Abstract
The numerous processes involved in the etiology of breast cancer such as cell survival, metabolism, proliferation, differentiation, and angiogenesis are currently being elucidated. However, underlying mechanisms that drive breast cancer progression and drug resistance are still poorly understood. As we discuss here in detail, the Notch signaling pathway is an important regulatory component of normal breast development, cell fate of normal breast stem cells, and proliferation and survival of breast cancer initiating cells. Notch exerts a wide range of critical effects through a canonical pathway where it is expressed as a type I membrane precursor heterodimer followed by at least two subsequent cleavages induced by ligand engagement to ultimately release an intracellular form to function as a transcriptional activator. Notch and its ligands are overexpressed in breast cancer, and one method of effectively blocking Notch activity is preventing its cleavage at the cell surface with γ-secretase inhibitors. In the context of Notch signaling, the application of clinically relevant anti-Notch drugs in treatment regimens may contribute to novel therapeutic interventions and promote more effective clinical response in women with breast cancer.
Introduction
In recent years, there have been many advances in deciphering critical cell signaling networks and their relationship to the driving forces of cancer onset, growth, and metastasis. Moreover, in the hierarchy of signaling pathways, several pathways are considered fundamental to regulation of cell fate and having widespread survival effects, namely the Notch, Wnt/Wingless (Wnt), and Hedgehog (HH) pathways.
This review will focus on the role of the canonical Notch signaling pathway in breast cancer etiology and progression. Furthermore, we will review the current therapeutic options available for inhibiting Notch. Blockade of an upregulated Notch signaling pathway can be achieved by inhibiting the formation of the main force of Notch activity, the Notch intracellular domain (NICD). Thus, a pharmacological approach using γ-secretase inhibitors (GSIs) to prevent the final cleavage step of the precursor form of Notch, ie, transmembrane Notch (Notch™) that will decrease levels of NICD could be a novel therapeutic strategy either as a single agent or in combination with targeted or cytotoxic chemotherapy for a subset of cancer patients.
Breast cancer subtypes
Breast cancer is a heterogeneous disease divided into four major subtypes: luminal A (estrogen receptor [ER]+/progesterone receptor [PR]+), luminal B (ER+/PR+/human epidermal growth factor receptor [HER]-2+), HER-2+/neu+, and triple negative (ER-PR-/HER-2+).Citation1,Citation2 Breast cancer of the luminal A or B subtype is derived from the luminal epithelium of the breast ducts, and these tumors express hormone receptors, ER and PR. These subtypes comprise 70%–80% of all breast cancers. The ER+/PR+ luminal A subtype is very sensitive to current antihormonal therapy such as tamoxifen, fulvestrant, or aromatase inhibitors. Luminal B breast tumors have a higher proliferative index than those of luminal A and are inherently more resistant to current antihormonal therapy. HER-2+/neu+ designates a breast cancer subtype that contains gene amplification for the ERBB2 proto-oncogene resulting in overexpression of the HER-2 receptor tyrosine kinase protein. The HER-2+/neu+ breast cancers are very sensitive to anti-HER-2 therapy such as trastuzumab or lapatinib. The final subtype of breast cancer is triple negative, which lacks expression of ER, PR, and HER-2. Triple negative breast tumors are the most aggressive, with poor prognosis and currently no approved targeted therapy. These triple negative breast tumors are treated with cytotoxic chemotherapy such as a DNA-damaging agent (cis- or carboplatin) or tubulin-destabilizing compounds (taxanes).
Although dramatic improvements have been made to cure breast cancer, one of the major problems that continue to plague both research scientists and clinicians is drug resistance. Therefore, elucidating the critical mechanisms that contribute to drug-resistant breast cancer will hopefully prevent tumor recurrence and disease progression and ultimately provide a “cure” to women with breast cancer.
Notch signaling
Over a century of research has revealed the mechanisms that regulate canonical Notch signaling in the context of cell-to-cell signaling that controls both embryonic and adult stem cell self-renewal, stem cell quiescence, cell fate and differentiation, cell survival, apoptosis, and tumorigenesis. Investigations elucidating the Notch pathway date back to the early 20th century, when in 1913 John Smith Dexter working in the laboratory of American geneticist Thomas Hunt Morgan observed the outcome of a mutation of a gene in Drosophila ampelophila, which resulted in a notch or indentation at the ends of the fly wings. He called them “perfect notched.”Citation3 Additional research in 1917 by Morgan identified the alleles of this fly gene which eventually became known as “Notch”Citation4 and he published his findings in The Physical Basis of HeredityCitation5 in 1919. The Notch gene was eventually cloned and identified for the first time in 1985–1986.Citation6,Citation7 Related research employing the nematode worm Caenorhabditis elegans further elucidated the Notch signaling pathway, cell-to-cell interactions, and lateral inhibition during embryogenesis. Presently, embryologists and cancer researchers are the largest groups of research scientists studying Notch signaling.
Developmental Notch
The Notch signaling pathway mediates cell fate determination in three ways: regulatory, inhibitory, and inductive action.Citation8 In regulatory signaling, during embryogenesis, Notch regulates the development and differentiation of many organ systems (angiogenesis, hematopoiesis, homeostasis, neurogenesis, nephrogenesis, myogenesis, and somatogenesis). The importance of this function was verified in Notch-1, Notch-2, Jagged-2, and Delta-1 knockout mice, which lack each Notch receptor or ligand regulatory components.Citation9–Citation12 The mice exhibited severe defects which resulted in embryonic or perinatal death. Notch activation in pluripotent stem cells initiates lateral inhibition so that a certain number of cells take on a specific cell fate and those adjacent are inhibited from differentiating.Citation13 This process is exemplified in the following experiment: nascent chick retinal neurons transiently overexpressing Deltex-1 were found to prevent adjacent neuroepithelial progenitor cells from differentiating into neurons.Citation14 Lastly, in inductive signaling, Notch promotes or induces the development of a certain cell type, usually amongst different (nonequivalent) cells. Such interactions are important for establishing demarcated boundaries between cell types, and the signaling is aptly referred to as boundary formation. The necessity of inductive signaling is evident in developmental studies. For example, a Notch-dependent localized signal affected the formation of the dorsoventral wing organizer in Drosophila,Citation15,Citation16 while the expression of Radical fringe determined the position of the dorsoventral boundary of vertebrate limbs.Citation17
Notch receptors
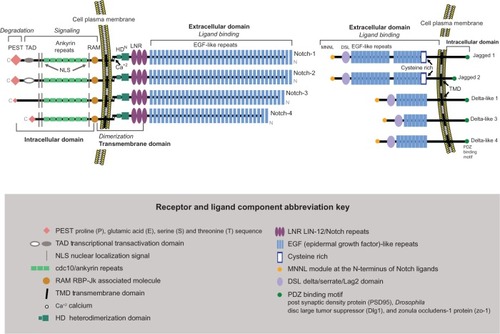
The Notch receptor is classified as a large single-pass type 1 transmembrane glycoprotein. It is expressed as a heterodimer at the cell membrane. There are four mammalian Notch receptors, Notch 1–4, with Notch-1 being the longest and Notch-4 the shortest.Citation13 They are comprised of three domains, extracellular, transmembrane, and intracellular (). Notch is synthesized as a single, relatively large (>300 kDa) polypeptide in the endoplasmic reticulum. There, it undergoes O-glycosylation. An initial addition of O-linked fucose to the epidermal growth factor (EGF)-like repeats is mediated by O-fucosyl transferase1. After the Notch preprotein is chaperoned by the guanosine triphosphate hydrolase (GTPase) Rab-protein 6 through the secretory pathway to the trans-Golgi network, it undergoes additional elongation of the O-fucose with carbohydrate chains on serine and threonine residues by the Fringe family O-fucose-specific β1,3-N acetylglucosaminyl-transferases Lunatic, Manic, or Radical.Citation18,Citation19 Modification of the Notch receptor by Fringe proteins controls ligand-mediated activation.Citation20 Next is cleavage by furin-like convertase into the N-terminus and C-terminus subunits and subsequent translocation of these mature entities to the cell plasma membrane.Citation21,Citation22 There, the cleaved subunits are assembled into the cell membrane as a fully functional heterodimeric receptor, noncovalently linked by a calcium cation awaiting engagement with a Notch ligand.
