Abstract
The carbon-based nanomaterial family consists of nanoparticles containing allotropes of carbon, which may have a number of interactions with biological systems. The objective of this study was to evaluate the toxicity of nanoparticles comprised of pristine graphene, reduced graphene oxide, graphene oxide, graphite, and ultradispersed detonation diamond in a U87 cell line. The scope of the work consisted of structural analysis of the nanoparticles using transmission electron microscopy, evaluation of cell morphology, and assessment of cell viability by Trypan blue assay and level of DNA fragmentation of U87 cells after 24 hours of incubation with 50 μg/mL carbon nanoparticles. DNA fragmentation was studied using single-cell gel electrophoresis. Incubation with nanoparticles containing the allotropes of carbon did not alter the morphology of the U87 cancer cells. However, incubation with pristine graphene and reduced graphene oxide led to a significant decrease in cell viability, whereas incubation with graphene oxide, graphite, and ultradispersed detonation diamond led to a smaller decrease in cell viability. The results of a comet assay demonstrated that pristine graphene, reduced graphene oxide, graphite, and ultradispersed detonation diamond caused DNA damage and were therefore genotoxic in U87 cells, whereas graphene oxide was not.
Introduction
Glioblastoma multiforme (GBM) is one of the most common and malignant primary tumors of the brain. Despite the use of very aggressive treatment methods, including surgical resection, local radiotherapy, and chemotherapy, its infiltrative and intensive growth makes it mostly incurable.Citation1,Citation2 Moreover, even the most promising chemotherapeutics may have toxic effects on healthy cells, which can be fatal in GBM given that the tumor is surrounded by healthy neurons and astroglial cells. The survival of a patient diagnosed with GBM is unlikely to exceed 12 months.Citation3 Therefore, it is important to develop new treatments that extend survival in patients with GBM.
Carbon nanoparticles have recently been identified as promising agents in cancer therapy,Citation4–Citation7 and are strong candidates for becoming viable materials in biomedical applications because of their biocompatibility when compared with metal nanoparticles.Citation8–Citation10 However, nanoparticles containing allotropes of carbon show different behavior, routes of distribution, and toxicity within a living organism.Citation11,Citation12
Graphene is a two-dimensional, one atom-thick layer of spCitation2 bonded carbon.Citation13 This means that carbon atoms, through hybridization between one s orbital and two p orbitals, form trigonal planar structures with σ bonds between them and, perpendicular to the planar structure, π bonds. Graphene has been investigated for use in a number of medical and biological areas, including drug/gene delivery, cancer therapy, biosensing, bioimaging, antibacterial materials, and scaffolds for cell culture.Citation14 However, different forms of graphene, such as graphene oxide (GO) and reduced graphene oxide (rGO), produced by chemical reactions,Citation15 or pristine graphene (GN), produced by physical methods, may have different properties.Citation16 Akhavan et alCitation17 showed that the genotoxicity and cytotoxicity of graphene sheets and nanoplatelets in human mesenchymal stem cells depended on their concentration, size, and duration of exposure. Graphene nanoplatelets with an average lateral dimension of 11±4 nm have a very high cytotoxic potential and ability to penetrate into the nucleus of human mesenchymal stem cells, causing DNA fragmentation and chromosomal aberrations, even at very low concentrations. It was shown that GO had no significant cytotoxic effects in A549 cells;Citation18 however, some modifications of GO, causing changes in surface charge, had cytotoxic and genotoxic effects in human lung fibroblasts.Citation19 Recently, Jaworski et alCitation20 demonstrated dose-dependent cytotoxicity of GN nanoplatelets on GBM cells. Further, they showed that flakes of graphene adhered to the cell membrane and induced apoptosis, with necrosis observed in a small number of cells. It seems that, due to the high diversity of graphene-related structures, there are still not enough studies comparing the bioactivity of the different graphenes to determine their potential application in cancer therapy.
