, as a result of incomplete medical records, the original dataset was resampled to exclusively contain records with complete ESBL profile data points (n=489).
The generation of classes in the dataset was extensive. The classes in question generated a larger amount of unbalanced data and yielded a higher number of instances, as seen by their sample size. In order to address the issue at hand, we implemented a strategy where we consolidated “similar sources” into a single class, thereby reducing the overall number of classes. As an illustration, the anatomical components of fingers, hands, and arms are collectively classified under the category of arms.
Prediction Algorithms
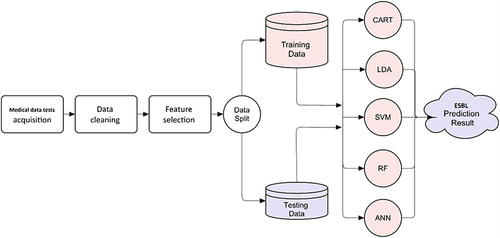
The primary goal of this work is to design a computerized system to predict ESBL based on a number of variables for each patient test (see ). The following stages comprise the prediction system: data acquisition, data cleaning and preprocessing, feature selection, model training, and model testing, as depicted in the graphical abstract.
Table 1 Statistical Characteristics of the Collected Antibiotic Dataset. Normal Factorization is Used for Categories
Feature Selection
In this study, we employed two feature selection methods: Feature correlationCitation17 and feature importance ranking (FIR).Citation18 A cross-analysis of the best qualities of both methods was conducted, assuming that a correlation of more than 10% is considered a strong correlation and considering only the top 50% of the ranked features. We selected only the features that are considered strong by both methods.
ML Model Training
We trained and tested six classifiers on the data using 5-fold cross-validation. Each of these ML approaches is a member of a distinct classifier family. Several experiments were conducted on the cleaned data to determine the best ML model for the proposed ESBL prediction system. The resultant models of CART, LDA, SVM, KNN, RF, and ANN were among the models explored in those experiments. CART uses decision trees for classification,Citation19 while LDA focuses on linear decision boundaries.Citation20 Support vector machine (SVM) employs support vectors to separate classes,Citation21 RF uses an ensemble of decision trees, Artificial neural network (ANN) mimics the structure and function of biological neural networks,Citation22 and k-nearest neighbor (k-NN) determines the class based on the feature space’s closest neighbors.Citation23 The implementation of the previous ML approach was developed using the CARET library developed under the R programming language.Citation24
This study intends to thoroughly analyze and compare the performance of several classifiers in the ESBL prediction system to find the best alternative for the proposed system. Because our results revealed two decision-based classifiers as the best performers, allowing further exploration of decision-based ML approaches, we added two additional classifiers, the Hoeffding Tree (HT)Citation25 and the Naive Bayes decision tree (NBTree).Citation26
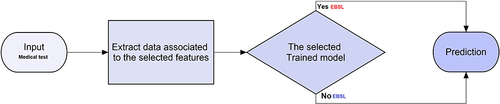
This study utilized default parameters for each classifier, which are pre-defined settings established by ML libraries or algorithm authors. These parameters are used to ensure fair comparisons between different classifiers, avoiding potential biases caused by manual parameter adjustment. However, default parameters may not always yield the best results for a specific dataset or situation. In practice, parameters are fine-tuned based on data characteristics and prediction system goals. In this study, default values were used to provide baseline performance for each classifier, avoiding potential biases caused by manual parameter adjustment. The system, after configuration, selecting the optimal feature set, and determining the optimal ML approach, is ready to predict whether a new medical test bacteria will produce ESBL as illustrated in .
Data Imbalance, Encoding, and Performance Evaluation
Utilizing 5-fold validation, the study determined the best-performing algorithm based on average accuracy. However, the dataset is class-imbalanced, with 291 (59.6%) positive EBSL instances and 198 (40.4%) negative EBSL instances. To assess the true efficacy of classifiers despite class imbalance, the F1-score and area under the curve (AUC) were used. Additionally, accuracy, precision, and recall metrics were applied, along with the factorized encoding method. Subsequently, a final experiment was conducted to identify the best-performing algorithm using the superior encoding method.
