Abstract
Niclosamide, an anthelmintic drug approved by the US Food and Drug Administration against cestodes, is used to treat tapeworm infection. In this study, we show that niclosamide can potentially inhibit signal transducer and activator of transcription 3 (STAT3) in colon cancer cell lines. Combined inhibition of epidermal growth factor receptor and STAT3 by erlotinib and niclosamide synergistically induces apoptosis and antiproliferation in colon cancer cell lines. Our findings suggest that erlotinib and niclosamide combination provides an effective therapeutic approach to improving the prognosis of colon cancer.
Introduction
Colon cancer is the third most common cancer worldwide and the second leading cause of cancer death in the US.Citation1 The mortality rate of patients with colon cancer has decreased owing to improved detection, but the incidence of colon cancer continues to increase.Citation2 Considerable progress in the development of chemotherapy for advanced colon cancer has been observed in the recent decade.Citation3 The epidermal growth factor receptor (EGFR) has been reported as a critical therapeutic target for the treatment of colon cancer.Citation4 However, an anti-EGFR agent used to treat colon cancer has exhibited moderate efficacy.Citation5 The mechanisms of this resistance have yet to be elucidated. Nevertheless, the potential of EGFR inhibitors to induce activation of the EGFR-independent pathway is under consideration.Citation6,Citation7
Signal transducer and activator of transcription (STAT) proteins significantly influence diverse biological processes, including cell proliferation, differentiation, survival, and inflammatory response.Citation8,Citation9 One of the well-studied STAT members, STAT3, has been shown to be an important molecule in various malignancies and verified as an effective target for cancer therapy, including endometrial, cervical, breast, brain, prostate, and colon cancer.Citation10–Citation13 A number of studies have indicated that aberrant STAT3 activation contributes to cell proliferation, differentiation, migration, and survival.Citation14,Citation15
Niclosamide, particularly effective against cestodes, has been used to treat tapeworm infection for approximately 50 years.Citation16 Finding a new use for traditional medicines is considerably easier than inventing a new drug because traditional medicines have known pharmacokinetics and have often been approved for human treatments.Citation17 In the past 5 years, niclosamide has been identified as a potential anticancer agent targeting multiple signaling pathways (eg, NF-κB, ROS, Notch, Wnt/b-catenin, and mTORc1).Citation18–Citation21 Recently, several studies have reported that niclosamide exhibits antiproliferative activity in head and neck, ovarian, breast, and hematologic cancer.Citation22–Citation25 Till date, no studies demonstrating the therapeutic efficacy of niclosamide in colon cancer are available. Taking this background into account, the present study aimed to investigate the effects of niclosamide on colon cancer and the molecular mechanisms underlying its therapeutic action.
Our outcomes demonstrated that niclosamide acted as a potent STAT3 inhibitor in colon cancer cell lines. The combination of niclosamide and erlotinib induced apoptosis and antiproliferation in colon cancer cell lines as well as sensitization of colon cancer cells to erlotinib. On the basis of our findings, we suggest that combination of niclosamide and erlotinib can potentially improve the prognosis of colon cancer.
Materials and reagents
Cell culture and antibodies
Human colon cancer cell lines (HCT116, SW620, and HT29) were obtained from Cell Resources center of the Shanghai Institutes for Biological Sciences (Chinese Academy of Sciences, Shanghai, People’s Republic of China). The cells were routinely cultured in RPMI-1640 (Gibco/BRL; Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT, USA) and 1% of antibiotic solution (100 units/mL penicillin and 100 µg/mL streptomycin) in a humidified atmosphere of 5% CO2 at 37°C.
Antibodies such as anti-BCL-2, anti-Bax, anti-cleaved PARP, anti-GAPDH, horseradish peroxidase (HRP)- conjugated goat anti-mouse IgG, and HRP-conjugated donkey anti-rabbit IgG were purchased from Santa Cruz Biotechnology (Dallas, TX, USA). Antibodies against Phospho-STAT3 and STAT3 were purchased from Cell Signaling Technology (Danvers, MA, USA).