The other promising carbon nanoparticles are those comprised of graphite and diamond. Previous studies suggested that graphite nanoparticles inhibit angiogenesis without affecting embryonic development.Citation21 These antiangiogenic effects indicate that graphite could be a potential anticancer agent or a supporting factor in cancer therapy. In comparison with graphene and graphite, diamond nanoparticles consist of spCitation3 bonded carbon atoms, forming a structure with tetrahedral symmetry. Diamond nanoparticles are highly biocompatible and may be effective agents for bioimaging, drug delivery, and cancer therapy.Citation8,Citation22,Citation23 However, diamond nanoparticles incubated with HeLa cells in serum-free medium were shown to be highly cytotoxic, with death of almost all cells after 6 hours of incubation.Citation22 It was also demonstrated that diamond nanoparticles may damage the DNA of embryonic stem cells.Citation24 Nevertheless, the specific mechanism of these interactions is still not clear.
Graphene and its related forms, graphite and diamond, being different carbon atom structures, may have cytotoxic effects that could be utilized as anticancer treatments, but their genotoxicity is unknown. Knowing the cytotoxicity related to genotoxic effects may be key information for applying graphene, graphite, and diamond nanostructures as agents or drug carriers in anticancer therapy. Moreover, until now, there has been insufficient information regarding the genotoxicity of graphene, diamond, and graphite nanoparticles towards GBM cells. We hypothesized that because of their physicochemical characteristics, different forms of graphene and related graphite and diamond nanoparticles may exert different toxic effects on GBM cells. Therefore, the objective of this study was to compare the genotoxic effects of nanoparticles containing different allotropes of carbon on GBM cells in vitro.
Materials and methods
Preparation and characterization of carbon nanoparticles
Graphene powders (purity >99.99%) were purchased as follows: GN from SkySpring Nanomaterials (Houston, TX, USA), and rGO and GO from the Institute of Electronic Materials Technology (Warsaw, Poland). GN was produced by liquid-phase exfoliation of graphite, whereas GO was produced by chemical oxidation of graphite and rGO by chemical reduction of GO. Graphite nanoparticles (purity >93%, synthesized by the detonation method) were purchased from SkySpring Nanomaterials. Ultradispersed detonation diamond (UDD, purity >95%) was purchased from SkySpring Nanomaterials. UDD was synthesized by the Danilenko method.Citation25 The shape and size of the nanoparticles were inspected using a JEM-2000EX transmission electron microscope operating at 80 keV (JEOL Ltd, Tokyo, Japan). The samples for transmission electron microscopy were prepared by placing hydrocolloid droplets onto Formvar-coated copper grids (Agar Scientific, Stansted, UK). Immediately after the droplets had dried in dry air, the grids were inserted into the transmission electron microscope. The test was performed in triplicate. The zeta potential was measured in water by a ZEN3500 Zetasizer Nano ZS (Malvern Instruments, Malvern, UK).
Prior to application, the carbon nanoparticles were dispersed in ultrapure water to prepare a 1.0 mg/mL solution. The solution was sonicated for 30 minutes and diluted to a concentration of 50 μg/mL with 1× Dulbecco’s Modified Eagle’s Medium (Sigma-Aldrich, St Louis, MO, USA).
Cell cultures
A human GBM U87 cell line was obtained from the American Type Culture Collection (Manassas, VA, USA) and cultured in Dulbecco’s Modified Eagle’s Medium with addition of 10% fetal bovine serum and 1% penicillin and streptomycin (Sigma-Aldrich) at 37°C in a humidified atmosphere of 5% CO2 and 95% air inside a NuAire DH AutoFlow CO2 air-jacketed incubator (Plymouth, MN, USA).
Cell morphology
The U87 cells were incubated on six-well plates (1×105 cells per well) and cultivated for 24 hours. GN, rGO, GO, graphite, and UDD were introduced to the cells separately at a constant concentration of 50 μg/mL. Cells cultured without the addition of nanoparticles were used as the control group. After 24 hours of exposure to the nanoparticles, the medium was removed and the cells were stained using the May-Grünwald-Giemsa (Sigma-Aldrich) method, and their morphology was investigated using a CKX 41 epifluorescent inverted microscope with a fluorescent filter (Olympus, Tokyo, Japan); images were captured using a ProgRes® c12 camera (Jenoptik, Jena, Germany).
