Abstract
Background and Objectives
Telomerase reverse transcriptase (TERT) promoter mutations are one of the most common mutations responsible for the development of hepatocellular carcinoma (HCC). Noncoding RNAs (ncRNAs) play a regulatory role in different cancers through the long noncoding RNA (lncRNA)/circular RNA (circRNA)-microRNA (miRNA)-mRNA axis. The aim of the study was to explore the influence of TERT promoter mutations on the ncRNA regulatory network in HCC.
Methods
Four tumor samples with a wildtype TERT promoter and four tumor samples with TERT promoter mutations (sequencing cohort) were collected from HCC patients for high-throughput next-generation sequencing. Selected ncRNAs and mRNAs were validated by qPCR in 15 HCC tumors with a wildtype TERT promoter and seven HCC tumors with TERT promoter mutations (validation cohort, including the sequencing cohort).
Results
In the mutant TERT promoter group, 536 lncRNAs, 21 circRNAs, 41 miRNAs, and 266 mRNAs were significantly up-regulated, while 1745 lncRNAs, 23 circRNAs, 32 miRNAs, and 1117 mRNAs were significantly down-regulated (P < 0.05) compared with the findings in wildtype group. AL360169.3-201, LINC02672-203, hsa_circ_0021412, hsa-miR-29b-1-5p, hsa-miR-4699-5p, hsa-miR-199a-5p, REG3A, SFRP5, and GSTM1 were verified at the RNA expression level to validate the sequencing results. A differentially expressed lncRNA/circRNA-miRNA-mRNA network was constructed to explore the effects of TERT promoter mutations on ncRNA regulation. Two ncRNA regulatory axes associated with TERT promoter mutations (hsa_circ_0003154/hsa_circ_0008952/IGLL5-AS1/LINC576/LINC575–hsa-miR-1260b –CLPTM1L/GSTM1 and hsa_circ_0031584/LINC2101–hsa-miR-214-3p–CD151) had carcinogenic potential.
Conclusion
This study provides novel insights into the role of TERT promoter mutations on ncRNAs regulatory network in HCC progression.
Introduction
Hepatocellular carcinoma (HCC) is the most common primary liver malignancy. In 2020, primary liver malignancy was the sixth most commonly diagnosed cancer and the third leading cause of cancer death worldwide.Citation1,Citation2 The incidence rate is extremely high in Eastern Asia, and China is one of the most high-risk HCC areas.Citation2 Multiple risk factors are responsible for the development of HCC, including chronic hepatitis B or C infection, cigarette smoking, and alcohol abuse. In addition, several mutations are associated with HCC, including telomerase reverse transcriptase (TERT) promoter mutation, which is one of the most-studied mutations in HCC.Citation3
Telomerase is a cellular ribonucleoprotein enzyme. It promotes cell proliferation by elongating the telomere. TERT is a subunit of telomerase that is responsible for its catalytic activity.Citation4 Typically, its expression is tightly limited in most normal cells, but up-regulated in multiple cancers.Citation5 TERT expression can be regulated at both the transcription and posttranslation levels. At the transcription level, some cancer-associated TERT promoter mutations function by generating de novo transcription-factor-binding sites. Two major TERT promoter mutations are −124C/T and −146C/T, which are named C228T and C250T, respectively. These two mutations generate putative ETS transcription-factor-binding sites (TTCC), thereby activating TERT transcription.Citation6,Citation7
TERT promoter mutations have been identified in several human cancers, including bladder cancer, glioblastoma, and melanoma.Citation8,Citation9 HCC has a high mutation frequency. Up to 60% of HCC patients carry TERT promoter mutations.Citation10,Citation11 However, in some cases, TERT promoter mutations are not accompanied by a high TERT expression.Citation12 No difference in TERT immunohistochemical expression levels was observed between skin tumors with and without TERT promoter mutations.Citation13 Moreover, there was no significant association between TERT mutation and expression in bladder cancer.Citation14 Furthermore, it was also reported that the TERT promoter mutation status did not affect the TERT mRNA expression level in HCC.Citation15 The findings of these studies indicate that TERT promoter mutations may have other unclear cancer-promoting roles as well.
Noncoding RNAs (ncRNAs) are a category of RNA without protein-coding function, which account for over 97% of the human transcriptome. Their regulatory role has been only recently discovered.Citation16 It has been extensively studied how microRNA (miRNA) regulate mRNA expression at the post-transcription level. MiRNA, a class of ncRNAs with approximately 22 nucleotides, promotes the degradation of complementary mRNA by binding to the 3’UTR of mRNA. Long-noncoding RNA (lncRNA), a group of ncRNAs with a length greater than 200 nucleotides, is also known as competitive endogenous RNA (ceRNA) as it competes against mRNA for miRNA binding.Citation17 Circular RNA (circRNA) is another group of ncRNAs with a head-to-tail closed loop structure, which protects them from degradation. CircRNA can also work as ceRNA. Both lncRNA and circRNA can down-regulate the miRNA–mRNA binding and therefore reduce mRNA degradation by competitive binding to miRNA-response elements.Citation18
Notably, lncRNA/circRNA-miRNA-mRNA regulatory networks play important roles in multiple cancers, including HCC. The tumor-promoting lncRNA MALAT1 has been reported to up-regulate the expression of FOXM1 by sponging miR-125a-3p.Citation19 Other lncRNAs, including HULC, HOTAIR, MEG-3, and H19, have also been identified as the indicators of HCC diagnosis and prognosis.Citation20 It has been reported that hsa_circRNA_104348 promotes HCC progression by modulating the miR-187-3p/RTKN2 axis.Citation21 However, it is still unclear how the lncRNA/circRNA-miRNA-mRNA regulatory networks differ in HCC with TERT promoter mutations.