Inhibitor drugs
Nifuroxazide, niclosamide, cryptotanshinone, and alantolactone were purchased from Selleck Chemicals (Houston, TX, USA). Nifuroxazide is an oral nitrofuran antibiotic not currently approved for use in the USA but is used elsewhere as an antidiarrheal agent.Citation26 Niclosamide has been used to treat tapeworm infection and also as a molluscicide for water treatment in schistosomiasis control programs.Citation27 Cryptotanshinone, a major lipophilic component isolated from Salvia miltiorrhiza Bunge, has been shown to possess chemotherapeutic properties against various types of cancer cells.Citation28 Alantolactone is used as an insect repellent, antibacterial, and anticancer agent.Citation29 The drug compounds were dissolved in sterile dimethyl sulfoxide (DMSO) to produce a 20 mM stock solution, which was then stored at −20°C and further diluted freshly with cell culture medium.
MTT assay
Cells were seeded into wells of a 96-well plate at 5×103 cells per well in 100 µL of the corresponding medium. The cells were then treated with drugs at different concentrations for 72 h. Subsequently, they were treated with a fresh solution of MTT (5 mg/mL) for 4 h at 37°C. The purple formazan crystals were finally solubilized with DMSO solution, and absorbance was recorded using a multi-well plate reader at 490 nm.
Western blot analysis
Cells were lysed in a lysis buffer containing a phosphatase inhibitor, and the lysates were clarified by centrifugation (12,000 rpm) at 4°C for 10 min. The supernatant was run on 10% and 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride membrane. After being blocked with 5% nonfat dry milk in Tris Buffer Solution Tween for 1.5 h, membranes were incubated with a specific primary antibody of 1:1,000 dilution overnight and a HRP-conjugated secondary antibody of 1:3,000 dilution for 1 h. Immunoreactive bands were visualized using enhanced chemiluminescence reagent.
Molecular docking
As one of the most widely used computational approaches for structure-based drug design, molecular docking study was used to predict the binding pose of compound in STAT3 SH2-binding site by using the software AutoDock (version 4.2.6).Citation30 The crystallographic coordinate for human STAT3 SH2 (Protein Data Bank [PDB] ID: 1BG1) was obtained from the PDB.Citation31 Prior to docking, protein structures were prepared by removing water molecules and other ligands using PyMol software.Citation32 A grid box size of 60×60×60 dimensions with a spacing of 0.375 Å between the grid points was implemented and covered almost the entire SH2-binding site. The grid parameter files were created setting up the map files directly. The Lamarckian genetic algorithm was applied to deal with the interactions of protein and inhibitors. The number of individuals in population was set to 300, and trials of 100 dockings and maximum number of energy evaluations were set as default along with other settings. AutoDockTools version 1.5.6 and PyMol were used to analyze the docking results.
Clonogenic assay
A total of 500 cells per well were seeded into a 6-well plate with 2 mL of RPMI-1640 and incubated overnight. The cells were then pretreated with nifuroxazide and erlotinib or DMSO for 8–12 h. After treatment, the cells were washed with phosphate buffer saline (PBS) twice and transferred to a fresh medium to grow for 7 days. Colonies were washed with PBS and then fixed with 4% methanol for 15 min at room temperature. The cells were washed with PBS twice and stained with 1% crystal violet (25% methanol) for 10 min at room temperature. Each experiment was conducted thrice.
Analysis of cell apoptosis
Cells (3×105) were seeded in 6-well plates and incubated overnight and then treated with nifuroxazide and erlotinib for 24 h. After treatment, the cells were harvested with trypsin and then washed with cold PBS twice. The cells were stained with Annexin V for 10 min under dark conditions and then with propidium iodide (PI) for 5 min. Apoptotic cells were counted using the FACS Calibur flow cytometer and quantified by flow cytometric analysis.
Statistical analyses
Data are represented as mean ± standard error of the mean of 3 independent experiments. Student’s t-test was performed to determine the statistical significance between 2 groups by using GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA, USA). Differences between groups were analyzed by the log-rank test using GraphPad Prism 6.0. P<0.05 was considered statistically significant.