Results
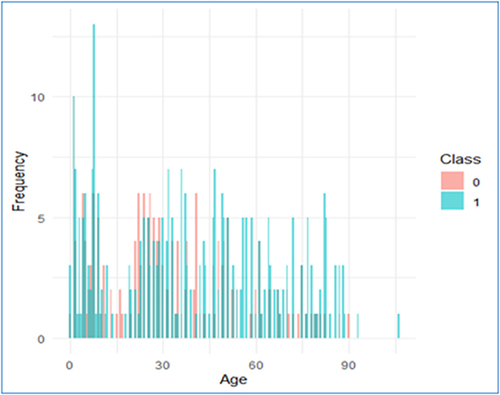
The study patients’ characteristics are shown in . The distribution of ages is shown in ranged from 106 years to less than a week. Notably, a total of 260 patients had ages greater than the sample mean, which came out to be 35.99 years old. The bulk of the patients who were impacted were female, comprising 77% (n=381). The most commonly observed diagnosis among these patients was UTI, which accounted for 68.7% (n=336) of the cases. The bacteria tested included Escherichia coli (E. coli), Klebsiella pneumoniae, Proteus mirabilis, and other species of Enterobacter. E. coli bacteria were detected in a total of 401 individuals, accounting for 82% of the patient population.
Figure 2 The distribution of the age variable over both classes, Minimum=7 days, Maximum= 106 years, Class= ESBL (0, 1). n= 489.
Two feature selection methods were employed to ascertain the key aspects in predicting the production of ESBL by bacteria. The correlation coefficients shed light on the relationship that exists between each feature and the target variable. Features with stronger correlations (either positive or negative) are likely to be more relevant for predicting the outcome when selecting features for an ESBL prediction automation system. The correlation test shown in and , suggests that cefuroxime, ceftazidime, and cefepime have the strongest positive correlation with correlation coefficients of 0.904, 0.767, and 0.514, respectively. However, it is worth noting that correlation does not indicate causation, and that other factors may influence prediction accuracy. As a result, for selecting features in the ESBL prediction system, we opted for a more robust method–the importance ranking of features using RF based on the Mean Decrease Gini index, as illustrated in .
Table 2 The Correlation Coefficient of Each Feature with the Class (ESBL)
Figure 3 Feature selection for an ESBL prediction automation system. (A) Heatmap of correlation coefficients between features and class (ESBL). Bluer colors indicate stronger positive correlations and redder colors indicate stronger negative correlations. (B) Feature importance ranking using Random Forest, based on the Mean Decrease Gini index. The features are sorted by importance, and the top features are visualized.
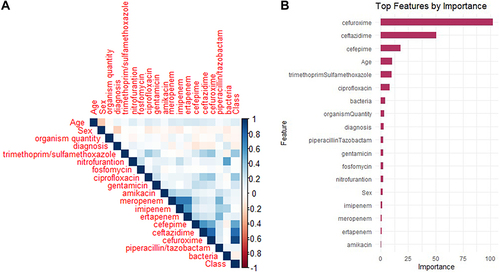
As can be seen from the cross-analysis shown in , we can select a set of strong features (SSF) with full agreement using both selection methods. These features include cefuroxime, ceftazidime, cefepime, trimethoprim/sulfamethoxazole, ciprofloxacin, gentamicin, and age. We can also identify the weakest features set (WFS), which includes features that were never selected by any method including piperacillin-tazobactam, amikacin, meropenem, imipenem, nitrofurantoin, fosfomycin, diagnosis, sex, organism quantity, and bacteria type.
Table 3 Cross-Analysis of the Best Feature Qualities of Both Feature Selection Methods in Both Approaches (Correlation and Feature Ranking), (✓) Means the Feature is Selected by the Method, and (×) Otherwise
The accuracy results of the first set of trials are presented in the supplementary table (S1) and . The trials were conducted utilizing all the features listed in . Upon examination of the data presented in , it becomes evident that the performance of the K-nearest neighbors (KNN) classifier was unsatisfactory. This can be attributed to the categorical nature of the data, which was encoded using a standard factorization method (1, 2, 3, …) for each category. However, it is important to note that calculating the mathematical distance, specifically the Euclidean distance in this case, does not necessarily capture the true similarity between these values. For instance, the distance between categories 1 and 2 is smaller than that between categories 1 and 5.
Table 4 ESBL Prediction Results Using 5-Fold Cross-Validation
Figure 4 A visual comparison of classifier performance across five runs. Box and whisker plots depict the accuracy range and KAPPA plots.