In the current study, high-throughput next-generation sequencing was performed to identify the expression profiles of lncRNAs, circRNA, miRNAs, and mRNAs in HCC tumors with or without TERT promoter mutations in order to compare the whole genome expression landscape. The aim of the study was to construct lncRNA/circRNA-miRNA-mRNA networks influenced by TERT promoter mutations and explore their roles in HCC carcinogenesis and development.
Materials and Methods
Patients with HCC
22 patients with newly diagnosed and histologically confirmed HCC were recruited from Shandong University affiliated Cheeloo Hospital, China. Tumor specimens were obtained from patients during surgical treatment, and for each patient, the specimen was collected from single HCC nodule. The presence of tumor cells within the collected specimens was verified by pathological examination, and the samples were frozen at −80 °C until use. The samples were divided into two groups based on the presence of TERT promoter mutations: seven HCC tumors with TERT promoter mutations and 15 HCC tumors with wildtype TERT promoter (validation cohort). Among them, four samples from mutant TERT promoter group and four samples from wildtype group were sent for sequencing (sequencing cohort). The study was approved by the Ethics Committee of Shandong University, China, with an ethics code SDULCLL2021-1-27. Written informed consent was obtained from all patients. The study complies with the Declaration of Helsinki. The clinical background is shown in Table S1.
DNA Extraction and Sanger Sequencing of the TERT Promoter
Genomic DNA from HCC frozen-tissues was extracted using TIANGEN DNA extraction kits and analyzed for TERT promoter mutations by Sanger sequencing. Two mutations defined as C228T and C250T in the TERT core promoter were found that correspond to positions 124 and 146 basepair upstream of the ATG site. The primer pair for TERT promoter sequencing was previously described: 5′-CAC CCG TCC TGC CCC TTC ACC TT-3′ (forward) and 5′-GGC TTC CCA CGT GCG CAG CAG GA-3′ (reverse).Citation22,Citation23 All the mutations were verified by sequencing from both directions.
LncRNA and mRNA Sequencing
Total tissue RNA was extracted with the TRIzol reagent kit (Life Technologies, Cat. # 15596-018, Carlsbad, USA). The quality of the total RNA was checked first using the NanoPhotometer® spectrophotometer (IMPLEN, CA, USA) with 1% agarose gels. The first strand of cDNA was synthesized using 3 μg of RNA by M-MuLV reverse transcriptase (RNase H) and random hexamer primer. After PCR, the products were purified by the AMPure XP system, and the sequencing library was generated by using the NEBNext® UltraTM RNA library prep kit. The library products were sequenced on Illumina HiSeq 4000 (Illumina, USA). The clean reads were first obtained after removing poly-N, adapter and low-quality reads and then used to calculate the FPKM of each gene. The sequencing was performed by Novogene Science and Technology Co. Ltd (Beijing, China).
Small RNA Sequencing
For each sample, 3 μg of RNA was used for sequencing library building. The libraries were generated using NEBNext® Multiplex Small RNA Library Prep Set (Illumina®, NEB, USA). PCR products obtained using LongAmp Taq were purified and subsequently sequenced on Illumina HiSeq 2500 (Novogene Science and Technology Co. Ltd., Beijing, China).
Quantitative PCR (qPCR)
qPCR was carried out in a Roche LightCycler 96 sequence detector (Roche, Basel, Switzerland) using SYBR Green and specific primers. GAPDH expression was used as a control for lncRNA, circRNA and mRNA, U6 expression was used as a control for miRNA. Levels of target RNAs were calculated based on the CT values and normalization to human GAPDH or U6 expression. Primers used for qPCR were listed in Table S2.
lncRNA-miRNAs-mRNA Network Analysis
The target genes of miRNA (mRNAs and lncRNAs) were predicted using miRanda and miRBase20.0. The differentially expressed (DE) mRNAs, miRNAs, and lncRNAs were analyzed by using the DESeq R package (1.8.3) with the transcriptome sequencing data. Using the results obtained after integrated analysis of DE mRNAs/miRNA/lncRNAs and miRanda and miRBase, a global network of DE lncRNAs-DE miRNAs-DE mRNAs was constructed and subsequently visualized by Cytoscape (Cytoscape_v3.5.1).
Statistical Analysis
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed for the functional annotation statistical enrichment of the DE genes in transcriptome sequencing. The data were collected and shown as mean ± standard deviation (SD). Statistical analysis was performed by Mann–Whitney test. P < 0.05 was considered to be statistically different.
Results
TERT Promoter Mutation Detection in 22 HCC Tumor Samples
The TERT promoter was sequenced and evaluated in the DNA from 22 HCC samples (validation cohort) by Sanger sequencing. Seven of these samples (31.8%) harbored TERT promoter mutations. C250T was found in one sample and C228T in the remaining six.