Results
Antiproliferative effects of niclosamide in human colon cancer cells
Nifuroxazide acts as a potent inhibitor of STAT3 signaling pathway in breast cancer cells, though it has little effect on cells lacking STAT3 activation.Citation33 Niclosamide has recently been identified to target multiple signaling pathways (eg, NF-κB, ROS, Notch, and STAT3).Citation16 Cryptotanshinone has previously been observed to possess the most powerful antibacterial, anti-inflammatory, and antitumor effect.Citation34 Alantolactone has an inhibitory effect on cancer cells migration, invasion, adhesion, and colony formation.Citation35 Therefore, we screened the antiproliferative effects of these 4 compounds in human colon cancer cells by MTT assay.
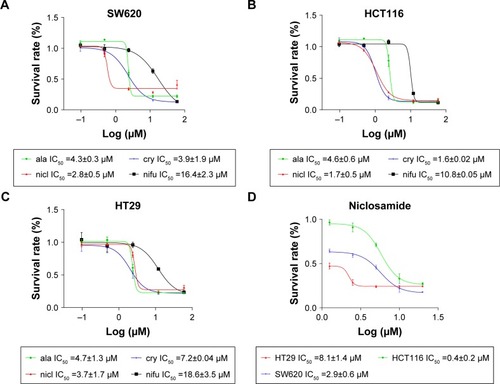
After the cells were treated with nifuroxazide, niclosamide, cryptotanshinone, and alantolactone for 72 h, MTT assay was employed. As shown in , administration of these 4 compounds in a dose-dependent manner reduced viability in SW620 (), HCT116 (), and HT29 cells (). Among these compounds, niclosamide exhibited the most potent antiproliferative effect against all tested human colon cancer cell lines. After exposure of the cells to niclosamide for 72 h, the half maximal inhibitory concentration (IC50) in SW620, HCT116, and HT29 cell lines was found to be 2.9, 0.4, and 8.1 µM, respectively (). This suggests that niclosamide can inhibit the proliferation of human colon cancer cells.
Figure 1 Antiproliferative effects of niclosamide in human colon cancer cells.
Abbreviation: IC50, half maximal inhibitory concentration.
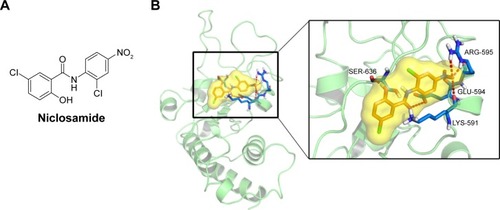
Molecular docking between STAT3 and niclosamide
STAT3, a member of the STAT family of transcription factors, has recently been verified as an attractive therapeutic target and shown to be an important molecule in cancer therapy, including colon cancer.Citation36 As the STAT3 SH2 domain is critical for the activation and biological function of STAT3,Citation37 we investigated interactions between the compound and the SH2 domain of STAT3 by molecular docking analysis of the active compound niclosamide (). We applied PyMol software to remove water molecules and other ligands to prepare the STAT3 SH2 domain protein structures and used the software AutoDock (version 4.2.6) to predict the binding pose of compound in STAT3 SH2-binding site to identify whether compound niclosamide acts as a STAT3 inhibitor that targets the STAT3 SH2 domain.Citation38 The molecular docking results indicated that niclosamide formed 4 hydrogen bonds with the side chain of ARG-595, SER-636, GLU-594, and LYS-591 as shown in . It was predicted that niclosamide could fit into the 2 major binding sites, the pTyr705 and the side pocket site, and so it could inhibit STAT3 phosphorylation. In summary, critical hydrogen bond interaction between niclosamide and STAT3 SH2 domain were predicted by a rational method combined with molecular docking. These methods clarified the interaction of niclosamide with STAT3. Further verification of co-crystallization of STAT3 and niclosamide is in progress.
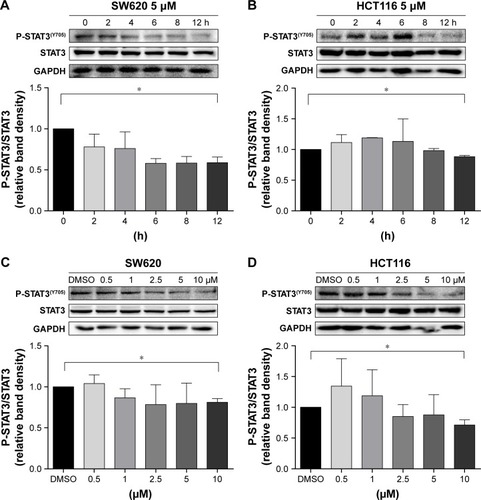
Inhibition of STAT3 phosphorylation by niclosamide in colon cancer cells HCT116 and SW620
We performed Western blot analysis on colon cancer cell lines to examine whether niclosamide could inhibit STAT3. Western blot analysis indicated that niclosamide inhibited STAT3 phosphorylation (). Notably, niclosamide inhibited STAT3 phosphorylation in a time- and dose-dependent manner in HCT116 and SW620 cell lines.