Cell viability
Cell viability was evaluated using Trypan blue (Sigma-Aldrich). U87 cells were cultivated in six-well plates (5×105 cells per well) and incubated for 24 hours. GN, rGO, GO, graphite, and UDD were then introduced separately to the cells at a concentration of 50 μg/mL for all types of nanoparticles. Cells without nanoparticles were used as the control. After 24 hours, the medium was removed, and the cells were detached using 0.005% trypsin. Next, 50 μL of a Trypan blue dye solution was mixed with 50 μL of cell suspension. The mixture was placed onto counting slides and counted using a TC10™ automated cell counter (BioRad, Hercules, CA, USA).
Single cell gel electrophoresis
The genotoxicity of GN, rGO, GO, graphite, and UDD was evaluated using single-cell gel electrophoresis (the comet assay).Citation26,Citation27 This procedure was based on the modified protocol described by Dhawan et al.Citation28 U87 cells were incubated for 24 hours in six-well plates (1×106 cells per well). GN, rGO, GO, graphite, and UDD were introduced to the cells separately at a constant concentration of 50 μg/mL. Cells grown without the addition of nanoparticles were used as the control group. After 24 hours of exposure to nanoparticles, the medium was removed, and the cells were rinsed with phosphate-buffered saline and trypsinized using 0.005% trypsin for 5 minutes. Higher concentrations of trypsin may increase DNA damage, so were not used. The cells were then washed and suspended in 1 mL of phosphate-buffered saline. Approximately 10,000 cells in 10 μL were mixed with low melting point agarose (A9414, Sigma-Aldrich) and immediately applied on a standard microscope glass slide, which had been coated a day earlier with 1% normal melting agarose (RM273, HiMedia Laboratories, Mumbai, India). The slides were immersed in cold lysing solution and stored for 90 minutes in the dark at 4°C. The slides were placed into DNA Comet electrophoretic system trays (Kucharczyk, Techniki Elektroniczne, Warsaw, Poland) and covered with fresh electrophoresis alkaline buffer (300 mM NaOH, 1 mM ethylenediaminetetraacetic acid, pH >13) for 20 minutes to allow unwinding of DNA and expression of alkali-labile damage. Next, electrophoresis was conducted at 24 V for 30 minutes. The slides were then gently lifted from the buffer, drained, and immersed in neutralization buffer (0.4 M Tris, pH 7.5) for 5 minutes. The slides were then drained, and this step was repeated twice. Next, the slides were stained using 50 μL of 4′, 6-diamidino-2-phenylindole (D9542, Sigma-Aldrich). All of the above steps were performed under dimmed light to prevent any DNA damage from fluorescent white light. The slides were viewed using a CKX 41 epifluorescent inverted microscope with a fluorescent filter, and the image was captured with a ProgRes c12 camera. At least 50 cells per group were randomly selected, photographed, and analyzed using Comet Assay Software Project software.Citation29
Statistical analysis
The data were analyzed using one-way analysis of variance followed by Tukey’s multiple range test. P-values <0.05 were considered to be statistically significant. Statgraphics Centurion software (StatPoint Technologies, Warrenton, VA, USA) was used for the statistical analysis.
Results
Characterization of nanoparticles containing carbon allotropes
The mean zeta potential for the nanoparticle samples was −9.61 for GN, −38.3 for rGO, −45.2 for GO, 28.7 for graphite, and −39.3 for UDD.