Profiles of DE lncRNAs, miRNAs, and mRNAs
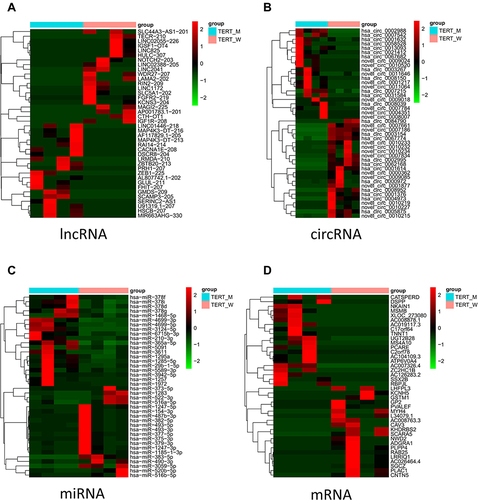
Four HCC tumors with TERT promoter mutations and four HCC tumors with wildtype TERT promoter (sequencing cohort) were selected from the validation cohort for high-throughput next-generation sequencing. After the analysis of the data, a total of 109,862 lncRNAs, 7005 circRNAs, 1496 miRNAs, and 83,725 mRNAs were detected. Compared to the wildtype group, the expression profiles showed that 536 lncRNAs, 21 circRNAs, 41 miRNAs, and 266 mRNAs were significantly up-regulated, while 1745 lncRNAs, 23 circRNAs, 32 miRNAs, and 1117 mRNAs were significantly down-regulated (P <0.05) in the mutant TERT promoter group (). The heatmaps of DE circRNAs, DE lncRNAs, DE miRNAs, and DE mRNAs are shown in . These findings demonstrated that TERT promoter mutations have a significant impact on the expression landscape changes of HCC.
Figure 1 Profiles of differentially expressed lncRNAs, circRNAs, miRNAs, and mRNAs. Volcano plots of (A) DE lncRNAs, (B) DE circRNAs, (C) DE miRNAs, and (D) DE mRNAs. Volcano plots show the expression variation of lncRNAs, circRNA, miRNAs, or mRNAs between the wildtype TERT promoter group and mutant TERT promoter group. Each lncRNA, circRNA, miRNA, or mRNA is represented by a dot; red dots indicate up-regulation and green dots indicate down-regulation. The black dots in the center area indicate no significant difference (P < 0.05).
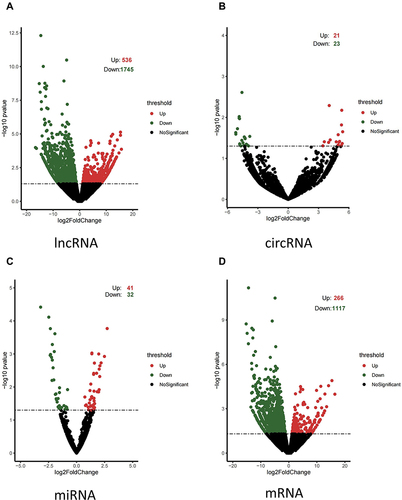
Figure 2 Heatmaps of DE lncRNAs, circRNAs, miRNAs, and mRNAs. The heatmap showing the hierarchical clustering of altered lncRNAs (A), circRNAs (B), miRNAs (C), and mRNAs (D) between the wildtype TERT promoter group and mutant TERT promoter group. Red represents up-regulation and green represents down-regulation.
Bioinformatics Analysis of DE mRNAs
GO and KEGG enrichment analyses of DE mRNAs were performed to evaluate gene functions. In GO enrichment analysis, the top 10 enrichments of biological processes (BP), cellular components (CC), and molecular functions (MF) are presented in . For DE mRNAs, the most enriched GO terms were single-organism process (GO ID: GO:0044699; type: BP; P < 0.01), multicellular organismal process (GO ID: GO:0032501; type: BP; P < 0.001), membrane (GO ID: GO:0016020; type: CC; P< 0.001), membrane part (GO ID: GO:0044425; type: CC; P < 0.001), receptor binding (GO ID: GO:0005102; type: MF; P < 0.001), and receptor activity (GO ID: GO:0004872; type: MF; P < 0.001).
Figure 3 Functional annotation of DE mRNAs. (A) Bar graphs of Gene Ontology (GO) enrichment analysis, molecular function (MF), biological process (BP), and cellular component (CC) terms of DE mRNAs between the wildtype TERT promoter group and mutant TERT promoter group. (B) Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of DE mRNAs between the wildtype TERT promoter group and mutant TERT promoter group.
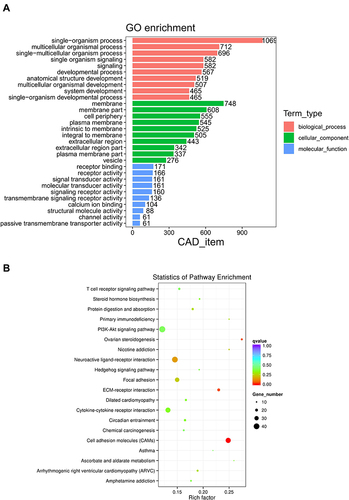
The top 20 pathways from KEGG analysis are shown in . The results indicated the main pathways involving DE mRNAs, such as cell adhesion molecules, extracellular matrix (ECM)-receptor interaction, T cell receptor signaling pathway, cytokine-cytokine receptor interaction, and neuroactive ligand-receptor interaction.
In GO analysis, top enrichment of CC included different aspects of the membrane. The most enriched MF terms were mostly related to receptors and transducers. KEGG analysis showed consistent results. Multiple receptor binding and signaling transduction pathways were mainly enriched. Integrated analyses of GO and KEGG showed that the receptor function and membrane component were the most affected parts in TERT promoter mutant HCC.