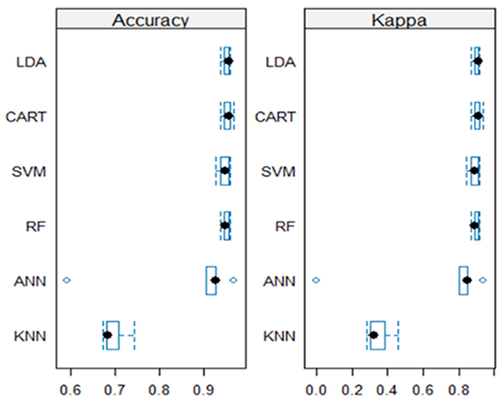
The LDA and the decision tree-based classifiers, namely CART and RF outperform the other classifiers. CART has an accuracy range of 0.938 to 0.969, suggesting constant good performance. Similarly, RF achieves accuracy between 0.938 and 0.960, demonstrating high overall performance equivalent to RF. This result is confirmed further by , in which the accuracy boxes for both classifiers are positioned towards the highest accuracy values, indicating narrower widths. This implies improved consistency and stability over the five runs. Furthermore, the KAPPA plots reveal that the mean for each classifier is relatively high, 0.905 and 0.898 for CART and RF respectively. The supplementary material (Table S2) show the results of a statistical test for pairwise differences in accuracy between different classifiers.
Assuming a 95% confidence interval, there are no statistically significant differences in accuracy observed between the classifiers in most cases, with the following exceptions: RF and KNN (p-value = 0.0005<0.05), CART and KNN (p-value = 0.0003<0.05), LDA and KNN (p-value = 0.0003<0.05), and KNN and SVM (p-value = 0.0011<0.05), indicating a significant difference in accuracy between these classifiers. These findings support the claims that CART and RF are the top performers, in addition to LDA. However, having two decision-based classifiers among the best performers on our data allows for further investigation of decision-based ML approaches, such as the Hoeffding Tree (HT) and naive Bayes decision tree (NBTree). This investigation incorporates more evaluation metrics, including precision, recall, F-score (F1), and ROC area under curve (AUC), in order to select the best approach for our proposed system. illustrates these results.
The decision tree-based classifiers exhibited strong performance across all evaluated measures. Furthermore, there was no discernible variation in performance among these classifiers. Once again, the CART model demonstrated superior performance in terms of accuracy, precision, recall, and F-score. On the other hand, the RF model outperformed others in terms of the AUC metric, obtaining a value of 0.976. There are two distinct encoding procedures that can be employed. In certain cases, these strategies provide comparable outcomes. Consequently, it is advisable to adhere to factorized encoding due to its ability to maintain a smaller training model size in comparison to the utilization of one-hot encoding. The RF algorithm continues to maintain its superiority in terms of F-score. The reason behind this high performance of the decision tree-based classifiers, particularly CART, lies in the output of the trained model, which is a decision tree. As evident from the output decision tree of CART illustrated in , this can be translated into only two rules:
Figure 5 The output decision tree of CART using all data (“<=0” represents = 0, and “>0” represents 1; because the values of cefuroxime are binary, zeros and ones).
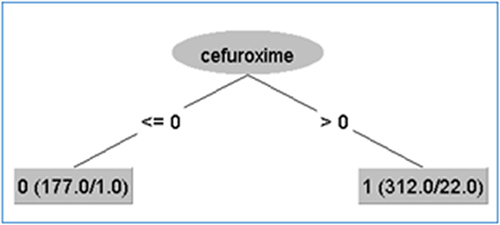
1- If cefuroxime=0 then ESBL=0, ie there is no ESBL produced by the bacteria in the tested sample. This rule is supposed to be true for 177 samples, however, one of them was erroneously identified as ESBL.
2- If cefuroxime=1 then ESBL=1, ie there is an ESBL produced by the bacteria in the tested sample. This rule is supposed to be true for 312 samples, however, 22 of them were erroneously identified as ESBL.
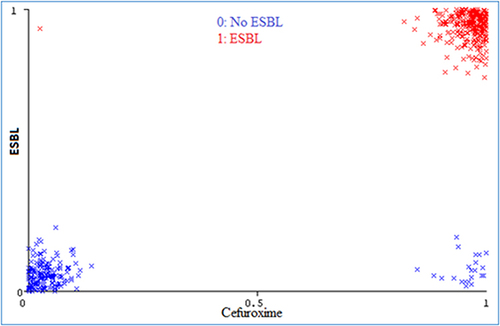
Surprisingly, the decision tree was constructed using only cefuroxime, which resulted in the best performance for the ESBL prediction system. This feature was identified in feature selection analysis as the most significant among all features in the dataset (see , and ), as it highly correlates with the existence/absence of ESBL. This is also evident from the distribution of cefuroxime values across both classes, as shown in .
Figure 6 Visualization of the data points for cefuroxime. All values are either 0 or 1, but noise is added to data for visualization purposes.
The previous results, thus far, are based on the ESBL dataset using all its features after cleaning the data by removing all examples with missing data and/or all features with a high percentage of missing data. In order to obtain better prediction results, we conducted another set of experiments using the winner classifiers CART and RF on a different set of features, including SSF, to improve results, and the WFS to justify omitting such weak features. shows the results of these experiments.