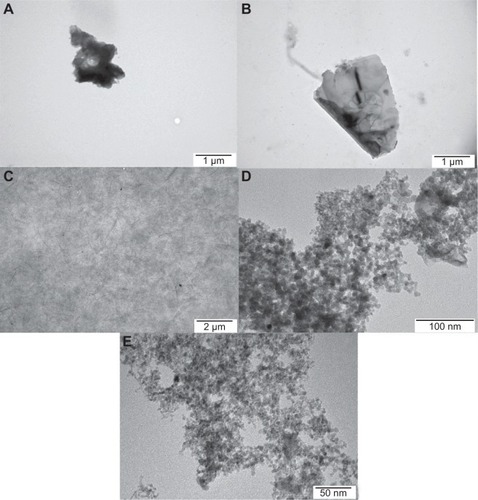
shows representative transmission electron micrographs of the nanoparticles. Although the thickness of the materials used was in the nanoscale range, the surface area of the graphene sheets was not, ranging from 450 nm to 1.5 μm and forming agglomerates over 5 μm in diameter. The variation in size of the GO nanoparticles was greater, ranging from 100 nm to agglomerates as large as 10 μm. Most of the sheets of graphene were visible as a single layer or a few layers. rGO showed the largest irregularities in shape, especially on the edges, forming sharp and rough structures. GN nanoparticles also had irregular and sharp edges. On the other hand, GO had more smooth edges and formed more regular structures. Both UDD and graphite had a size distribution between 4 nm and 5 nm and showed a tendency to form agglomerates.
Cell morphology
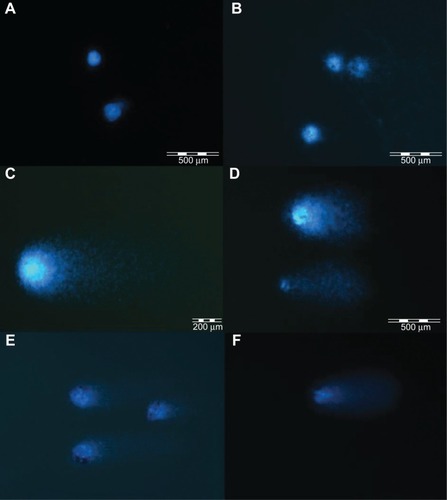
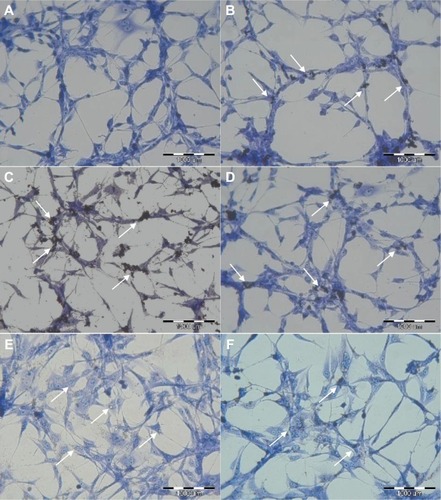
Images from May-Grünwald-Giemsa () showed high affinity of graphene for the cell membrane, specifically for the body of the cell rather than its protrusions. Moreover, the images show very high uptake of UDD and graphite (). However, when compared with the control group, none of the nanoparticles altered the cell morphology.
Figure 2 Evaluation of U87 cell morphology using May-Grünwald-Giemsa staining. Images show control cells (A) and cells incubated with graphene oxide (B), reduced graphene oxide (C), pristine graphene (D), ultradispersed detonation diamond (E), and graphite (F). Arrows point to graphene (B–D), ultradispersed detonation diamond (E), and graphite (F).
Cell viability
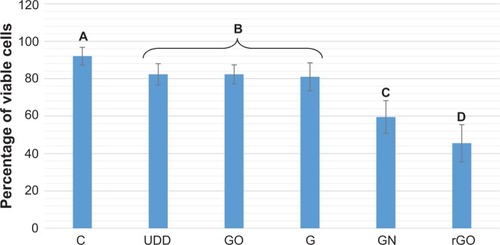
The viability assay () revealed that incubation of U87 cells with rGO, GN, GO, graphite, and UDD resulted in an increase in cell mortality. Incubation with GN and rGO markedly decreased cell viability, whereas incubation with GO, graphite, and UDD led to a small decrease in viability.