The N-terminal extracellular domain of each Notch receptor is the ligand-binding component and consists of 29–36 multiple EGF-like repeats in tandem. From each extracellular domain extend six cysteine residues, which form three intra-domain disulfide bridges. Adjacent to the extracellular domain and closer to the cell membrane is the transmembrane domain, a dual hybrid moiety. The two components of this domain are: (1) the juxtamembrane RAM23 section (the negative regulatory region) made of three Lin-12/Notch repeats, which prevent ligand-independent interactions, plus two conserved cysteine residues; and (2) the heterodimerization section, which maintains the Notch receptor in a nonactivated state. The third part of the Notch receptor is the intracellular domain (C-terminus), which extends from the inner cell membrane into the cytoplasm. It contains four separate entities: (1) the DNA-binding protein recombination signal-binding protein for immunoglobulin kappa J (RBP-JK associated molecule or RAM domain), followed by a linker with a nuclear localization sequence; (2) seven iterated cdc10/ankyrin repeats; (3) a transcription activation domain (TAD) with an additional nuclear localization sequence; and (4) polypeptide proline, glutamate, serine, and threonine-rich motifs (PEST) with degradation signals or degrons that stabilize NICD in the nucleus and target it for rapid proteolytic degradation. Lastly, TAD is found in Notch-1 and Notch-2, but not in Notch-3 and Notch-4.
Notch ligands and activation
In vertebrates, the Notch ligands are known as Delta-like 1, 3, and 4Citation13,Citation23–Citation26 and Jagged-1 and 2Citation13,Citation26 (). They are single-pass Type 1 transmembrane proteins that bind and activate the Notch receptor “in trans” (at the surface of a neighboring cell). They have extracellular and intracellular domains. The Jagged ligands are longer than the Delta-like ligands, the length determined by the 6–16 EGF-like repeats in the extracellular domain. A cysteine-rich area is located at the end of the EGF-like repeats, with Jagged ligands having an additional cysteine-rich area. The intracellular domain of each ligand has a shorter cytoplasmic tail than the extracellular domain and contains a PDZ (post synaptic density protein [PSD95], Drosophila disc large tumor suppressor [Dlg1], and zonula occludens-1 protein [zo-1])-binding motif which aids in intracellular protein–protein interactions. The ligand-activated cell-surface receptor initiates a cascade of events with two subsequent proteolytic cleavages that result in NICD entry into the nucleus to function as a transcriptional activator.Citation27–Citation29
Cell-cell contact mediates Notch ligand to receptor binding which initiates short-range cell-to-cell communication, a mono-directional cascade of events beginning at the cell membrane and ultimately activating the CSL (C promoter binding factor-1 [CBF-1], suppressor of hairless, Lag-1) family of transcription factors in the nucleus. The ligand engages the Notch receptor through its cognate high affinity EGF-like repeats (). Ligand-mediated endocytosis in the ligand-expressing cell (trans-endocytosis) provides a force to pull the extracellular domain of the Notch receptor from the transmembrane domain. This mechanical pull exposes the S2 cleavage site for the α-secretases of “A disintegrin and metalloprotease” family ADAM17 (tumor necrosis factor-α-converting enzyme TACE) or ADAM10, leading to ectodomain shedding of the extracellular portion of the transmembrane portion of the Notch receptor at approximately 12 amino acids outside the transmembrane domain.Citation30,Citation31 This proteolytic ectodomain “release” or shedding forms a carboxyterminal fragment called Notch extracellular truncation (NEXT).Citation32 The ligand-Notch extracellular portion undergoes trans-endocytosis into the ligand-expressing, signal-sending cell, followed by endosomal-mediated degradation or recycling. Monoubiquitination by E3 ligases Mindbomb-1 and -2 or Neuralized-1 and -2 marks the ligand for endocytosis.
Figure 2 Significant components in the Notch signaling pathway.
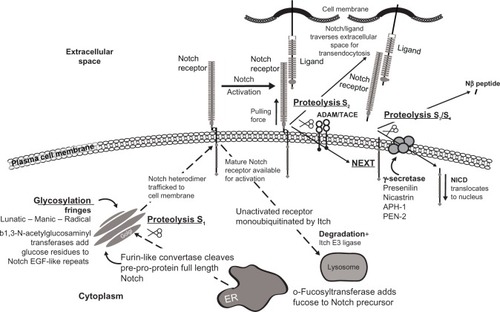
The remaining NEXT portion now exposes the S3 and S4 cleavage sites that are mediated by the γ-secretase complex.Citation33 Interestingly, there are many γ-secretase substrates, a great number having relevance in breast cancer.Citation34,Citation35 This transmembrane aspartyl proteinase, considered a large complex, is comprised of a catalytic subunit designated presenilin 1 or presenilin 2, a seven-pass transmembrane protein, and accessory subunits comprised of the transmembrane proteins nicastrin (NCT), anterior pharynx-defective 1 (APH1), and presenilin enhancer 2 (PEN-2), a two-pass transmembrane protein. Nicastrin and APH1 stabilize PEN-2, which induces endoproteolysis of presenilin.Citation36 Following receptor activation, NICD that is still attached to the inner cell membrane is marked for proteosomal degradation by E3 ubiquitin ligases Numb and Itch. γ-secretase severs NICD from the inside of the cell membrane, allowing it to enter the cytoplasmCitation37 and eventually translocate to the nucleus ().Citation38
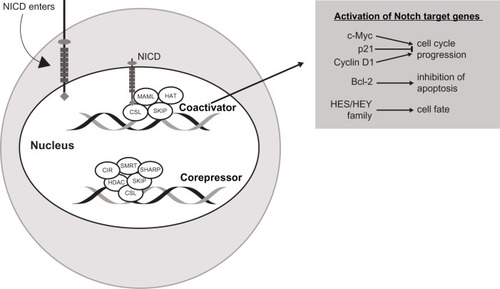
NICD forms a transcriptional activation complex with CSL in the nucleus once the ankyrin-repeat motif of NICD docks with the Rel homology region of the DNA-binding factor CSL. Thus, CSL changes from a transcriptional repressor to a transcriptional activator. There occurs a release of transcription factor co-repressors (CoRs) like class I or II histone deacetylases, CBF-1-interacting repressor (CIR), SKI-interacting protein (SKIP), silencing mediator of retinoid and thyroid hormone receptor (SMRT), and SMRT/HDAC (histone deacetylase)-1-associated repressor protein (SHARP), and a recruitment of transcription factor co-activators (CoAs) such as mastermind-like 1–3 (MAML) protein. MAML further recruits the histone acetyltransferases, cyclic AMP (adenosine monophosphate) response element-binding (CREB) protein CBP/p300 and p300/CBP-associated factor or general control non-depressible 5 (GCN5), to acetylate histone tails for the unwinding of nucleosomes within chromatin for active transcription. This leads to an increased expression of specific genes. Some of the Notch gene targets that can be activated are: c-Myc, p21, and cyclin D1 (cell cycle progression), Bcl-2 (inhibition of apoptosis), and hairy and enhancer of split basic helix-loop-helix HES 1, 5, 6, and 7, and HEY 1 and 2, and HEY-L family of proteins (transcriptional repressors).Citation37 NICD activity in the nucleus ends with phosphorylation triggered by cyclin c-cyclin-dependent kinase 8 (C-CDK8). Subsequently, glycogen synthase kinase 3β phosphorylates the PEST domain of the C-terminus of the NICD, which is then targeted for polyubiquitination by E3 ligase SEL10/FWB7 in the proteosome.Citation39 depicts Notch-mediated nuclear transcription.