Table 5 CART and RF Prediction Results on the Strongest and Weakest Subsets of Features
As indicated in , the performance of the CART algorithm remains the highest. However, no improvement or reduction is observed when utilizing the strongest features. This outcome is anticipated since the resulting decision tree is constructed solely based on the strongest feature, cefuroxime, as depicted in . Consequently, even if the experiment were conducted using only this feature, the results would remain unchanged. The performance of the RF is negatively affected when it is solely applied to the most prominent features. This is due to the nature of RF, which utilizes an ensemble approach by constructing multiple small decision trees to create the trained model. It is possible that some of these decision trees do not incorporate the strongest feature (Cefuroxime), leading to a decrease in accuracy from 0.941 to 0.926, as well as a decline in other metrics as presented in . In contrast, the classifiers that have been deemed the most effective demonstrate an inability to accurately predict ESBL when relying solely on the least informative features. In fact, the prediction outcomes occasionally fall below random chance, and at best, achieve a success rate of approximately 50%. This suggests a significant inconsistency within the data provided by these particular features.
Based on the aforementioned results, it is advisable to incorporate a comprehensive set of features in the proposed ESBL prediction system. Specifically, if the CART classifier is utilized, it is recommended to include the most influential feature, cefuroxime. Alternatively, when employing the RF classifier, it is suggested to incorporate all the available feature as given in .
Discussion
Predicting the most important features of ESBL can be achieved using ML algorithms based on medical records, even when patient data is limited. Previous studies exhibited a deficiency in diverse datasets, which failed to encompass patients of varying demographics, clinical history, and antibiotic regimens. These features significantly impede the advancement of prediction systems. Prediction models were built in this study, which yielded correct outcomes even in the presence of data heterogeneity. The inclusion of a larger sample size would have yielded enhanced statistical power for the predictive model. However, this analysis incorporated a relatively bigger patient cohort compared to the majority of previous studies that investigated ESBL-producing bacteria patterns.
In our trials, the LDA and the decision tree-based classifiers, namely CART and RF, outperformed the other classifiers. On the other hand, the performance of the KNN classifier was unsatisfactory. This can be attributed to the categorical nature of the data, which was encoded using a standard factorization method (1, 2, 3, …) for each category. We might improve the performance of KNN by using another type of encoding, such as one-hot encoding,Citation27 and other distance measures, such as Hasanat distance, which was proven to be unaffected by outliers and data noise.Citation28 However, such improvement is beyond the scope of this manuscript.
AUC is a statistic often employed to assess a classifier’s overall performance in binary classification situations. Meanwhile, the F1 score is a measure that combines precision and recall, which are crucial for evaluating a classifier’s performance in cases involving class imbalance or where both false positives and false negatives are critical. The F1 score is especially beneficial when attempting to achieve a compromise between precision and recall because it provides a single metric that takes both into account. As a result, considering the classifier’s performance on imbalanced data, even when it is slightly imbalanced as in our case, and recognizing the importance in a medical application where administrators must trade-off between false positives and false negatives, the F1 score emerges as a suitable metric for classifier comparisons in our case.Citation29 Therefore, CART is identified as the best classifier for our ESBL prediction system.
The identification of key features is crucial in elucidating the underlying causes of antimicrobial resistance associated with ESBL synthesis. Nevertheless, it is important to note that the risk variables associated with resistance phenotypes should not be interpreted as causal. This is because the models utilized were designed to predict antimicrobial resistance rather than estimate causal effects. Genome-centric investigations are necessary to enhance comprehension of the determinants influencing the emergence and progression of ESBL development. In this study, we used two feature selection methods: feature correlation and feature importance ranking using RF. These methods helped us build a more accurate and efficient model for predicting ESBL production in bacteria. We used cross-analysis of the best features across both approaches (correlation and feature ranking) which entails investigating the chosen features and their usefulness in predicting ESBL using our data. It aids in assessing the consistency and reliability of the features chosen across different methodologies, as well as identifying features with good predictive value across numerous methods.Citation27,Citation30
With a 95% confidence interval, our analysis reveals that in most cases, there are no statistically significant differences in accuracy observed between the classifiers. However, exceptions are noted: RF and KNN (p-value = 0.0005<0.05), CART and KNN (p-value = 0.0003<0.05), LDA and KNN (p-value = 0.0003<0.05), and KNN and SVM (p-value = 0.0011<0.05). These instances indicate a significant difference in accuracy between these specific pairs of classifiers. Accordingly, these findings confirm the assertions that CART and RF are the leading performers, alongside LDA.