Genotoxicity
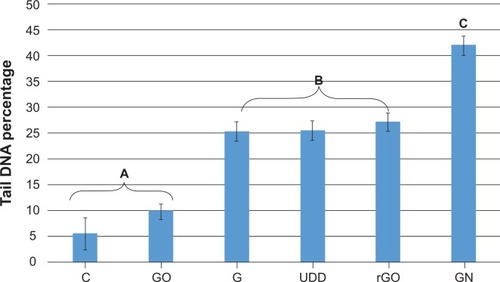
The comet assay showed formation of comets in cells incubated with GN, rGO, graphite, and UDD, indicating genotoxic properties. In contrast, cells treated with GO did not form comets, which was similar to cells from the control group (). Statistical analysis showed that rGO, GN, graphite, and UDD were highly genotoxic towards U87 cells, whereas GO was not ().
Discussion
In the present studies GN, rGO, GO, graphite, and UDD were introduced to the cells at the concentration of 50 μg/mL. This concentration was chosen based on our earlier finding that at 50 μg/mL (comparing with lower and higher concentrations) most cells were viable.Citation20 Our measurements were carried out only in the U87 cell line, and the responses to graphene are only specific for this line.Citation20 Although this limits the generalizability of our results, they are still indicative of the responses of cancer cells to graphene and other carbon allotropes.
Visualization of GN, rGO, and GO () showed that the thickness of the graphene-based materials used was consistent with their characteristic thin layer of bonded carbon atoms. The high affinity of graphene for cell membranes (), specifically the body of the cell rather than its protrusions, is in agreement with previous reports.Citation20 However, graphene was not able to penetrate the cell membrane caused by the high surface area. In contrast with graphene, UDD penetrated the cell membranes (). According to previous studies, diamond nanoparticles up to 100 nm in size are internalized by cells via macropinocytosis and clathrin-mediated pathways,Citation12,Citation30 and localize mainly in endocytic and clathrin-coated vesicles.Citation12,Citation30 Smaller particles (5–10 nm) are able to escape the vesicles or penetrate into the cells via passive transport.Citation12 In our study, graphite nanoparticles also penetrated cell membranes (). The exact mechanism of this process is unknown, although the size of graphite, which is similar to the size of UDD, may play a role.
Incubation with GN and rGO markedly decreased cell viability, and may be attributable to the physical and chemical properties of graphene. rGO showed a high affinity for the cell membranes, but the irregular and sharp edges of rGO and GN could be damaging the integrity of the membrane.Citation31 Another explanation is that rGO and GN activate receptors present on the cell membrane, which may cause apoptosis of cells by activation of the mitochondrial pathway associated with the combined role of transforming growth factor-beta and mitogen-activated protein kinases.Citation20,Citation32 On the other hand, GO, which has a high content of oxygen, regular edges, a smaller size, and hydrophilic properties, increased cell mortality only slightly. Recent studies also showed that GO had very low cytotoxicity and could not induce apoptosis in RPMI-8226 cells.Citation33 However, further studies of internalization and localization of graphene within cells are required. UDD uptake, as previously mentioned, did not cause any significant membrane damage and therefore did not increase cell mortality. Other studiesCitation8,Citation23 have shown that the presence of diamond nanoparticles inside cells is not cytotoxic. It is possible that graphite nanoparticles, which are similar in size to UDD, also penetrated the cell membrane without inducing membrane damage or affecting cell mortality.