Figure 3 Notch-mediated nuclear transcription.
Notch and cancer: general overview
One of the earliest associations between Notch signaling and cancer occurred in 1991 in human T-cell acute lymphoblastic leukemia, where the Notch-1 gene was associated with the t(7;9)(q34;q34.3) chromosomal translocation.Citation40 Notch cell signaling defects were detected in the form of alterations in the Notch-1 negative regulatory region and a loss of the C-terminus PEST domain, both of which lead to increased Notch-1 intracellular domain (N-1ICD) activity.Citation41 In B-cell malignancies such as chronic lymphocytic leukemia, Notch-1 mutations were linked to increased disease progression and resistance to chemotherapy.Citation42 Inconsistencies in the role of Notch in malignant B-cells became apparent as some data indicated that Notch signaling inhibited B-cell growth,Citation43–Citation45 while other data reported a Notch-induced increase in B-cell proliferation.Citation46–Citation48 In mantle cell lymphoma, Notch-1Citation49 or Notch-2Citation50 PEST domain mutations were reported. In addition to the presence of dysfunctional Notch receptors in leukemia, the ligand Jagged-2 was found to be significantly overexpressed in multiple myeloma.Citation51 In addition to hematologic malignancies, aberrant Notch signaling has been found in solid tumors; for example, cervical,Citation52,Citation53 colon,Citation54,Citation55 liver,Citation56,Citation57 lung,Citation58 pancreatic,Citation59–Citation62 prostate,Citation63,Citation64 ovarian,Citation65,Citation66 and renal.Citation67 Indeed, based on the numerous reports on the role of Notch signaling in cancer development and progression, Notch signaling has become a major target for novel therapeutic strategies.Citation68–Citation72 The role of Notch signaling in cancer could possibly be a double-edge sword. It was reported that Notch receptors and ligands were both oncogenic and tumor-suppressive in the same tumor.Citation73 The possibility that Notch promotes or suppresses tumor growth has also been put forth by others.Citation74–Citation76 Some discrepancies in Notch signaling in cancer may be explained in part by “cell context, dose, and timing,”Citation77 as well as Notch cross-talk with other signaling pathways, the micro-tumor environment, and the stage of cancer at the time of detection.
Notch and breast cancer
There is strong evidence that Notch signaling is dysregulated in solid tumors,Citation28,Citation76,Citation78 though as reported in leukemia, it may be both a tumor oncogene and suppressor in breast and other cancers.Citation79 In mouse studies, tissue specific expression of N-1ICD induces spontaneous mammary tumors.Citation80–Citation82 Furthermore, transgenic (Tg) mice expressing mammary specific N-4ICD also form spontaneous mammary tumors.Citation83 In fact, Notch-1 and Notch-4 are categorized as bonafide breast oncogenes.Citation84 Further studies showed that overexpression of Notch-3Citation81 and Notch-4Citation85–Citation87 also leads to murine mammary tumor formation. Studies from human breast cancer cell lines show deregulated expression of Notch and Notch ligand messenger RNA (mRNA).Citation88 Results from a human xenograft model for inflammatory breast carcinoma (MARY-X) implicate altered Notch-3 signaling specificalxly.Citation89 In a study of 200 Greek women from differing breast cancer subtypes, Notch-4 mRNA levels were found to be higher in the hormone receptor and HER-2-positive breast cancers, while Notch-1 and Notch-3 mRNA levels were higher in triple-negative specimens compared with normal tissue.Citation90 When Notch-1 and Notch-4 and Jagged-1 and Delta-like-1 expression were measured by immunohistochemistry in breast hyperplasia and carcinomas, high levels of Notch-1 were found in the hyperplasias, ductal carcinoma in situ, infiltrating ductal carcinomas (IDCs), and infiltrating lobular carcinomas (ILCs), as well as elevated expression of Notch-4 and Jagged-1 in IDCs and ILCs.Citation91 Moreover, Notch-1 and Notch-3 NICD levels were increased in both human breast cancer specimens and cell lines, and Notch-3 activated nuclear transcription in those specimens and cells.Citation92 Further evidence for altered Notch-1 in human breast cancer was found in the form of aberrant Notch-1 activation in various breast cancer subtypes.Citation93 In addition, samples from breast cancer patients showed co-overexpression of the Notch-1 receptor and its ligand Jagged-1 predicting the poorest patient survival.Citation94,Citation95 Lastly, examination of almost 100 breast cancer specimens by immunohistochemistry and quantitative polymerase chain reaction (PCR) showed the expression of Notch-1 also correlated with poor outcomes.Citation96
Nonetheless, Notch receptors are not a homogeneous group functionally. When the transcriptional activities of N-1ICD, N-2ICD, and N-3ICD on HES-1 and HES-5 promoters were measured using a luciferase reporter assay, some of the differences were related to the combination of receptors used and expression level of RBP-Jκ (CSL or CBF-1). Also, inhibitory Notch-2 activity was confirmed, as co-expression of N-2ICD with N-1ICD or N-3ICD reduced their activity.Citation97 In a xenograft study using MDA-MB-231 cells, Notch-2 inhibited tumor growth.Citation98 Similarly, a clinical study which examined Notch-2 expression in breast cancer tissue by immunohistochemistry and qualitative and quantitative PCR concluded it may function as a tumor suppressor.Citation96
In assessing the role of Notch signaling in breast cancer stem cells, it was concluded from in vitro and in vivo experiments that Notch-4 activity was eightfold higher in breast cancer stem cells than in differentiated cells, and inhibition of Notch-4 resulted in suppression of tumor growth.Citation99 Moreover, breast cancer stem cells exhibited increased Notch signaling as compared with bulk tumor cells, especially in levels of HES-1 mRNA, and GSIs effectively blocked mammosphere formation, which is an assay to measure survival and self-renewal of breast cancer stem cells.Citation100
Notch and tumorigenesis