The features deemed pertinent to the occurrence of ESBL in this study encompassed cephalosporin medicines, specifically cefuroxime (a second-generation cephalosporin), ceftazidime (a third-generation cephalosporin), and cefepime (the most recent addition to the fourth generation of cephalosporins). ESBLs have been found to reduce the effectiveness of extended-spectrum cephalosporins.Citation3 Following this, a new class of β-lactam antibiotics with expanded-spectrum properties was newly introduced, rendering them resistant to hydrolysis by ESBL enzymes. The oxyimino-cephalosporins, notably ceftazidime, gained significant popularity and widespread utilization.Citation31 However, the emergence of novel β-lactamases capable of hydrolyzing these recently developed medicines occurred. This property is ascribed to numerous ESBL enzymes.Citation3,Citation4,Citation31 One illustrative instance involves the characterization of oxacillinase enzymes, which have been classified as possessing extended-spectrum activity. The substrates of these enzymes encompass early cephalosporins as well as third- and/or fourth-generation cephalosporins.Citation32
The ESBL prediction method under consideration demonstrates the capability to utilize a variety of feature sets, encompassing both clinical and demographic factors. Specifically, when employing the CART classifier, the inclusion of the most influential feature, cefuroxime, is crucial. Alternatively, while utilizing the RF classifier, it is advisable to incorporate all accessible features including cefuroxime. The clinical significance of this discovery lies in the potential use of cefuroxime as an initial laboratory diagnostic tool for detecting ESBL-positive bacteria. Although the Clinical and Laboratory Standards Institute (CLSI) offers guidelines for the detection of ESBLs, it is important to note that these guidelines are based on the general assumption that the addition of clavulanic acid enhances the efficacy of the laboratory diagnosis.Citation33
Our investigation identified UTIs as the primary source of infection. The oral therapy options for UTIs due to ESBL isolates are limited, with trimethoprim-sulfamethoxazole being one of the drugs available.Citation34 There has been an observed increase in the prevalence of UTIs caused by bacteria that produce ESBLs, leading to the need for the implementation of carbapenems as a broad treatment approach.Citation35 Nevertheless, it was anticipated that this medication would exhibit a limited degree of antibacterial efficacy against ESBL isolates within this cohort. The results of our investigation are consistent with previous research, such as the study conducted by Schwaber et al, which provides evidence that trimethoprim-sulfamethoxazole exhibits diminished effectiveness against ESBL isolates.Citation36 This discovery highlights the challenges involved with managing patients in our specific geographical area who are afflicted with UTIs caused by ESBL bacteria.
Ciprofloxacin, a fluoroquinolone, and gentamycin have been identified as notable predictors of ESBL in the models employed. The obtained results exhibit resemblances to prior research conducted among patients from different locations.Citation37–39 Gentamicin is frequently utilized as the main antibacterial antibiotic in the management of UTIs. Ciprofloxacin is a widely utilized antimicrobial agent with broad-spectrum activity, employed for the treatment of many bacterial illnesses. The increased resistance demonstrated by ESBL isolates against gentamicin and ciprofloxacin is a matter of concern due to its potential impact on limiting the range of possible treatment options.
The outcome of the ML models revealed a significant association between the age of the patients and the presence of ESBL. The present findings align with the conclusions drawn in earlier conducted studies.Citation40,Citation41 We found a noteworthy trend wherein a significant proportion of ESBL isolates are derived from patients diagnosed with UTIs. This observation, coupled with the well-established association between contracting a UTI caused by E. coli and a decline in immune function with advancing age, offers a plausible explanation for our findings. Specifically, our results indicate that age plays a pivotal role in predicting the emergence of the ESBL phenotype in the context of the antibiotics commonly employed for UTI treatment. The significance of age suggests that elderly individuals with pre-existing health conditions remain susceptible to the impact of resistant pathogens. Nevertheless, the correlation between age and ESBL is still unclear. The observed pattern has been ascribed by certain researchers to the mechanism of action of antibiotics.Citation42 Research conducted on the population of Jordan indicates that age is a factor that exhibits variability in antibiotic utilization.Citation43 This variability is believed to have a direct impact on the emergence of ESBLs and the development of MDR. The analysis of ESBL patterns in relation to age within a certain region can potentially enhance treatment efficacy, leading to improved cure rates. This study represents the inaugural investigation that elucidates the incidence of ESBL in relation to age.