DE lncRNA, circRNA, miRNA, and mRNA Validation by qPCR
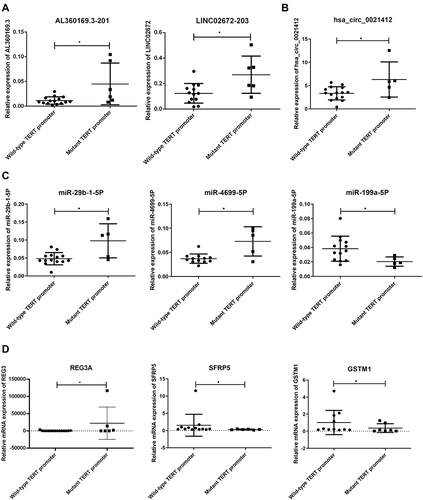
To validate the high-throughput sequencing results, qPCR was performed in the validation cohort to confirm the expression change of the selected DE lncRNAs, circRNAs, miRNAs, and mRNAs between the two groups. The validation cohort included 15 wildtype TERT promoter HCC tumors and seven mutant TERT promoter HCC tumors.
The results demonstrated that lncRNA AL360169.3-201 and LINC02672-203 were up-regulated in the mutant TERT promoter group (). With regard to circRNAs, hsa_circ_0021412 was up-regulated in the mutant TERT promoter group (). With regard to miRNAs, hsa-miR-29b-1-5p and hsa-miR-4699-5p were up-regulated, and hsa-miR-199a-5p was down-regulated in the mutant TERT promoter group (). With regard to mRNAs, regenerating family member 3 alpha (REG3A) was up-regulated, and secreted frizzled related protein 5 (SFRP5) and glutathione S-transferase mu 1 (GSTM1) were down-regulated (). These results were consistent with our sequencing data.
Figure 4 Differentially expressed lncRNA, circRNA, miRNA, and mRNA validation by qPCR. The expression of selected lncRNA (A), circRNA (B), miRNA (C), and mRNA (D) was detected in the validation cohort. (A) AL360169.3–201 and LINC02672-203. (B) hsa_circ_0021412. (C) hsa-miR-29b-1-5p and hsa-miR-4699-5p. (D) REG3A, SFRP5 and GSTM1. Statistical analysis was performed by Mann–Whitney test. P < 0.05 was considered to be statistically different. *P < 0.05.
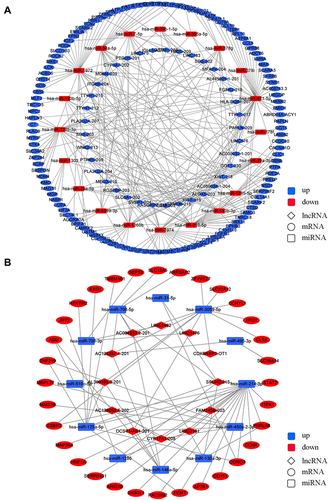
LncRNA–miRNA–mRNA Regulatory Network Analysis
By sponging, lncRNAs influence miRNA-mRNA binding and therefore regulate mRNA expression. One lncRNA can bind to different miRNAs and one miRNA can bind to different mRNAs. As a result, multiple lncRNA–miRNA–mRNA axes form that constitute the regulatory network. The analysis of DE lncRNA-targeted DE miRNAs and DE miRNA-targeted DE mRNAs was performed for the lncRNA–miRNA–mRNA network in the TERT promoter mutant HCC. The DE RNAs were divided into “down-up-down” () and “up-down-up” () two groups based on whether they were up-regulated or down-regulated in TERT promoter mutant HCC. 37 down-regulated lncRNAs, 20 up-regulated miRNAs, and 117 down-regulated mRNAs were used for the “down-up-down” network construction. 12 up-regulated lncRNAs, 12 down-regulated miRNAs, and 32 up-regulated mRNAs were used for the “up-down-up” network construction (FPKM ≥ 1.0, fold change ≥ 1.0, P < 0.05). The details of the genes in networks are shown in Tables S3 and S4.
CircRNA-miRNA-mRNA Regulatory Network Analysis
CircRNAs play a similar role to lncRNAs in miRNA-mRNA binding regulation by working as ceRNA. The “down-up-down” () and “up-down-up” () networks of DE circRNA-targeted DE miRNAs and DE miRNA-targeted DE mRNAs were constructed. 10 down-regulated circRNAs, 12 up-regulated miRNAs, and 70 down-regulated mRNAs were used for the “down-up-down” network construction, while 8 up-regulated circRNAs, 11 down-regulated miRNAs, and 34 up-regulated mRNAs were used for the “up-down-up” network construction (FPKM ≥ 1.0, fold change ≥ 1.0, P < 0.05). The details of the genes in networks are shown in Tables S5 and S6.
Figure 6 CircRNA-miRNA-mRNA regulatory network. (A) Network of down-regulated circRNA-up-regulated miRNA-down-regulated mRNA. (B) Network of up-regulated circRNA-down-regulated miRNA-up-regulated mRNA.