Figure 3 Inhibition of STAT3 phosphorylation by niclosamide in colon cancer cells HCT116 and SW620.
Abbreviation: DMSO, dimethyl sulfoxide.
Sensitization of colon cancer cells to erlotinib by treatment of cells with niclosamide
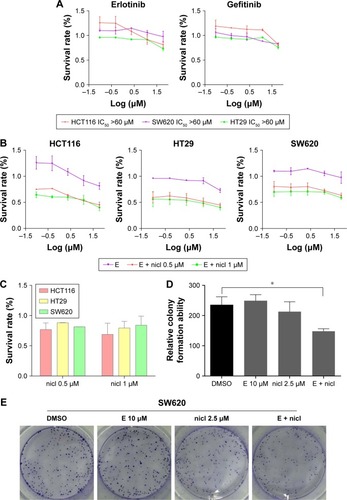
Erlotinib or gefitinib, EGFR tyrosine kinase inhibitors, have been shown to benefit patients with non-small-cell lung cancer and pancreatic cancer. However, almost all patients develop a progressive disease during therapy. As shown in , after treatment of cells with erlotinib and gefitinib for 72 h, the IC50 in SW620, HCT116, and HT29 cells exceeded 60 µM. These results indicate that erlotinib and gefitinib could not inhibit the proliferation of SW620, HCT116, and HT29 cell lines. We thus examined the anti-proliferative effects of erlotinib as well as erlotinib combined with niclosamide by MTT assay. As shown in , treatment of cells with niclosamide sensitized colon cancer cells to erlotinib, while niclosamide had no effect on SW620, HCT116, and HT29 cells at 0.5 µM and 1 µM concentrations (). To verify the anticancer properties of the combination, we performed colony-forming experiments. SW620 cells (5000) were seeded in a 6-well plate overnight and then treated with erlotinib (10 µM) or/and niclosamide (2.5 µM) for 1 week. We then removed the drugs and washed the cells with PBS twice. The cell colonies were stained with crystal violet and then counted. The combination of niclosamide and erlotinib suppressed the colony formation of colon cancer cells, as shown in . Colony formation assay demonstrated that the synergistic effect of niclosamide and erlotinib efficiently inhibits the growth of colon cancer cells.
Figure 4 Sensitization of colon cancer cells to erlotinib by niclosamide.
Abbreviations: DMSO, dimethyl sulfoxide; E, erlotinib, nicl, niclosamide.
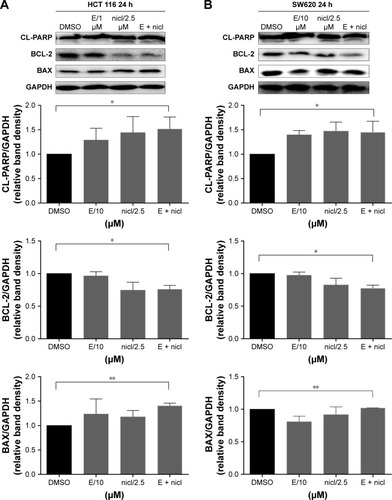
Induction of apoptosis in colon cancer cells with combined niclosamide and erlotinib
Compared with single-agent treatment, the combination of erlotinib and niclosamide enhanced the inhibition of cell viability in all tested cell lines (). We sought to investigate whether the combination of erlotinib and niclosamide could also induce cell apoptosis. We first employed Western blot analysis. HCT116 and SW620 cells were treated with erlotinib and niclosamide alone, or with both for 24 h. The combination significantly increased cleaved PARP and BAX, as shown in . BCL-2 expression was markedly decreased.
Figure 5 Induction of apoptosis in colon cancer cells by combined niclosamide and erlotinib.