Based on the results of correlation analysis and feature ranking, it is suggested that amikacin, meropenem, and imipenem, along with a piperacillin-tazobactam combination could serve as important treatment alternatives for individuals suffering from UTIs and bladder infections caused by ESBL-producing bacteria. Specifically, the combination of piperacillin-tazobactam which is a potent antimicrobial agent that exhibits a wide range of activity against both Gram-positive and Gram-negative bacteria, including aerobic and anaerobic strains. This is achieved by its dual mechanism of action as a beta-lactam antibiotic and a beta-lactamase inhibitor.Citation44 Moreover, prior research has established a notable efficacy of amikacin and meropenem therapy in the treatment of UTIs caused by ESBL bacteria.Citation45 It was shown that fosfomycin and nitrofurantoin, both non-β-lactam agents, exhibited the most pronounced inverse relationship with ESBL. Therefore, they have the potential to demonstrate great efficacy against ESBL-producing bacteria.
Conclusion
We endeavored to address the inquiry regarding the important features of bacteria that produce ESBL and their multidrug resistance to commonly prescribed antibiotics through the utilization of ML models and observational data. One advantage of our methodology is the incorporation of widely used ML models. We have developed an optimal ESBL prediction system utilizing the CART classifier. This system incorporates a comprehensive collection of features, with a particular emphasis on the strongest feature, cefuroxime. Additionally, we have implemented the RF classifier, which encompasses all features examined in this study. The application of these models enabled us to assess the efficacy of commonly used antibiotics for the treatment of ESBL infections. When ESBL infections are discovered, cefuroxime, ceftazidime, cefepime, trimethoprim/sulfamethoxazole, ciprofloxacin, and gentamicin may not be the best medications. Conversely, in the context of Jordanian patients, amikacin, meropenem, imipenem, piperacillin/tazobactam, and fosfomycin nitrofurantoin are suitable options. This information is crucial for informing public health initiative about appropriate antibiotic therapy.
Ethical Approval
This study complies with the Declaration of Helsinki; involved the analysis of retrospective data. Prior to analysis, all patient information was anonymized and de-identified, then transferred to Excel files. These files used unique study-specific patient numbers to maintain confidentiality, ensuring that any linkage to patient names, medical record numbers, or other personally identifiable information was securely protected. This study was approved by the Administration and Scientific Research at Al-Balqa Applied University and Jordan’s Al-Hussein/Salt Hospital and waived the requirement for informed consent. Furthermore, the compliance officer of the Al-Hussein/Salt Hospital authorized the data exchange and file transfer procedures; Approval number: 35/4/1205.
Disclosure
The authors report no conflicts of interest in this work.
Acknowledgments
We thank Jordan’s AL-Hussein/Salt Hospital for allowing access to medical records from the microbiology laboratory section.
Additional information
Funding
References
- de Kraker MEA, Stewardson AJ, Harbarth S. Will 10 million people die a year due to antimicrobial resistance by 2050? PLoS Med. 2016;13(11):e1002184. doi:10.1371/journal.pmed.1002184
- WHO global priority pathogens list of antibiotic-resistant bacteria - Combat AMR. Available from: https://www.combatamr.org.au/news-events/who-global-priority-pathogens-list-of-antibiotic-resistant-bacteria. Accessed August 24, 2023.
- Castanheira M, Simner PJ, Bradford PA. Extended-spectrum β-lactamases: an update on their characteristics, epidemiology and detection. JAC-Antimicrob Resist. 2021;3(3):dlab092. doi:10.1093/jacamr/dlab092
- Patel HB, Lusk KA, Cota JM. The role of cefepime in the treatment of extended-spectrum beta-lactamase infections. J Pharm Pract. 2019;32(4):458–463. doi:10.1177/0897190017743134
- Gniadkowski M. Evolution and epidemiology of extended-spectrum β-lactamases (ESBLs) and ESBL-producing microorganisms. Clin Microbiol Infect. 2001;7(11):597–608. doi:10.1046/j.1198-743x.2001.00330.x
- ESBL-producing Enterobacterales | HAI | CDC; 2021. Available from: https://www.cdc.gov/hai/organisms/ESBL.html. Accessed October 16, 2023.