Of the more than 300 breast cancer cases examined, approximately 50% showed a loss of Numb-mediated inhibition of Notch signaling by ubiquitination and proteosomal degradation.Citation101 Of particular interest are two germline alterations (R62H and R71W) of presenilin-2 (PS-2) that have been reported in breast cancer patients with axillary node-negative disease, resulting in PS-2 being more susceptible to degradation.Citation102 Furthermore, nicastrin-knockout mice, which have decreased proteolytic cleavage of Notch and consequently lower NICD, developed myeloproliferative disease,Citation103 and Notch-1 knockout mice formed spontaneous basal cell carcinoma as they grew older.Citation104
Possible mechanisms of action for Notch-driven tumor propagation are: gain of function mutation, ligand-mediated activation of Notch, and downregulation of Notch.Citation105 Nonetheless, Notch tumorigenicity may be organ-dependent. In self-renewing systems such as skin, intestine, and bone marrow, Notch interacts with multiple signaling pathways. Oncogenesis derails these interactions such that Notch becomes a tumor suppressor in the skin and an oncogene in the bone marrow.Citation75 Manipulation of gene expression has been useful to study Notch receptors and ligands in tumorigenic systems. For example, MCF-10A cells, considered nonmalignant and noninvasive, when transfected with Notch-4, grew in a soft agar assay, suggesting that Notch-4 is a breast oncogene.Citation106 Similarly, mice bred to express Notch-1ICD and Notch-3ICD in mammary epithelial cells developed mammary tumors.Citation81 Nonetheless, Notch receptors may not be equivalent in their capacity to induce cancer. Notch-2 may suppress tumorigenicity, as MDA-MB-231 cells with constitutively expressing N-2ICD showed increased apoptosis and did not form xenograft tumors in mice.Citation98 Notch signaling is also responsive to hormonal drivers of tumorigenicity, since estrogen was found to upregulate Notch-1 and Jagged-1 in MCF-7 cells.Citation107 In contrast, Rizzo et al demonstrated that estrogen-mediated ER activation suppresses Notch activation, and the combination of anti-estrogen therapy with a GSI was more effective in inhibiting ER+ breast tumor growth than either therapy alone.Citation91 Specifically, the same group identified that Notch-1ICD activates ER-responsive genes under low estrogen conditions, suggesting that Notch-1ICD could mediate activation of the ER in an estrogen-independent manner.Citation108
Furthermore, loss of negative regulatory mechanisms contributes to neoplastic metastasis. For example, expression levels of the negative regulator of Notch signaling Numb inversely correlated with tumor aggressiveness.Citation101 In in-vitro and in-vivo experiments examining osteolytic bone metastasis of human breast cancer cells, osteoblasts together with secretion of transforming growth factor β1 enhanced Notch-3 expression in the breast cancer cells and mediated their metastasis; this effect was inhibited by GSI L-685458.Citation109
Notch and oncogenic crosstalk
The oncogenic reach of the Notch signaling pathway is partly due to its communication or crosstalk with other signaling pathways. Hurlbut et alCitation110 proposed more than 50 connections for the Notch crosstalk network; for example, receptor tyrosine kinases (RTKs), HH, Janus kinase/signal transducers and activators of transcription (Jak/STAT), transforming growth factor-β/decapentaplegic (TGF-β), and Wnt pathways. In addition to HH, Wnt, and TGF-β, amongst others pertinent to Notch crosstalk are platelet-derived growth factor (PDGF/PDGFR), vascular endothelial growth factor (VEGF), phosphatidylinositol 3-kinase (PI3K/Akt), Ras, mammalian target of rapamycin (mTOR), nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kB), hypoxia-inducible factor (HIF), and cytokines interleukin-6 (IL-6), IL-1, and leptin plus ER signaling as well as microRNAs considered operationally important for Notch crosstalk in breast cancer.Citation111 The majority of functions which include cell proliferation, differentiation, and development, tumor angiogenesis, morphogenesis, and somitogenesis are all important during oncogenesis. One of the most critical pathways necessary for survival of cancer cells is the NF-κB pathway. It has been shown that NF-κB regulates Notch and is regulated by Notch. For example, N-1ICD or N-3ICD has been shown to activate NF-κB signaling components such as IKK (inhibitor of kappa B kinase).Citation112 Furthermore, NF-κB has also been shown to regulate Notch indirectly by inducing Jagged-1, HES-5, and/or Deltex-1.Citation113
The existence of Notch and RTK crosstalk in breast and other solid tumors has been established by our research group and many others since. We and others have shown that Notch-1 signaling is decreased in ErbB-2 overexpressing BT-474, SkBr3, and MCF-7/HER2 breast cancer cells and that anti-HER-2 therapy using trastuzumab or a small molecule tyrosine kinase inhibitor similar to lapatinib reactivated Notch-1. More importantly, a GSI or specifically Notch-1 knockdown increased the sensitivity of ErbB-2+ breast cancer cells to anti-HER-2-mediated growth inhibition, indicating that Notch-1 signaling might contribute to trastuzumab resistance in vitro.Citation114 Moreover, tumor recurrence was prevented in mice injected with trastuzumab-sensitive BT-474 cells following treatment with trastuzumab and MRK-003 GSI or significantly reduced with trastuzumab and LY-411575 GSI; additionally, BT-474 breast tumors that were resistant to trastuzumab were re-sensitized by addition of MRK-003 GSI.Citation115 An overview of the role and significance of Notch signaling in trastuzumab resistant breast cancer is reviewed by Mehta and Osipo.Citation116
The regulation and activation of Notch signaling in triple negative breast cancer was recently eludicated by Clementz et al.Citation117 Specifically, the investigators demonstrated that PEA3, an Ets family transcription factor, activates transcription of Notch-1 and Notch-4.Citation117 It was identified that enrichment of PEA3 on the Notch-1 promoter was independent of AP-1 while PEA3 recruitment to the Notch-4 promoter was dependent on c-JUN and Fra-1, but negatively regulated by c-Fos. The findings from this study also showed that knockdown of PEA3 was potent in inhibiting triple negative breast cancer growth in vitro and in vivo.
GSIs: mode of action and side effects
The more than 100 GSIs synthesized to date can be divided into three classes: peptide isosteres, azepines, and sulfonamides.Citation118,Citation119 They are oral agents, the azepines and sulfonamides being the most popular. A list of select GSIs is presented in . GSIs currently undergoing US clinical trials are listed in .