Abbreviations: DMSO, dimethyl sulfoxide; E, erlotinib, nicl, niclosamide.
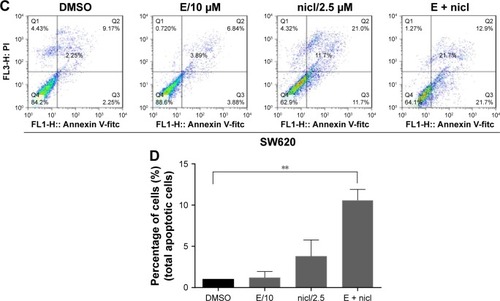
We also used flow cytometry to examine whether the combination induced cell apoptosis in SW620 cells. SW620 cells were treated with erlotinib (10 µM) or/and niclosamide (2.5 µM) for 24 h and then stained with Annexin V and PI for apoptosis analysis. The combination resulted in a significant increase in early (Annexin V+, PI−) and late apoptotic cells (Annexin V+, PI+), as shown in .
Discussion
Colon cancer is one of the most prevalent tumors worldwide.Citation39 Despite conventional therapies such as surgery, radiation, and chemotherapy, the overall survival rate for colon cancer has not significantly improved.Citation40 The human EGFR is overexpressed in various gastrointestinal cancer types, including 60%–80% of colon cancers, correlating with poor prognosis and early disease progression.Citation41 In addition, cancer cell also upregulates other important downstream genes such as STAT3, which contributes to cell proliferation, cell survival, and angiogenesis in colon cancer.Citation42 Therefore, more effective approaches need to be developed to improve the prognosis of colon cancer.
Here, our results showed that niclosamide reduces cell viability in a dose-dependent manner. This finding suggests that niclosamide could be effective against colon cancer. Several groups have independently revealed the activity of niclosamide against cancer cells;Citation16,Citation27,Citation43 however, the precise mechanism underlying this antitumor activity has yet to be elucidated. Therefore, we first used molecular docking study to verify whether there is interaction between niclosamide and STAT3. In our study, we found critical hydrogen bond interaction between niclosamide and STAT3 SH2 domain. We further investigated the correlation between niclosamide and STAT3 in SW620 and HCT116 cell lines, and the results showed that niclosamide suppressed STAT3 phosphorylation in a time- and dose-dependent manner. Several studies have revealed that inhibition of EGFR by erlotinib induces activation of STAT3, which contributes to erlotinib resistance.Citation44,Citation45 Our data showed that when erlotinib and niclosamide were combined, they synergistically induced apoptosis of colon cancer cells. It has been reported that multiple signal transduction pathway inhibitors, including the mitogen-activated protein kinase, fibroblast growth factor receptor inhibitors, and chemotherapy drugs, can induce activation of STAT3 survival signaling pathway, leading to resistance.Citation38,Citation46 It is possible that, in addition to erlotinib, niclosamide may also sensitize these target drugs. Based on our findings, we conclude that niclosamide may represent a novel and more effective drug for colon cancer.
Drug development process from initial lead discovery to final medication is costly, lengthy, and incremental.Citation33 Finding new uses for old or natural products is much easier and more economical than inventing a new drug from scratch. The present study demonstrates that niclosamide can inhibit the cell growth of colon cancer by inhibiting STAT3 phosphorylation. We also investigated the combined inhibition of EGFR and STAT3 by using erlotinib and niclosamide to synergistically repress colon cancer. This study was conducted only in vitro; some compounds may exhibit potent antitumor activities in vitro but show no antitumor activity in vivo. Therefore, further molecular investigations in preclinical and clinical settings are warranted to evaluate anticancer potential in a synergistic manner to substantiate niclosamide as an effective therapeutic candidate for colon cancer.