Images of DNA comets indicate that GN, rGO, graphite, and UDD, but not GO, have a strong genotoxic effect in U87 cells (). Statistical analysis of the study data confirmed that, after 24 hours of incubation with nanoparticles at a concentration of 50 μg/mL, GN, rGO, graphite, and UDD, severe genotoxicity occurred in U87 cells, whereas GO did not seem to have any genotoxic effects (). The genotoxicity of GN and rGO may be explained by the high mobility and sharp edges of graphene. Penetration of graphene nanosheets can partially damage the membrane of the cell, resulting in destruction of the cell. GN and rGO can then interact directly with the DNA in the nucleus of the cell. This fact is confirmed by experimental data showing that the threshold for graphene genotoxicity is significantly lower than the threshold of its cytotoxicity, which can be explained by the direct interaction between graphene and nuclear DNA.Citation17,Citation31 The difference in genotoxicity between GN, rGO, and GO might be attributable to the different structures of rGO and GO. rGO and GN, which are hydrophobic and have many more rough edges, might be more potent in damaging cell membranes and penetrating into the cell. They can then interact directly with nuclear DNA, with significant genotoxic effects. GO has smoother edges and hydrophilic properties, and seems to be less potent in terms of penetrating cell compartments and interacting with DNA, resulting in the absence of genotoxic effects. Furthermore, it has been demonstrated that GO particles smaller than 500 nm may be internalized via clathrin-mediated endocytosis by interaction with proteins.Citation34 On the other hand, Yue et alCitation35 showed that 350 nm GO are wrapped by active filopodia of macrophages, while 2 μm GO enter cells nearly perpendicularly. This mechanism may trap GO inside vesicles, making it impossible for GO to penetrate into the nucleus and interact directly with DNA.
UDD was very potent in penetrating the cell membrane. Previous studies have demonstrated that diamond nanoparticles have a high level of biocompatibility, and are located in the cytoplasm and perinuclear region but do not enter the nucleus.Citation12,Citation30 However, our study indicated otherwise. UDD induced a high genotoxic effect in U87 cells. The mechanism of this interaction is still unclear, but is possibly due to the ability of diamond to bind with proteins and somehow interrupt cellular DNA repair mechanisms. Graphite nanoparticles had genotoxic effects similar to those of UDD. This may be explained by the small size of graphite. It is possible that these small nanoparticles may penetrate directly into the nucleus or disrupt the function of proteins essential for DNA repair.
It has to be underlined that this preliminary study indicated different effects of the applied carbon allotropes on cell viability and DNA damage, thereby, showing directions for further research on mechanisms of action of carbon allotropes on GBM cell responses.
Conclusion
Incubation of U87 cells for 24 hours with 50 μg/mL GN, rGO, GO, graphite, or UDD resulted in decreased cell viability. GN, rGO, graphite, and UDD showed high levels of genotoxicity in U87 cells, whereas GO did not. These preliminary findings indicate the potential use of GN, rGO, graphite, and UDD in the direct elimination of GBM cells or GO as a drug nanocarrier with minimal toxicity.
Acknowledgments
This work was supported by a grant from the Polish National Research Council (NCN OPUS 2011/03/B/NZ9/03387).
Disclosure
The authors report no conflicts of interest in this work.
References
- ZhangXZhangWCaoW-DChengGZhangY-QGlioblastoma multiforme: molecular characterization and current treatment strategy (Review)Exp Ther Med20123191422969836
- PreusserMde RibaupierreSWöhrerACurrent concepts and management of glioblastomaAnn Neurol201170192121786296
- VerhaakRGHoadleyKAPurdomEIntegrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1Cancer Cell20101719811020129251
- KimGCKimGJParkSRAir plasma coupled with antibody-conjugated nanoparticles: a new weapon against cancerJ Phys D Appl Phys2008423032005
- BriggerIDubernetCCouvreurPNanoparticles in cancer therapy and diagnosisAdv Drug Deliv Rev200254563165112204596
- FarokhzadOCChengJTeplyBATargeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivoProc Natl Acad Sci U S A2006103166315632016606824
- ChoKWangXNieSChenZGShinDMTherapeutic nanoparticles for drug delivery in cancerClin Cancer Res20081451310131618316549
- ZhuYLiJLiWThe biocompatibility of nanodiamonds and their application in drug delivery systemsTheranostics20122330231222509196
- LiuZRobinsonJTSunXDaiHPEGylated nanographene oxide for delivery of water-insoluble cancer drugsJ Am Chem Soc200813033108761087718661992
- StarkWNanoparticles in biological systemsAngew Chemie Int Ed201150612421258
- SasidharanAPanchakarlaLSChandranPDifferential nano-bio interactions and toxicity effects of pristine versus functionalized grapheneNanoscale2011362461246421562671
- FaklarisOJoshiVIrinopoulouTPhotoluminescent diamond nanoparticles for cell labeling: study of the uptake mechanism in mammalian cellsACS Nano20093123955396219863087
- NovoselovKSGeimAKMorozovSVElectric field effect in atomically thin carbon filmsScience2004306569666666915499015
- ShenHZhangLLiuMZhangZBiomedical applications of grapheneTheranostics20122328329422448195
- ParkSRuoffRSChemical methods for the production of graphenesNat Nanotechnol20094421722419350030
- HernandezYNicolosiVLotyaMHigh-yield production of graphene by liquid-phase exfoliation of graphiteNat Nanotechnol20083956356818772919
- AkhavanOGhaderiEAkhavanASize-dependent genotoxicity of graphene nanoplatelets in human stem cellsBiomaterials201233328017802522863381
- ChangYYangS-TLiuJ-HIn vitro toxicity evaluation of graphene oxide on A549 cellsToxicol Lett2011200320121021130147
- WangAPuKDongBRole of surface charge and oxidative stress in cytotoxicity and genotoxicity of graphene oxide towards human lung fibroblast cellsJ Appl Toxicol201333101156116423775274
- JaworskiSSawoszEChwalibogAIn vitro evaluation of the effects of graphene platelets on glioblastoma multiforme cellsInt J Nanomedicine20138141342023378763
- WierzbickiMSawoszEGrodzikMCarbon nanoparticles downregulate expression of basic fibroblast growth factor in the heart during embryogenesisInt J Nanomedicine201383427343524039425
- LiJZhuYLiWZhangXPengYHuangQNanodiamonds as intracellular transporters of chemotherapeutic drugBiomaterials201031328410841820692696
- KaurRBadeaINanodiamonds as novel nanomaterials for biomedical applications: drug delivery and imaging systemsInt J Nanomedicine2013820322023326195
- XingYXiongWZhuLOsawaEHussinSDaiLDNA damage in embryonic stem cells caused by nanodiamondsACS Nano2011532376238421370893
- DanilenkoVVSynthesizing and sintering of diamond by explosionMoscowEnergoatomizdat2003 Russian
- SinghNPMcCoyMTTiceRRSchneiderELA simple technique for quantitation of low levels of DNA damage in individual cellsExp Cell Res198817511841913345800
- TiceRRAgurellEAndersonDSingle cell gel/comet assay: guidelines for in vitro and in vivo genetic toxicology testingEnviron Mol Mutagen200035320622110737956
- DhawanABajpayeeMPandeyAKParmarDProtocol for the single cell gel electrophoresis/comet assay for rapid genotoxicity assessmentLucknow, IndiaIndustrial Toxicology Research Centre2003110 Available from: http://www.cometassayindia.org/protocolforcometassay.pdfAccessed December 21, 2013
- KońcaKLankoffABanasikAA cross-platform public domain PC image-analysis program for the comet assayMutat Res20035341–2152012504751
- LiuK-KWangC-CChengC-LChaoJ-IEndocytic carboxylated nanodiamond for the labeling and tracking of cell division and differentiation in cancer and stem cellsBiomaterials200930264249425919500835
- AkhavanOGhaderiEEmamyHAkhavanFGenotoxicity of graphene nanoribbons in human mesenchymal stem cellsCarbon N Y201354419431
- LiYLiuYFuYThe triggering of apoptosis in macrophages by pristine graphene through the MAPK and TGF-beta signaling pathwaysBiomaterials201233240241122019121
- WuSZhaoXCuiZCytotoxicity of graphene oxide and graphene oxide loaded with doxorubicin on human multiple myeloma cellsInt J Nanomedicine2014911413142124672235
- MuQSuGLiLSize-dependent cell uptake of protein-coated graphene oxide nanosheetsACS Appl Mater Interfaces2012442259226622409495
- YueHWeiWYueZThe role of the lateral dimension of graphene oxide in the regulation of cellular responsesBiomaterials201233164013402122381473