Table 1 Chemical structure of γ-secretase inhibitors
Table 2 Clinical trials employing γ-secretase inhibitors in the treatment of breast cancer
The GSIs are classified into two types, depending on structure and binding sites: (1) aspartyl proteinase transition-state analogs as peptide isosteres that mimic the transition state of a substrate cleavage by γ-secretase and bind competitively to the catalytic active site of presenilins; and (2) small molecule non-transition-state inhibitors where the binding site is different from the active site, possibly at the interface of the γ-secretase complex dimer.Citation120 Well known side effects of GSIs occur within the gastrointestinal tract. For example, acute treatment of TgCRND8 mice with LY-411575 for 15 days caused an increase in the number of mucin-containing goblet cells in the small and large intestines and changes in the tissue architecture of the gastrointestinal tract which resulted in severe diarrhea.Citation121 The GSIs also cause various off-target effects in breast cancer cells and Notch signaling. An early transition-state analog GSI, IL X (cbz-IL-CHO), produced a decrease in mRNA and protein levels of HES-1, induced G0–G1 cell cycle arrest, and inhibited human tongue carcinoma Tca8113 cell growth.Citation122 Dipeptide GSI XII (z-Ile-Leu-CHO) induced apoptosis in breast cancer cell lines by inducing Noxa, a pro-apoptotic protein.Citation123 A later generation GSI, LY-294002 suppressed angiogenesis by blocking the epidermal growth factor (EGF)-induced upregulation of Jagged-1 in squamous cell carcinoma, thereby inhibiting EFG-Notch crosstalk.Citation124 Tripeptide GSI I (z-Leu-Leu-Nle-CHO) suppressed cell proliferation and induced apoptosis in Notch-3 overexpressing ovarian cell lines.Citation65
An early generation non-transition state analog is DAPT, N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester, a dipeptide inhibitor of the benzodiazepine type, also known as GSI IX and Compound 3.Citation125 It is the most widely used in the laboratory setting.Citation126 DAPT potentiated the apoptotic effects of the DNA-damaging drug melphalan in MCF-7 breast cancer cells.Citation127 In cell lines with chromosomal translocations, DAPT inhibited the proliferation of truncated Notch-1 expressing an ADAM cleavage site but not of truncated Notch-2 which was without the cleavage site.Citation128 From DAPT, numerous other GSIs have been developed that are even more effective, ie, LY-411,575 (Compound 5) 100-fold stronger than DAPT, LY-450,139 (Semagacestat or Compound 6),Citation129 and RO-4929097 (Roche, Nutley, NJ, USA).
The small-molecule GSI classified as a tetralin imidazole PF-03084014 (Pfizer Inc., Groton, CT, USA) is in a Phase I trial to treat advanced breast cancer and other solid tumors.Citation130 It is considered a selective or Notch-sparing GSI or GS (gamma secretase) modulator. When evaluated for Notch activity, PF-03084014 significantly decreased tumor cell migration and mammosphere formation in vitro, reduced tumor cell self-renewal ability in vivo, and decreased mRNA expression of Notch target genes HES-1, HES-4, Notch-1, and HEY-2 in HCC1599 xenograft tumors.Citation131
Another small-molecule GSI, RO-4929097, was used in a multicenter Phase I clinical dose escalating study and continued on to Phase II and combination therapy studies. Derived from LY-411575 and containing a dibenzazepinone core, it is being tested for the treatment of breast cancer and other solid tumors.Citation132,Citation133 Patients with low basal levels of plasma IL-6 and IL-8 responded well, indicating that cytokines may be predictive biomarkers for response to therapy.Citation134 In in vitro studies using a colon cancer cell line A549, RO-4929097 produced a significant decrease in mRNA levels of Notch target genes HES-1, HES-4, and HEY-1.Citation135
Another GSI, the sulfonamide-containing non-transition-state GSI analog MK-0752 (Merck and Co, Inc, Whitehouse Station, NJ, USA) is in a Phase I study to treat metastatic or locally advanced breast cancer.
Novel combination strategies
Since single-drug therapy is ineffective for long-term use, combination therapy oftentimes becomes necessary. Such a treatment regimen is applicable to “endocrine therapy, targeted therapies, chemotherapy, or possibly even radiation therapy.”Citation72 There are several clinical studies using RO-4929097 in combination therapy. One Phase I study is using RO-4929097 with capecitabine for patients with refractory solid tumors and another with paclitaxel and carboplatin for patients with Stage I or III triple negative breast cancer. A Phase I clinical trial is presently underway testing the efficacy of RO-4929097 and a potent HH antagonist GDC-0449 vismodegib in patients with advanced breast cancer.Citation136 These patients may have been selected on the basis of upregulated crosstalk between Notch and the self-renewal pathway (targeted therapy). Another Phase I study with RO-4929097 is adding cediranib for post-menopausal patients with advanced solid tumors (targeted therapy). In a Phase Ib clinical trial, RO-4929097 is combined with letrazole for patients with ER+/PR+ Stage II or III breast cancer (endocrine therapy). Another GSI in combination therapy is MK-0752. In a Pilot study, MK-0752 is being combined with tamoxifen or letrazole for patients with early stage breast cancer (endocrine therapy). In a Phase I study, MK-0752 (or MK-2206 Akt inhibitor) is being combined with ridaforolimus (MK-8669) in patients with advanced solid tumors (targeted therapy). A Phase I/II study is combining MK-0752 with docetaxel in patients presenting with locally advanced or metastatic breast cancer (chemotherapy).
Gastrointestinal toxicity is a major side-effect with GSI use.Citation137 Nonetheless, careful monitoring of treatment protocols, whether by modulating expression of Notch receptors with receptor antibody pretreatment before GSI treatmentCitation138 or development of a practical combination therapyCitation139 should minimize problematic side-effects. Notch activation must be assessed prior to GSI treatment (mutations and/or overexpression), since GSIs are more effective against tumors with upregulated Notch signaling. In addition, close attention must be paid to the therapeutic window so that the minimally active dose needed to inhibit Notch is employed, thereby reducing adverse side effects.
Conclusion
Much progress has been made in understanding Notch signaling in breast cancer. Molecular profiling of patients, fast becoming standard of care, identifies the type and location of signaling dysfunction. Moreover, pharmacological innovations are helping produce more selective GSIs with fewer side effects. A “one problem–one solution” type of cure to breast cancer seems unlikely. Inhibition of Notch signaling with pharmacodynamically active drugs such as the GSIs is preventing metastasis and recurrence and increasing disease-free survival. The next level of care for determining the molecular signature of a breast tumor will develop therapeutic combinatorial protocols that effectively target crosstalk pathways, tumor microenvironment, tumor-initiating cells (or cancer stem cells), developmental factors, non-canonical signaling components, and possibly other additional modulating factors still unknown. Breast cancer management will require a multidisciplinary team to prepare and optimize the anticancer drug regimen, conduct the therapy, and even interpret results and treatment progress. Overall, targeting the Notch signaling pathway in breast cancer therapy and attempting its downregulation with GSIs looks promising.
Acknowledgments
The authors thank Rima M Rusinas, MDes, for the design of the figures and tables.
Disclosure
The authors report no conflicts of interest in this work.