Acknowledgments
This research was supported by the National Natural Science Fund of China (21472142), the Zhejiang Provincial Natural Science Fund (LY16H160052, LY17H160050, and LY17H160055), Medical Scientific Research Fund of Zhejiang Province (2017192276), and Wenzhou Science and Technology Project (Y20160052). The authors thank Dr Jianzhang Wu for helpful discussions and assistance in writing the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
- SiegelRLMillerKDJemalACancer statistics, 2016CA Cancer J Clin201666173026742998
- ZahidAYoungCJHow to decide on stent insertion or surgery in colorectal obstruction?World J Gastrointest Surg201681848926843916
- YoungJIMongoue-TchokoteSWieghardNTreatment and survival of small-bowel adenocarcinoma in the United States: a comparison with colon cancerDis Colon Rectum201659430631526953989
- de Castro-CarpeñoJBelda-IniestaCCasado SáenzEHernández AgudoEFeliu BatlleJGonzález BarónMEGFR and colon cancer: a clinical viewClin Transl Oncol200810161318208787
- KuwaiTNakamuraTSasakiTPhosphorylated epidermal growth factor receptor on tumor-associated endothelial cells is a primary target for therapy with tyrosine kinase inhibitorsNeoplasia200810548950018472966
- LiRYouSHuZInhibition of STAT3 by niclosamide synergizes with erlotinib against head and neck cancerPLoS One201389e7467024019973
- KangYHuWBaiECurcumin sensitizes human gastric cancer cells to 5-fluorouracil through inhibition of the NFkB survival-signaling pathwayOnco Targets Ther201697373738427980427
- YuHPardollDJoveRSTATs in cancer inflammation and immunity: a leading role for STAT3Nat Rev Cancer200991179880919851315
- ShodeindeALBartonBEPotential use of STAT3 inhibitors in targeted prostate cancer therapy: future prospectsOnco Targets Ther2012511912522815644
- LeeHDengJKujawskiMSTAT3-induced S1PR1 expression is crucial for persistent STAT3 activation in tumorsNat Med201016121421142821102457
- LesinaMKurkowskiMULudesKStat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancerCancer Cell201119445646921481788
- GaoSPMarkKGLeslieKMutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomasJ Clin Invest2007117123846385618060032
- HauraEBZhengZSongLCantorABeplerGActivated epidermal growth factor receptor-Stat-3 signaling promotes tumor survival in vivo in non-small cell lung cancerClin Cancer Res200511238288829416322287
- CrescenzoRAbateFLasorsaEEuropean T-Cell Lymphoma Study Group, T-Cell Project: Prospective Collection of Data in Patients with Peripheral T-Cell Lymphoma and the AIRC 5xMille Consortium “Genetics-Driven Targeted Management of Lymphoid Malignancies”Convergent mutations and kinase fusions lead to oncogenic STAT3 activation in anaplastic large cell lymphomaCancer Cell201527451653225873174
- WakeMSWatsonCJSTAT3 the oncogene – still eluding therapy?FEBS J2015282142600261125825152
- PanJXDingKWangCYNiclosamide, an old antihelminthic agent, demonstrates antitumor activity by blocking multiple signaling pathways of cancer stem cellsChin J Cancer201231417818422237038
- ChongCRSullivanDJJrNew uses for old drugsNature2007448715464564617687303
- JinYLuZDingKAntineoplastic mechanisms of niclosamide in acute myelogenous leukemia stem cells: inactivation of the NF-kappaB pathway and generation of reactive oxygen speciesCancer Res20107062516252720215516
- WangAMKuHHLiangYCChenYCHwuYMYehTSThe autonomous notch signal pathway is activated by baicalin and baicalein but is suppressed by niclosamide in K562 cellsJ Cell Biochem2009106468269219160421
- OsadaTChenMYangXYAntihelminth compound niclosamide downregulates Wnt signaling and elicits antitumor responses in tumors with activating APC mutationsCancer Res201171124172418221531761
- BalgiADFonsecaBDDonohueEScreen for chemical modulators of autophagy reveals novel therapeutic inhibitors of mTORC1 signalingPLoS One200949e712419771169
- ZouMZhangXXuCIL6-induced metastasis modulators p-STAT3, MMP-2 and MMP-9 are targets of 3,3′-diindolylmethane in ovarian cancer cellsCell Oncol (Dordr)2016391475726510945