- Chen Y, Liu Z, Zhang Y, Zhang Z, Lei L, Xia Z. Increasing prevalence of esbl-producing multidrug resistance Escherichia coli from diseased pets in Beijing, china from 2012 to 2017. Front Microbiol. 2019;10:2852. doi:10.3389/fmicb.2019.02852
- Nikaido H. Multidrug Resistance in Bacteria. Annu Rev Biochem. 2009;78(1):119–146. doi:10.1146/annurev.biochem.78.082907.145923
- Bush K, Jacoby GA. Updated functional classification of β-lactamases. Antimicrob Agents Chemother. 2010;54(3):969–976. doi:10.1128/AAC.01009-09
- Al-khlifeh EM, Hassanat AB. Predicting the distribution patterns of antibiotic-resistant microorganisms in the context of Jordanian cases using machine learning techniques. J Appl Pharm Sci. 2024;14(6):174–183. doi:10.7324/JAPS.2024.177584
- Chen Y, Chen X, Liang Z, et al. Epidemiology and prediction of multidrug-resistant bacteria based on hospital level. J Glob Antimicrob Resist. 2022;29:155–162. doi:10.1016/j.jgar.2022.03.003
- Cánovas-Segura B, Campos M, Morales A, Juarez JM, Palacios F. Development of a clinical decision support system for antibiotic management in a hospital environment. Prog Artif Intell. 2016;5(3):181–197. doi:10.1007/s13748-016-0089-x
- Moran E, Robinson E, Green C, Keeling M, Collyer B. Towards personalized guidelines: using machine-learning algorithms to guide antimicrobial selection. J Antimicrob Chemother. 2020;75(9):2677–2680. doi:10.1093/jac/dkaa222
- Feucherolles M, Nennig M, Becker SL, et al. Combination of MALDI-TOF mass spectrometry and machine learning for rapid antimicrobial resistance screening: the case of campylobacter spp. Front Microbiol. 2022;12:804484. doi:10.3389/fmicb.2021.804484
- Lechowicz L, Urbaniak M, Adamus-Białek W, Kaca W. The use of infrared spectroscopy and artificial neural networks for detection of uropathogenic Escherichia coli strains’ susceptibility to cephalothin. Acta Biochim Pol. 2013;60(4):713–718.
- Faron ML, Buchan BW, Hyke J, et al. Multicenter evaluation of the bruker maldi biotyper CA system for the identification of clinical aerobic Gram-negative bacterial isolates. PLoS One. 2015;10(11):e0141350. doi:10.1371/journal.pone.0141350
- Huang L, Tang J, Chen S, Ding C, Luo B. an efficient algorithm for feature selection with feature correlation. In: Yang J, Fang F, Sun C editors. Intelligent Science and Intelligent Data Engineering. Lecture Notes in Computer Science. Springer; 2013:639–646. doi:10.1007/978-3-642-36669-7_78.
- Wojtas MA, Chen K Feature importance ranking for deep learning. In: Proceedings of the 34th International Conference on Neural Information Processing Systems. NIPS’20. Curran Associates Inc.; 2020:5105–5114.
- Krzywinski M, Altman N. Classification and regression trees. Nature Methods. 2017;14(8):757–758. doi:10.1038/nmeth.4370
- Izenman AJ. Linear discriminant analysis. In: Izenman AJ editor. Modern Multivariate Statistical Techniques: Regression, Classification, and Manifold Learning. Springer Texts in Statistics. Springer; 2008:237–280. doi:10.1007/978-0-387-78189-1_8.
- Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20(3):273–297. doi:10.1007/BF00994018
- Tian Y, Shu M, Jia Q. Artificial neural network. In: Daya sagar BS, Cheng Q, McKinley J, Agterberg F editors. Encyclopedia of Mathematical Geosciences. Encyclopedia of Earth Sciences Series. Springer International Publishing; 2020:1–4. doi:10.1007/978-3-030-26050-7_44-1.
- Mucherino A, Papajorgji PJ, Pardalos PM. k-nearest neighbor classification. In: Mucherino A, Papajorgji PJ, Pardalos PM editors. Data Mining in Agriculture. Springer Optimization and Its Applications. Springer; 2009:83–106. doi:10.1007/978-0-387-88615-2_4.
- Max K. Building predictive models in r using the caret package. J Stat Softw. 2008;28. doi:10.18637/jss.v028.i05
- Muallem A, Shetty S, Pan JW, Zhao J, Biswal B. hoeffding tree algorithms for anomaly detection in streaming datasets: a survey. JIS. 2017;08(04):339–361. doi:10.4236/jis.2017.84022
- Wang LM, Li XL, Cao CH, Yuan SM. Combining decision tree and naive bayes for classification. Knowledge-Based Syst. 2006;19(7):511–515. doi:10.1016/j.knosys.2005.10.013
- Alkhawaldeh I, Al-Jafari M, Abdelgalil M, Tarawneh A, Hassanat A. A machine learning approach for predicting bone metastases and its three-month prognostic risk factors in hepatocellular carcinoma patients using SEER data. Ann Oncol. 2023;34:140. doi:10.1016/j.annonc.2023.04.414
- Hassanat AB, Tarawneh AS, Abed SS, Altarawneh GA, Alrashidi M, Alghamdi M. RDPVR: random data partitioning with voting rule for machine learning from class-imbalanced datasets. Electronics. 2022;11(2):228. doi:10.3390/electronics11020228
- Hassanat A, Altarawneh G, Alkhawaldeh IM, et al. The jeopardy of learning from over-sampled class-imbalanced medical datasets. In: 2023 IEEE Symposium on Computers and Communications (ISCC).; 2023:1–7. doi:10.1109/ISCC58397.2023.10218211.