References
- PerouCMSørlieTEisenMBMolecular portraits of human breast tumoursNature2000406679774775210963602
- HuZFanCOhDSThe molecular portraits of breast tumors are conserved across microarray platformsBMC Genomics2006719616643655
- DexterJSThe analysis of a case of continuous variation in drosophila by a study of its linkage relationsAm Nat191448576712758
- MorganTHThe theory of the geneAm Nat1917513544
- MorganTHThe Physical Basis of HeredityPhiladelphiaJB Lippincott1919
- KiddSKelleyMRYoungMWSequence of the Notch locus of Drosophila melanogaster: relationship of the encoded protein to mammalian clotting and growth factorsMol Cell Biol198669309431083097517
- WhartonKAJohansenKMXuTArtavanis-TsakonasSNucleotide sequence from the neurogenic locus Notch implies a gene product that shares homology with proteins containing EGF-like repeatsCell1985433 Pt 25673935325
- LaiECNotch signaling: control of cell communication and cell fateDevelopment2004131596597314973298
- SwiatekPJLindsellCEDel AmoFFWeinmasterGGridleyTNotch1 is essential for postimplantation development in miceGenes Dev1994867077197926761
- ConlonRAReaumeAGRossantJNotch1 is required for the coordinate segmentation of somitesDevelopment19951215153315457789282
- JiangRLanYChapmanHDDefects in limb, craniofacial, and thymic development in Jagged2 mutant miceGenes Dev1998127104610579531541
- Hrabe de AngelisMMcIntyreIJGosslerAMaintenance of somite borders in mice requires the Delta homologue DII1Nature19973867177219109488
- Artavanis-TsakonasSRandMDLakeRJNotch signaling: cell fate control and signal integration in developmentScience1999284541577077610221902
- HenriqueDHirsingerEAdamJMaintenance of neuroepithelial progenitor cells by Delta–Notch signalling in the embryonic chick retinaCurr Biol1997796616709285721
- NeumannCJCohenSMA hierarchy of cross-regulation involving Notch, wingless, vestigial and cut organizes the dorsal/ventral axis of the Drosophila wingDevelopment199612211347734858951063
- de CelisJFGarcia-BellidoABraySJActivation and function of Notch at the dorsal-ventral boundary of the wing imaginal discDevelopment199612213593698565848
- GauntSJDevelopmental biology-chick limbs, fly wings and homology at the fringeNature199738666233243259121545
- MoloneyDJPaninVMJohnstonSHFringe is a glycosyltransferase that modifies NotchNature2000406679436937510935626
- HaltiwangerRSStanleyPModulation of receptor signaling by glycosylation: fringe is an O-fucose-β1, 3-N-acetylglucosaminyltransferaseBiochim Biophys Acta20021573332833512417415
- HainesNIrvineKDGlycosylation regulates Notch signallingNat Rev Mol Cell Biol200341078679714570055
- BlaumuellerCMQiHZagourasPArtavanis-TsakonasSIntracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membraneCell19979022812919244302
- LogeatFBessiaCBrouCThe Notch1 receptor is cleaved constitutively by a furin-like convertaseProc Natl Acad Sci USA19989514810881129653148
- BettenhausenBde AngelisMHSimonDGuénetJLGosslerATransient and restricted expression during mouse embryogenesis of Dll1, a murine gene closely related to Drosophila DeltaDevelopment19951218240724187671806
- DunwoodieSLHenriqueDHarrisonSMBeddingtonRMouse Dll3: a novel divergent Delta gene which may complement the function of other Delta homologues during early pattern formation in the mouse embryoDevelopment199712416306530769272948
- ShutterJRScullySFanWDll4, a novel Notch ligand expressed in arterial endotheliumGenes Dev200014111313131810837024
- LindsellCEShawberCJBoulterJWeinmasterGJagged: a mammalian ligand that activates Notch1Cell19958069099177697721
- BraySJNotch signalling: a simple pathway becomes complexNat Rev Mol Cell Biol20067967868916921404
- MieleLNotch signalingClin Cancer Res20061241074107916489059
- KopanRIlaganMXGThe canonical Notch signaling pathway: unfolding the activation mechanismCell2009137221623319379690
- MummJSKopanRNotch signaling: from the outside inDev Biol2000228215111112321
- BrouCLogeatFGuptaNA novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACEMol Cell20005220721610882063
- ReissKSaftigPThe “a disintegrin and metalloprotease”(ADAM) family of sheddases: physiological and cellular functionsSemin Cell Dev Biol200920212613719049889
- MummJSSchroeterEHSaxenaMTA ligand-induced extracellular cleavage regulates-secretase-like proteolytic activation of Notch1Mol Cell20005219720610882062
- LleoMActivity of gamma-secretase on substrates other than APPCurr Top Med Chem20088191618220928
- De StrooperBAph-1, Pen-2, and nicastrin with presenilin generate an active gamma-secretase complexNeuron200338191212691659
- LaiECNotch cleavage: nicastrin helps presenilin make the final cutCurr Biol2002126R200R20211909545
- SchroeterEHKisslingerJAKopanRNotch-1 signalling requires ligand-induced proteolytic release of intracellular domainNature199839366833823869620803
- StruhlKHistone acetylation and transcriptional regulatory mechanismsGenes Dev19981255996069499396
- WuGLyapinaSDasISEL-10 is an inhibitor of Notch signaling that targets Notch for ubiquitin-mediated protein degradationMol Cell Biol200121217403741511585921
- EllisenLWBirdJWestDCTAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasmsCell19916646496611831692
- WengAPFerrandoAALeeWActivating mutations of NOTCH1 in human T cell acute lymphoblastic leukemiaScience2004306569426927115472075
- FabbriGRasiSRossiDAnalysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activationJ Exp Med201120871389140121670202
- MorimuraTGoitsukaRZhangYSaitoIRethMKitamuraDCell cycle arrest and apoptosis induced by Notch1 in B cellsJ Biol Chem200027547365233653110967117
- RomerSSaundersUJäckHJehnBNotch1 enhances B-cell receptor-induced apoptosis in mature activated B cells without affecting cell cycle progression and surface IgM expressionCell Death Differ200310783384412815466
- NefedovaYChengPAlsinaMDaltonWSGabrilovichDIInvolvement of Notch-1 signaling in bone marrow stroma-mediated de novo drug resistance of myeloma and other malignant lymphoid cell linesBlood200410393503351014670925
- HubmannRSchwarzmeierJDShehataMNotch2 is involved in the overexpression of CD23 in B-cell chronic lymphocytic leukemiaBlood200299103742374711986231
- JundtFAnagnostopoulosIFörsterRMathasSSteinHDörkenBActivated Notch1 signaling promotes tumor cell proliferation and survival in Hodgkin and anaplastic large cell lymphomaBlood20029993398340311964309
- JundtFPröbstingKSAnagnostopoulosIJagged1-induced Notch signaling drives proliferation of multiple myeloma cellsBlood200410393511351514726396
- KridelRMeissnerBRogicSWhole transcriptome sequencing reveals recurrent NOTCH1 mutations in mantle cell lymphomaBlood201211991963197122210878
- LeeSKumanoKNakazakiKGain-of-function mutations and copy number increases of Notch2 in diffuse large B-cell lymphomaCancer Sci2009100592092619445024