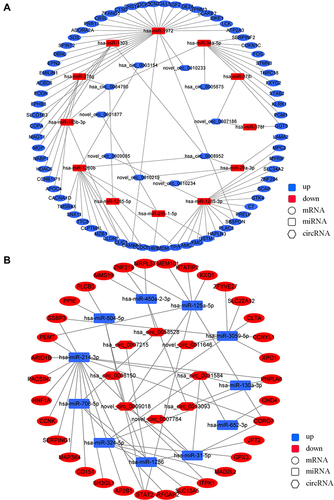
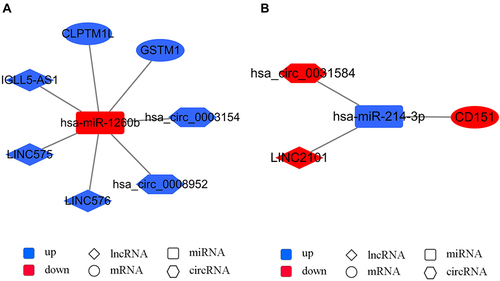
Two representative axes in the network are shown in . The up-regulated miRNA hsa-miR-1260b was regulated by down-regulated circRNA (hsa_circ_0003154/hsa_circ_0008952) and lncRNA (IGLL5-AS1/LINC576/LINC575). The abovementioned RNAs and hsa-miR-1260b target mRNAs palate transmembrane 1 like (CLPTM1L) and GSTM1 constituted the down-up-down axis hsa_circ_0003154/hsa_circ_0008952/IGLL5-AS1/LINC576/LINC575 - CLPTM1L/GSTM1 (). The down-regulated miRNA hsa-miR-214-3p was regulated by up-regulated circRNA hsa_circ_0031584 and lncRNA LINC2101. The abovementioned RNAs and hsa-miR-214-3p target mRNA cluster of differentiation 151 (CD151) constituted the up-down-up axis hsa_circ_0031584/LINC2101 - hsa-miR-214-3p - CD151 ().
Discussion
Telomerase is aberrantly activated and TERT is highly expressed in different cancers.Citation3,Citation4 Many cancers, such as glioblastoma, bladder cancer, and melanoma, also show a high frequency of TERT promoter mutations, and it is suggested that TERT promoter mutations help with telomerase activation by promoting TERT transcription.Citation8,Citation9 However, not all cancers show this high frequency. TERT promoter mutations are rare in some tumors, including prostate and breast cancer, although these two cancers show increased TERT expression.Citation24–27 In cancers with a high frequency of TERT promoter mutations, compared with adjacent normal tissues, TERT expression levels are higher in tumor tissues regardless of the presence or absence of TERT promoter mutations. Research indicates that there should be alternative pathways for TERT expression activation, and one of them is the DNA methylation pattern on TERT promoter. Unlike the canonical role of promoter hypermethylation in the transcription of most tumor suppressors, hypermethylation of TERT promoter can increase TERT expression.Citation28 Beyond that, ncRNAs also play a role in regulating TERT transcription. It has been reported that hsa-miR-195-5p works as a tumor suppressor in thyroid cancer by targeting TERT mRNA.Citation29 On the other hand, many studies failed to observe significant correlations between TERT promoter mutations and increased TERT expression.Citation12–14 It has been reported that the increase in TERT expression is not significant in HCC samples with TERT promoter mutations.Citation15,Citation30 Moreover, in the current study, no significant difference on TERT expression was found in the sequencing or validation cohort. The above evidence suggests that the understanding of how TERT promoter mutations promote HCC progression is still incomplete.
The malignant transformation of cirrhotic hepatocytes is a multistep process, and TERT promoter mutations are shown to be the earliest recurrent somatic genetic alterations, as well as the “gatekeeper” for HCC.Citation3 Existing research on TERT promoter mutations mainly focuses on their roles in TERT transcription stimulation, and research on their roles besides transcription stimulation is rare. In view of the fact that TERT promoter mutations are not always associated with higher TERT expression, potential roles beyond transcription should be explored. Another predominant mutation in HCC, CTNNB1 activating mutation, has been shown to promote HCC partly through influencing miRNA expression.Citation31 It has been reported that TERT promoter mutated follicular thyroid carcinomas exhibit a distinct miRNA expressional profile.Citation32 Additionally, it has been reported that ncRNA can regulate TERT expression in cancer; however, it is still unclear whether TERT promoter mutations can influence ncRNA regulation.Citation29 The evidence indicates that ncRNA might be a novel prospect of TERT promoter mutation research in HCC. Since TERT promoter mutations are located in the noncoding part, relative sequencing data are lacking in previous studies. The current study aimed to explore the potential role of TERT promoter mutations in the ncRNA regulation network in HCC.
The GO and KEGG analyses of DE mRNAs revealed the role of TERT promoter mutations in the overall gene function in HCC. GO analysis showed that DE mRNAs were primarily involved in the membrane and receptor binding/activity in TERT promoter mutant HCC. KEGG analysis showed similar results. The cell adhesion molecules of the membrane and focal adhesion of the extracellular region were the top two enriched pathways. Several receptor signaling pathways were also highly enriched, including T cell receptor signaling pathway, ECM-receptor interaction, and cytokine-cytokine receptor interaction. In addition, cancer-related pathways involving PI3K-Akt and the Hedgehog signaling pathway were enriched in TERT promoter mutant HCC. These results indicate that TERT promoter mutations might play a role in the membrane component, including receptors in signal transduction and extracellular matrix connection.