- Hassanat A, Alkafaween E, Tarawneh AS, Elmougy S Applications review of hassanat distance metric. In: 2022 International Conference on Emerging Trends in Computing and Engineering Applications (ETCEA).; 2022:1–6. doi:10.1109/ETCEA57049.2022.10009844.
- Paterson DL, Bonomo RA. Extended-Spectrum β-Lactamases: a Clinical Update. Clin Microbiol Rev. 2005;18(4):657–686. doi:10.1128/CMR.18.4.657-686.2005
- Yoon EJ, Jeong SH. Class D β-lactamases. J Antimicrob Chemother. 2021;76(4):836–864. doi:10.1093/jac/dkaa513
- Guideline: Wayne, PA: clinical and Laboratory Standards. - Google Scholar. Available from: https://scholar.google.com/scholar_lookup?title=Wayne+PA:+Clinical+and+Laboratory+Standards+Institute&publication_year=2009&. Accessed October 17, 2023.
- Karaiskos I, Giamarellou H. Carbapenem-sparing strategies for ESBL producers: when and how. Antibiotics. 2020;9(2):61. doi:10.3390/antibiotics9020061
- Shi HJ, Wee JH, Eom JS. Challenges to early discharge of patients with upper urinary tract infections by esbl producers: tmp/smx as a step-down therapy for shorter hospitalization and lower costs. Infect Drug Resist. 2021;14:3589–3597. doi:10.2147/IDR.S321888
- Schober P, Vetter TR. Logistic regression in medical research. Anesth Analg. 2021;132(2):365–366. doi:10.1213/ANE.0000000000005247
- Mwakyoma AA, Kidenya BR, Minja CA, et al. Allele distribution and phenotypic resistance to ciprofloxacin and gentamicin among extended-spectrum β-lactamase-producing Escherichia coli isolated from the urine, stool, animals, and environments of patients with presumptive urinary tract infection in Tanzania. Front Antibiot. 2023;2:1164016
- Johnson TJ, Hargreaves M, Shaw K, et al. Complete genome sequence of a carbapenem-resistant extraintestinal pathogenic Escherichia coli strain belonging to the sequence type 131 h30r subclade. Genome Announc. 2015;3(2). doi:10.1128/genomea.00272-15
- Wiener ES, Heil EL, Hynicka LM, Johnson JK. Are fluoroquinolones appropriate for the treatment of extended-spectrum β-lactamase-producing gram-negative bacilli? J Pharm Technol. 2016;32(1):16–21. doi:10.1177/8755122515599407
- Seyedjavadi SS, Goudarzi M, Sabzehali F. Relation between blaTEM, blaSHV and blaCTX-M genes and acute urinary tract infections. J Acute Dis. 2016;5(1):71–76. doi:10.1016/j.joad.2015.07.007
- Deku JG, Duedu KO, Ativi E, Kpene GE, Feglo PK. Occurrence and distribution of extended-spectrum β-lactamase in clinical Escherichia coli isolates at ho teaching hospital in Ghana. Ghana Med J. 2021;55(4):298–307. doi:10.4314/gmj.v55i4.11
- Garcia A, Delorme T, Nasr P. Patient age as a factor of antibiotic resistance in methicillin-resistant Staphylococcus aureus. J Med Microbiol. 2017;66(12):1782–1789. doi:10.1099/jmm.0.000635
- Abdelmalek S, AlEjielat R, Rayyan WA, Qinna N, Darwish D. Changes in public knowledge and perceptions about antibiotic use and resistance in Jordan: a cross-sectional eight-year comparative study. BMC Public Health. 2021;21(1):750. doi:10.1186/s12889-021-10723-x
- Gin A, Dilay L, Karlowsky JA, Walkty A, Rubinstein E, Zhanel GG. Piperacillin-tazobactam: a beta-lactam/beta-lactamase inhibitor combination. Expert Rev Anti Infect Ther. 2007;5(3):365–383. doi:10.1586/14787210.5.3.365
- Kuti JL, Wang Q, Chen H, Li H, Wang H, Nicolau DP. Defining the potency of amikacin against Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii derived from Chinese hospitals using CLSI and inhalation-based breakpoints. Infect Drug Resist. 2018;11:783–790. doi:10.2147/IDR.S161636