- HoudeCLiYSongLOverexpression of the NOTCH ligand JAG2 in malignant plasma cells from multiple myeloma patients and cell linesBlood2004104123697370415292061
- ZagourasPStifaniSBlaumuellerCMCarcangiuMLArtavanis-TsakonasSAlterations in Notch signaling in neoplastic lesions of the human cervixProc Natl Acad Sci USA19959214641464187604005
- WeijzenSZlobinABraidMMieleLKastWMHPV16 E6 and E7 oncoproteins regulate Notch-1 expression and cooperate to induce transformationJ Cell Physiol2003194335636212548555
- MayRRiehlTEHuntCSurebanSMAnantSHouchenCWIdentification of a novel putative gastrointestinal stem cell and adenoma stem cell marker, doublecortin and CaM kinase-like-1, following radiation injury and in adenomatous polyposis coli/multiple intestinal neoplasia miceStem Cells200726363063718055444
- AkiyoshiTNakamuraMYanaiKγ-Secretase inhibitors enhance taxane-induced mitotic arrest and apoptosis in colon cancer cellsGastroenterology2008134113114418166351
- GaoBJeongWITianZLiver: an organ with predominant innate immunityHepatology200747272973618167066
- QiRAnHYuYNotch1 signaling inhibits growth of human hepatocellular carcinoma through induction of cell cycle arrest and apoptosisCancer Res200363238323832914678992
- KonishiJKawaguchiKSVoHγ-Secretase inhibitor prevents Notch3 activation and reduces proliferation in human lung cancersCancer Res200767178051805717804716
- TerrisBBlaveriECrnogorac-JurcevicTCharacterization of gene expression profiles in intraductal papillary-mucinous tumors of the pancreasAm J Pathol200216051745175412000726
- MiyamotoYMaitraAGhoshBNotch mediates TGFα-induced changes in epithelial differentiation during pancreatic tumorigenesisCancer Cell20033656557612842085
- WangZZhangYLiYBanerjeeSLiaoJSarkarFHDown-regulation of Notch-1 contributes to cell growth inhibition and apoptosis in pancreatic cancer cellsMol Cancer Ther20065348349316546962
- KimuraKSatohKKannoAActivation of Notch signaling in tumorigenesis of experimental pancreatic cancer induced by dimethylbenzanthracene in miceCancer Sci200798215516217297654
- SantagataSDemichelisFRivaAJAGGED1 expression is associated with prostate cancer metastasis and recurrenceCancer Res200464196854685715466172
- RamdasLGiriUAshornCLmiRNA expression profiles in head and neck squamous cell carcinoma and adjacent normal tissueHead Neck200931564265419260130
- ParkJTLiMNakayamaKNotch3 gene amplification in ovarian cancerCancer Res200666126312631816778208
- RoseSLKunnimalaiyaanMDrenzekJSeilerNNotch 1 signaling is active in ovarian cancerGynecol Oncol2010117113013320060575
- SjolundJJohanssonMMannaSSuppression of renal cell carcinoma growth by inhibition of Notch signaling in vitro and in vivoJ Clin Invest2008118121722818079963
- MieleLMiaoHNickoloffBNOTCH signaling as a novel cancer therapeutic targetCurr Cancer Drug Targets20066431332316848722
- JoutelATournier-LasserveENotch signalling pathway and human diseasesSemin Cell Dev Biol19989661962510075489
- NickoloffBJOsborneBAMieleLNotch signaling as a therapeutic target in cancer: a new approach to the development of cell fate modifying agentsOncogene200322426598660814528285
- MieleLOsborneBArbiter of differentiation and death: Notch signaling meets apoptosisJ Cell Physiol1999181339340910528225
- Al-HussainiHSubramanyamDReedijkMSridharSSNotch signaling pathway as a therapeutic target in breast cancerMol Cancer Ther201110191520971825
- LeongKGKarsanARecent insights into the role of Notch signaling in tumorigenesisBlood200610762223223316291593
- RadtkeFRajKThe role of Notch in tumorigenesis: oncogene or tumour suppressor?Nat Rev Cancer200331075676714570040
- WilsonARadtkeFMultiple functions of Notch signaling in self-renewing organs and cancerFEBS Lett2006580122860286816574107
- RoyMPearWSAsterJCThe multifaceted role of Notch in cancerCurr Opin Genet Dev2007171525917178457
- MaillardIPearWSNotch and cancer: best to avoid the ups and downsCancer Cell20033320320512676578
- MieleLGoldeTOsborneBNotch signaling in cancerCurr Mol Med20066890591817168741
- RanganathanPWeaverKLCapobiancoAJNotch signalling in solid tumours: a little bit of everything but not all the timeNat Rev Cancer201111533835121508972
- KiarisHPolitiKGrimmLMModulation of Notch signaling elicits signature tumors and inhibits hras1-induced oncogenesis in the mouse mammary epitheliumAm J Pathol2004165269570515277242
- HuCDiévartALupienMCalvoETremblayGJolicoeurPOverexpression of activated murine Notch1 and Notch3 in transgenic mice blocks mammary gland development and induces mammary tumorsAm J Pathol2006168397399016507912
- KlinakisASzabolcsMPolitiKKiarisHArtavanis-TsakonasSEfstratiadisAMyc is a Notch1 transcriptional target and a requisite for Notch1-induced mammary tumorigenesis in miceProc Natl Acad Sci USA2006103249262926716751266
- PolitiKFeirtNKitajewskiJNotch in mammary gland development and breast cancerSemin Cancer Biol200414534134715288259
- DiévartABeaulieuNJolicoeurPInvolvement of Notch1 in the development of mouse mammary tumorsOncogene199918445973598110557086
- SmithGHGallahanDDiellaFJhappanCMerlinoGCallahanRConstitutive expression of a truncated INT3 gene in mouse mammary epithelium impairs differentiation and functional developmentCell Growth Differ1995655635777544153
- GallahanDCallahanRThe mouse mammary tumor associated gene INT3 is a unique member of the NOTCH gene family (NOTCH4)Oncogene19971416188318909150355
- GallahanDJhappanCRobinsonGExpression of a truncated Int3 gene in developing secretory mammary epithelium specifically retards lobular differentiation resulting in tumorigenesisCancer Res1996568177517858620493
- RobinsonDRKalyana-SundaramSWuYMFunctionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancerNat Med201117121646165122101766
- XiaoYYeYZouXThe lymphovascular embolus of inflammatory breast cancer exhibits a Notch 3 addictionOncogene201030328730020838375
- KonstantinosTExpression of Notch receptors in primary breast cancer and correlation with pathological featuresClin Exp Pharmacol201221000109
- RizzoPMiaoHD’SouzaGCross-talk between Notch and the estrogen receptor in breast cancer suggests novel therapeutic approachesCancer Res200868135226523518593923
- YamaguchiNOyamaTItoENOTCH3 signaling pathway plays crucial roles in the proliferation of ErbB2-negative human breast cancer cellsCancer Res20086861881188818339869
- StylianouSClarkeRBBrennanKAberrant activation of Notch signaling in human breast cancerCancer Res20066631517152516452208
- ReedijkMOdorcicSChangLHigh-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survivalCancer Res200565188530853716166334
- DicksonBCMulliganAMZhangHHigh-level JAG1 mRNA and protein predict poor outcome in breast cancerMod Pathol200720668569317507991