The sequencing result was validated in a validation cohort consisting of 15 wildtype TERT promoter HCC tumors and seven mutant TERT promoter HCC tumors. Two lncRNAs, one circRNA, three miRNAs, and three mRNAs were validated by qPCR; the change was consistent with the sequencing results. Among them, miR-29b-1-5P, which was up-regulated in the mutant TERT promoter group, has been reported to show oncogenic activity.Citation33 In addition, miR-199a-5P, another validated miRNA, was down-regulated in the mutant TERT promoter group. Studies have demonstrated its tumorigenesis suppressing role in HCC.Citation34 REG3A mRNA, up-regulated in the mutant TERT promoter group, has been reported to promote HCC via tumor-stroma crosstalk.Citation35 SFRP5 mRNA, down-regulated in the mutant TERT promoter group, has been reported to inhibit the progression of multiple cancers by suppressing the Wnt/β-catenin pathway.Citation36,Citation37 GSTM1 is a member of the glutathione S-transferases (GSTs) family, which plays an important role in Phase II detoxification. In the current study, GSTM1 was down-regulated in the mutant TERT promoter group. The homozygous deletion (null genotype) and polymorphisms of GSTM1 have been demonstrated to increase the HCC risk. The null genotype of GSTM1 is suggested to be associated with the complete absence of its enzyme activity, and therefore, it increases the susceptibility to cancer.Citation38 AL360169.3-201, LINC02672-203, hsa_circ_0021412, and hsa-miR-4699-5p are novel RNAs that lack research in cancers. Among the RNAs validated, the up-regulated RNAs in the mutant TERT promoter group have a tumor-promoting role, and the down-regulated RNAs can suppress tumors.
A comprehensive analysis of wildtype and mutant TERT promoter groups of the lncRNA/circRNA-miRNA-mRNA network revealed several lncRNA/circRNA-miRNA-mRNA axes. The down-up-down axis was hsa_circ_0003154/hsa_circ_0008952/IGLL5-AS1/LINC576/LINC575 - hsa-miR-1260b - CLPTM1L/GSTM1. The regulatory circRNAs (hsa_circ_0003154/hsa_circ_0008952) and lncRNAs (IGLL5-AS1/LINC576/LINC575) in this axis are novel ncRNAs in cancers. The up-regulated miRNA hsa-miR-1260b has been reported to promote HCC, colorectal cancer, and non-small cell lung cancer.Citation39–41 The current study revealed that in the mutant TERT promoter group, hsa-miR-1260b down-regulated the mRNA expression of CLPTM1L and GSTM1. As mentioned above, decreased GSTM1 promotes HCC progression.Citation38 The CLPTM1L gene is present next to the TERT gene, which is also involved in the synthesis of telomerase and apoptotic response, and is therefore associated with cisplatin-resistance in cancer cells.Citation42 Moreover, both CLPTM1L and TERT are located in the human 5p15.33 locus, and single nucleotide polymorphisms (SNPs) in the TERT-CLPTM1L locus have been revealed to be significantly associated with the risk of developing different cancers.Citation43–45 Among these SNPs, rs401681, in the noncoding region, was reported to be associated with the increased risk of HCC.Citation46 It might regulate TERT expression as an enhancer by interacting with the TERT promoter in lung cancer.Citation44 A previous study proposed that TERT promoter mutations might increase the expression of CLPTM1L because of its adjacent location.Citation12 However, in our study, CLPTM1L was down-regulated in the mutant TERT promoter group. The result suggested that the influence of TERT promoter-related ncRNA on CLPTM1L might be different from the TERT promoter itself. Further research is necessary to clarify the association between TERT, TERT promoter, and CLPTM1L.
The representative up-down-up axis was hsa_circ_0031584/LINC2101 - hsa-miR-214-3p - CD151. The role of upstream ncRNAs hsa_circ_0031584 and LINC2101 are not clear in cancers. However, it is known that hsa-miR-214-3p is an anti-tumor miRNA and down-regulated in HCC.Citation47 CD151 is involved in maintaining cellular integrity and angiogenesis, which promote carcinogenesis and tumor metastasis. CD151 is also reported to be positively associated with the invasiveness of HCC.Citation48
Limitations
The current study mainly focused on revealing the noncanonical function of TERT promoter mutations and highlighted a number of interesting ncRNAs for future studies. However, further research is needed to explore the mechanism of how TERT promoter mutations influence the ncRNAs.
Conclusions
In summary, TERT promoter mutations can affect HCC in various aspects. The GO and KEGG pathway analyses indicated that TERT promoter mutations appear to be involved in the regulation of the membrane and its receptors. An integrated analysis of lncRNAs, circRNAs, miRNAs, and mRNAs revealed that TERT promoter mutations may affect the lncRNA/circRNA-miRNA-mRNA regulatory network in HCC and play a dual role in CLPTM1L regulation. The results indicate potential regulatory pathways in HCC associated with TERT promoter mutations.
Data Sharing Statement
The datasets presented in this study can be found in online repositories. The names of the repository and accession number can be found below: Sequence Read Archive (SRA) PRJNA808183.
Disclosure
The authors declare no conflicts of interest.
Acknowledgments
The authors would like to thank the Shandong University affiliated Cheeloo Hospital.