- EiringAMPageBDKraftILCombined STAT3 and BCR-ABL1 inhibition induces synthetic lethality in therapy-resistant chronic myeloid leukemiaLeukemia201529358659725134459
- LiuJChenXWardTPegramMShenKCombined niclosamide with cisplatin inhibits epithelial-mesenchymal transition and tumor growth in cisplatin-resistant triple-negative breast cancerTumour Biol20163779825983526810188
- JinBWangCLiJDuXDingKPanJAnthelmintic Niclosamide Disrupts the Interplay of p65 and FOXM1/β-catenin and Eradicates Leukemia Stem Cells in Chronic Myelogenous LeukemiaClin Cancer Res201723378980327492973
- ZhuYYeTYuXNifuroxazide exerts potent anti-tumor and anti-metastasis activity in melanomaSci Rep201662025326830149
- YeTXiongYYanYThe anthelmintic drug niclosamide induces apoptosis, impairs metastasis and reduces immunosuppressive cells in breast cancer modelPLoS One201491e8588724416452
- ZhuZZhaoYLiJCryptotanshinone, a novel tumor angiogenesis inhibitor, destabilizes tumor necrosis factor-α mRNA via decreasing nuclear-cytoplasmic translocation of RNA-binding protein HuRMol Carcinog201655101399141026310813
- KhanMYiFRasulAAlantolactone induces apoptosis in glioblastoma cells via GSH depletion, ROS generation, and mitochondrial dysfunctionIUBMB Life201264978379422837216
- MorrisGMHueyRLindstromWAutoDock4 and AutoDockTools4: automated docking with selective receptor flexibilityJ Comput Chem200930162785279119399780
- BeckerSGronerBMüllerCWThree-dimensional structure of the Stat3-beta homodimer bound to DNANature199839466891451519671298
- AlexanderNWoetzelNMeilerJBcl::Cluster: a method for clustering biological molecules coupled with visualization in the Pymol Molecular Graphics SystemIEEE Int Conf Comput Adv Bio Med Sci201120111318
- YangFHuMLeiQNifuroxazide induces apoptosis and impairs pulmonary metastasis in breast cancer modelCell Death Dis20156e170125811798
- YuHJParkCKimSJChoNPChoSDSignal transducer and activators of transcription 3 regulates cryptotanshinone-induced apoptosis in human mucoepidermoid carcinoma cellsPharmacogn Mag201410Suppl 3S622S62925298683
- ChunJLiRJChengMSKimYSAlantolactone selectively suppresses STAT3 activation and exhibits potent anticancer activity in MDA-MB-231 cellsCancer Lett2015357139340325434800
- ZhaoCXiaoHWuXRational combination of MEK inhibitor and the STAT3 pathway modulator for the therapy in K-Ras mutated pancreatic and colon cancer cellsOncotarget2015616144721448725961376
- ZhaoCLiHLinHJYangSLinJLiangGFeedback activation of STAT3 as a cancer drug-resistance mechanismTrends Pharmacol Sci2016371476126576830
- van KesselKEZuiverloonTCAlbertsARBoormansJLZwarthoffECTargeted therapies in bladder cancer: an overview of in vivo researchNat Rev Urol2015121268169426390971
- ChorostMIDattaRSantiagoRCColon cancer screening: where have we come from and where do we go?J Surg Oncol200485171314696082
- Azcárate-PerilMASikesMBruno-BárcenaJMThe intestinal microbiota, gastrointestinal environment and colorectal cancer: a putative role for probiotics in prevention of colorectal cancer?Am J Physiol Gastrointest Liver Physiol20113013G401G42421700901
- PohlMStrickerISchoeneckAAntitumor activity of the HER2 dimerization inhibitor pertuzumab on human colon cancer cells in vitro and in vivoJ Cancer Res Clin Oncol2009135101377138619340455
- WenWWuJLiuLSynergistic anti-tumor effect of combined inhibition of EGFR and JAK/STAT3 pathways in human ovarian cancerMol Cancer20151410025928246
- SackUWaltherWScudieroDNovel effect of antihelminthic Niclosamide on S100A4-mediated metastatic progression in colon cancerJ Natl Cancer Inst2011103131018103621685359
- WangXGoldsteinDCrowePJOvercoming resistance of targeted EGFR monotherapy by inhibition of STAT3 escape pathway in soft tissue sarcomaOncotarget2016716214962150926909593
- KraljevicSStambrookPJPavelicKAccelerating drug discoveryEMBO Rep20045983784215470377
- SakamotoKMGrantSSaleiroDTargeting novel signaling pathways for resistant acute myeloid leukemiaMol Genet Metab2015114339740225533111