- ParrCWatkinsGJiangWThe possible correlation of Notch-1 and Notch-2 with clinical outcome and tumour clinicopathological parameters in human breast cancerInt J Mol Med200414577978615492845
- ShimizuKChibaSSaitoTKumanoKHamadaYHiraiHFunctional diversity among Notch1, Notch2, and Notch3 receptorsBiochem Biophys Res Commun2002291477577911866432
- O’NeillCFUrsSCinelliCNotch2 signaling induces apoptosis and inhibits human MDA-MB-231 xenograft growthAm J Pathol200717131023103617675579
- HarrisonHFarnieGHowellSJRegulation of breast cancer stem cell activity by signaling through the Notch4 receptorCancer Res201070270971820068161
- GrudzienPLoSAlbainKSInhibition of Notch signaling reduces the stem-like population of breast cancer cells and prevents mammosphere formationAnticancer Res201030103853386721036696
- PeceSSerresiMSantoliniELoss of negative regulation by Numb over Notch is relevant to human breast carcinogenesisJ Cell Biol2004167221522115492044
- ToMGokgozNDoyleTFunctional characterization of novel presenilin-2 variants identified in human breast cancersOncogene200625253557356416474849
- KlinakisALobryCAbdel-WahabOA novel tumour-suppressor function for the Notch pathway in myeloid leukaemiaNature2011473734623023321562564
- NicolasMWolferARajKNotch1 functions as a tumor suppressor in mouse skinNat Genet200333341642112590261
- AllenspachEJMaillardIAsterJCPearWSNotch signaling in cancerCancer Biol Ther200215460460
- ImataniACallahanRIdentification of a novel NOTCH-4/INT-3 RNA species encoding an activated gene product in certain human tumor cell linesOncogene200019222323110645000
- SoaresRBaloghGGuoSGärtnerFRussoJSchmittFEvidence for the Notch signaling pathway on the role of estrogen in angiogenesisMol Endocrinol20041892333234315192074
- HaoLNotch-1 Activates Estrogen Receptor Alpha-Dependent Transcription via IKK AlphaLoyola University Chicago2010
- ZhangZWangHIkedaSNotch3 in human breast cancer cell lines regulates osteoblast-cancer cell interactions and osteolytic bone metastasisAm J Pathol201017731459146920651241
- HurlbutGDKankelMWLakeRJArtavanis-TsakonasSCrossing paths with Notch in the hyper-networkCurr Opin Cell Biol200719216617517317139
- GuoSLiuMGonzalez-PerezRRRole of Notch and its oncogenic signaling crosstalk in breast cancerBiochim Biophys Acta20111815219721321193018
- SongLLPengYYunJNotch-1 associates with IKK alpha and regulates IKK activity in cervical cancer cellsOncogene200827445833584418560356
- OsipoCGoldeTEOsborneBAMieleLAOff the beaten pathway: the complex cross talk between Notch and NF-κBLab Invest2007881111718059366
- OsipoCPatelPRizzoPErbB-2 inhibition activates Notch-1 and sensitizes breast cancer cells to a γ-secretase inhibitorOncogene200827375019503218469855
- PandyaKMeekeKClementzATargeting both Notch and ErbB-2 signalling pathways is required for prevention of ErbB-2-positive breast tumour recurrenceBr J Cancer2011105679680621847123
- MehtaKOsipoCTrastuzumab resistance: role for Notch signalingSci World J2009914381448
- ClementzAGRogowskiAPandyaKMieleLOsipoCNOTCH-1 and NOTCH-4 are novel gene targets of PEA3 in breast cancer: novel therapeutic implicationsBreast Cancer Res2011133R6321679465
- OlsonREAlbrightCFRecent progress in the medicinal chemistry of-secretase inhibitorsCurr Top Med Chem200881173318220929
- KreftAFMartoneRPorteARecent advances in the identification of γ-secretase inhibitors to clinically test the Aβ oligomer hypothesis of Alzheimer’s diseaseJ Med Chem200952206169618819694467
- ClarkeEEChurcherIEllisSIntra-or intercomplex binding to the γ-secretase enzymeJ Biol Chem200628142312793128916899457
- WongGTManfraDPouletFMChronic treatment with the γ-secretase inhibitor LY-411,575 inhibits β-amyloid peptide production and alters lymphopoiesis and intestinal cell differentiationJ Biol Chem200427913128761288214709552
- YaoJDuanLFanMWuXγ-secretase inhibitors exerts antitumor activity via down-regulation of Notch and Nuclear factor kappa B in human tongue carcinoma cellsOral Dis200713655556317944672
- SévenoCLoussouarnDBréchetSCamponeMJuinPBarillé-NionSγ-Secretase inhibition promotes cell death, Noxa upregulation, and sensitization to BH3 mimetic ABT-737 in human breast cancer cellsBreast Cancer Res2012143R9622703841
- ZengQLiSChepehaDBCrosstalk between tumor and endothelial cells promotes tumor angiogenesis by MAPK activation of Notch signalingCancer Cell200581132316023595
- DoveyHJohnVAndersonJFunctional gamma-secretase inhibitors reduce beta-amyloid peptide levels in brainJ Neurochem200976117318111145990
- PurowBNotch inhibition as a promising new approach to cancer therapyAdv Exp Med Biol201272730531922399357
- MeuretteOStylianouSRockRColluGMGilmoreAPBrennanKNotch activation induces Akt signaling via an autocrine loop to prevent apoptosis in breast epithelial cellsCancer Res200969125015502219491273
- RobinsonDRKalyana-SundaramSWuY-MFunctionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancerNat Med201117121646165122101766
- SiemersESkinnerMDeanRASafety, tolerability, and changes in amyloid beta concentrations after administration of a gamma-secretase inhibitor in volunteersClin Neuropharmacol200528312613215965311
- WeiPWallsMQiuMEvaluation of selective γ-secretase inhibitor PF-03084014 for its antitumor efficacy and gastrointestinal safety to guide optimal clinical trial designMol Cancer Ther2010961618162820530712
- ZhangCCPavlicekAZhangQBiomarker and pharmacologic evaluation of the γ-secretase inhibitor PF-03084014 in breast cancer modelsClin Cancer Res201218185008501922806875
- TolcherAMikulskiSMessersmithWA phase I study of R04929097, a novel gamma secretase inhibitor, in patients with advanced solid tumorsJ Clin Oncol (Meeting Abstracts)201028Suppl 152502
- TolcherAWMessersmithWAMikulskiSMPhase I study of RO4929097, a gamma secretase inhibitor of Notch signaling, in patients with refractory metastatic or locally advanced solid tumorsJ Clin Oncol201230192348235322529266
- HeWLuistroLCarvajalDHigh tumor levels of IL6 and IL8 abrogate preclinical efficacy of the γ-secretase inhibitor, RO4929097Mol Oncol20115329230121315665
- LuistroLHeWSmithMPreclinical profile of a potent γ-secretase inhibitor targeting Notch signaling with in vivo efficacy and pharmacodynamic propertiesCancer Res200969197672768019773430
- WuJLoRussoPMMatherlyLHLiJImplications of plasma protein binding for pharmacokinetics and pharmacodynamics of the γ-secretase inhibitor RO4929097Clin Cancer Res20121872066207922351688
- BartenDMMeredithJEZaczekRHoustonJGAlbrightCFGamma-secretase inhibitors for Alzheimers disease: balancing efficacy and toxicityDrugs R D200672879716542055
- WuYCain-HomCChoyLTherapeutic antibody targeting of individual Notch receptorsNature201046472911052105720393564
- RizzoPOsipoCForemanKGoldeTOsborneBMieleLRational targeting of Notch signaling in cancerOncogene200827385124513118758481