Additional information
Funding
References
- Llovet JM, Kelley RK, Villanueva A, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7(1):6. doi:10.1038/s41572-020-00240-3
- Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
- Zucman-Rossi J, Villanueva A, Nault JC, Llovet JM. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology. 2015;149(5):1226–1239.e1224. doi:10.1053/j.gastro.2015.05.061
- Blackburn EH, Epel ES, Lin J. Human telomere biology: a contributory and interactive factor in aging, disease risks, and protection. Science. 2015;350(6265):1193–1198. doi:10.1126/science.aab3389
- Wu RA, Upton HE, Vogan JM, Collins K. Telomerase mechanism of telomere synthesis. Annu Rev Biochem. 2017;86:439–460. doi:10.1146/annurev-biochem-061516-045019
- Heidenreich B, Kumar R. TERT promoter mutations in telomere biology. Mutat Res Rev Mutat Res. 2017;771:15–31. doi:10.1016/j.mrrev.2016.11.002
- Liu T, Yuan X, Xu D. Cancer-specific Telomerase Reverse Transcriptase (TERT) promoter mutations: biological and clinical implications. Genes. 2016;7:7. doi:10.3390/genes7070038
- Liu X, Wu G, Shan Y, Hartmann C, von Deimling A, Xing M. Highly prevalent TERT promoter mutations in bladder cancer and glioblastoma. Cell Cycle. 2013;12(10):1637–1638. doi:10.4161/cc.24662
- Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339(6122):957–959. doi:10.1126/science.1229259
- Nault JC, Zucman-Rossi J. TERT promoter mutations in primary liver tumors. Clin Res Hepatol Gastroenterol. 2016;40(1):9–14. doi:10.1016/j.clinre.2015.07.006
- Nault JC, Calderaro J, Di Tommaso L, et al. Telomerase reverse transcriptase promoter mutation is an early somatic genetic alteration in the transformation of premalignant nodules in hepatocellular carcinoma on cirrhosis. Hepatology. 2014;60(6):1983–1992. doi:10.1002/hep.27372
- Fredriksson NJ, Ny L, Nilsson JA, Larsson E. Systematic analysis of noncoding somatic mutations and gene expression alterations across 14 tumor types. Nat Genet. 2014;46(12):1258–1263. doi:10.1038/ng.3141
- Pópulo H, Boaventura P, Vinagre J, et al. TERT promoter mutations in skin cancer: the effects of sun exposure and X-irradiation. J Invest Dermatol. 2014;134(8):2251–2257. doi:10.1038/jid.2014.163
- Allory Y, Beukers W, Sagrera A, et al. Telomerase reverse transcriptase promoter mutations in bladder cancer: high frequency across stages, detection in urine, and lack of association with outcome. Eur Urol. 2014;65(2):360–366. doi:10.1016/j.eururo.2013.08.052
- Chen YL, Jeng YM, Chang CN, et al. TERT promoter mutation in resectable hepatocellular carcinomas: a strong association with hepatitis C infection and absence of hepatitis B infection. Int J Surg. 2014;12(7):659–665. doi:10.1016/j.ijsu.2014.05.066
- Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18(1):5–18. doi:10.1038/nrc.2017.99
- Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661–5667. doi:10.1038/onc.2017.184
- Shen H, Liu B, Xu J, et al. Circular RNAs: characteristics, biogenesis, mechanisms and functions in liver cancer. J Hematol Oncol. 2021;14(1):134. doi:10.1186/s13045-021-01145-8
- Liu S, Qiu J, He G, et al. LncRNA MALAT1 acts as a miR-125a-3p sponge to regulate FOXM1 expression and promote hepatocellular carcinoma progression. J Cancer. 2019;10(26):6649–6659. doi:10.7150/jca.29213
- Abbastabar M, Sarfi M, Golestani A, Khalili E. lncRNA involvement in hepatocellular carcinoma metastasis and prognosis. Excli J. 2018;17:900–913. doi:10.17179/excli2018-1541
- Huang G, Liang M, Liu H, et al. CircRNA hsa_circRNA_104348 promotes hepatocellular carcinoma progression through modulating miR-187-3p/RTKN2 axis and activating Wnt/β-catenin pathway. Cell Death Dis. 2020;11(12):1065. doi:10.1038/s41419-020-03276-1
- Liu T, Wang N, Cao J, et al. The age- and shorter telomere-dependent TERT promoter mutation in follicular thyroid cell-derived carcinomas. Oncogene. 2014;33(42):4978–4984. doi:10.1038/onc.2013.446
- Wang N, Liu T, Sofiadis A, et al. TERT promoter mutation as an early genetic event activating telomerase in follicular thyroid adenoma (FTA) and atypical FTA. Cancer. 2014;120(19):2965–2979. doi:10.1002/cncr.28800
- Stoehr R, Taubert H, Zinnall U, et al. Frequency of TERT promoter mutations in prostate cancer. Pathobiology. 2015;82(2):53–57. doi:10.1159/000381903
- Sabaliauskaite R, Jarmalaite S, Petroska D, et al. Combined analysis of TMPRSS2-ERG and TERT for improved prognosis of biochemical recurrence in prostate cancer. Genes Chromosomes Cancer. 2012;51(8):781–791. doi:10.1002/gcc.21963
- Tanaka M, Yamashita SI, Yoshinaga Y, et al. Combination of DYRK2 and TERT expression is a powerful predictive marker for early-stage breast cancer recurrence. Anticancer Res. 2022;42(4):2079–2085. doi:10.21873/anticanres.15689
- Shimoi T, Yoshida M, Kitamura Y, et al. TERT promoter hotspot mutations in breast cancer. Breast Cancer. 2018;25(3):292–296. doi:10.1007/s12282-017-0825-5
- Zhu J, Zhao Y, Wang S. Chromatin and epigenetic regulation of the telomerase reverse transcriptase gene. Protein Cell. 2010;1(1):22–32. doi:10.1007/s13238-010-0014-1
- Liu Z, Zhang L, Chen W, et al. miR-195-5p regulates cell proliferation, apoptosis, and invasion of thyroid cancer by targeting telomerase reverse transcriptase. Bioengineered. 2021;12(1):6201–6209. doi:10.1080/21655979.2021.1963908
- Jang JW, Kim JS, Kim HS, et al. Significance of TERT genetic alterations and telomere length in hepatocellular carcinoma. Cancers. 2021;13:9. doi:10.3390/cancers13092160
- Xiao X, Mo H, Tu K. CTNNB1 mutation suppresses infiltration of immune cells in hepatocellular carcinoma through miRNA-mediated regulation of chemokine expression. Int Immunopharmacol. 2020;89(Pt A):107043. doi:10.1016/j.intimp.2020.107043
- Paulsson JO, Zedenius J, Juhlin CC. TERT Promoter Mutated Follicular thyroid carcinomas exhibit a distinct microRNA expressional profile with potential implications for tumor progression. Endocr Pathol. 2021;32(4):513–516. doi:10.1007/s12022-021-09695-w
- Kurihara-Shimomura M, Sasahira T, Shimomura H, Nakashima C, Kirita T. The oncogenic activity of miR-29b-1-5p induces the epithelial-mesenchymal transition in oral squamous cell carcinoma. J Clin Med. 2019;8:2. doi:10.3390/jcm8020273
- Liu P, Xia P, Fu Q, et al. miR-199a-5p inhibits the proliferation of hepatocellular carcinoma cells by regulating CDC25A to induce cell cycle arrest. Biochem Biophys Res Commun. 2021;571:96–103. doi:10.1016/j.bbrc.2021.07.035
- Cho Y, Park MJ, Kim K, et al. Tumor-stroma crosstalk enhances REG3A expressions that drive the progression of hepatocellular carcinoma. Int J Mol Sci. 2020;21:2. doi:10.3390/ijms21020472
- Sheng W, Zhang ZC, Shi DY, et al. Epigenetic silencing of SFRP5 promotes the metastasis and invasion of chondrosarcoma by expression inhibition and Wnt signaling pathway activation. Chem Biol Interact. 2018;296:1–8. doi:10.1016/j.cbi.2018.08.020
- Zhou W, Ye C, Li L, et al. Adipocyte-derived SFRP5 inhibits breast cancer cells migration and invasion through Wnt and epithelial-mesenchymal transition signaling pathways. Chin J Cancer Res. 2020;32(3):347–360. doi:10.21147/j.issn.1000-9604.2020.03.06
- Wang B, Huang G, Wang D, et al. Null genotypes of GSTM1 and GSTT1 contribute to hepatocellular carcinoma risk: evidence from an updated meta-analysis. J Hepatol. 2010;53(3):508–518. doi:10.1016/j.jhep.2010.03.026
- Li X, Song H, Liu Z, Bi Y. miR-1260b promotes cell migration and invasion of hepatocellular carcinoma by targeting the regulator of G-protein signaling 22. Biotechnol Lett. 2018;40(1):57–62. doi:10.1007/s10529-017-2455-6
- Liu DR, Guan QL, Gao MT, Jiang L, Kang HX. miR-1260b is a potential prognostic biomarker in colorectal cancer. Med Sci Monit. 2016;22:2417–2423. doi:10.12659/MSM.898733
- Xu L, Xu X, Huang H, et al. MiR-1260b promotes the migration and invasion in non-small cell lung cancer via targeting PTPRK. Pathol Res Pract. 2018;214(5):776–783. doi:10.1016/j.prp.2018.02.002
- Yamamoto K, Okamoto A, Isonishi S, Ochiai K, Ohtake Y. A novel gene, CRR9, which was up-regulated in CDDP-resistant ovarian tumor cell line, was associated with apoptosis. Biochem Biophys Res Commun. 2001;280(4):1148–1154. doi:10.1006/bbrc.2001.4250
- Carkic J, Nikolic N, Radojevic-Skodric S, et al. The role of TERT-CLPTM1L SNPs, hTERT expression and telomere length in the pathogenesis of oral squamous cell carcinoma. J Oral Sci. 2016;58(4):449–458. doi:10.2334/josnusd.16-0108
- Yang YC, Fu WP, Zhang J, Zhong L, Cai SX, Sun C. rs401681 and rs402710 confer lung cancer susceptibility by regulating TERT expression instead of CLPTM1L in East Asian populations. Carcinogenesis. 2018;39(10):1216–1221. doi:10.1093/carcin/bgy084
- Zhang Y, Zhang X, Zhang H, et al. Common variations in TERT-CLPTM1L locus are reproducibly associated with the risk of nasopharyngeal carcinoma in Chinese populations. Oncotarget. 2016;7(1):759–770. doi:10.18632/oncotarget.6397
- Lee HW, Park WJ, Heo YR, Park TI, Park SY, Lee JH. TERT-CLPTM1 locus polymorphism (rs401681) is associated with the prognosis of hepatocellular carcinoma. Onco Targets Ther. 2017;10:4853–4858. doi:10.2147/OTT.S138956
- Zhan M, He K, Xiao J, et al. LncRNA HOXA11-AS promotes hepatocellular carcinoma progression by repressing miR-214-3p. J Cell Mol Med. 2018;22(8):3758–3767. doi:10.1111/jcmm.13633
- Ke AW, Shi GM, Zhou J, et al. Role of overexpression of CD151 and/or c-Met in predicting prognosis of hepatocellular carcinoma. Hepatology. 2009;49(2):491–503. doi:10.1002/hep.22639