Abstract
Background:
Prohibitin 3′ untranslated region 1630 C>T (rs6917) polymorphism creates a variant T allele that lacks the antiproliferative activity of the more common functional C allele. Previous studies indicate that women carrying the prohibitin T allele have an increased susceptibility to breast cancer. However, the role of 1630 C>T polymorphism in mRNA expression of prohibitin and its contribution to carcinogenesis in the breast remains controversial.
Methods:
Using mRNA expression data from the HapMap online database, we sought an association between prohibitin 1630 C>T polymorphism and its mRNA expression, then conducted a meta-analysis of prohibitin 1630 C>T polymorphism and risk of breast cancer.
Results:
Although no significant association was found between prohibitin 1630 C>T polymorphism and mRNA expression in lymphoblastoid cell lines from the HapMap database (Ptrend = 0.543), the present meta-analysis involving 5072 cases and 4796 controls demonstrated that prohibitin 1630 C>T polymorphism was significantly correlated with breast cancer risk in allele contrast model T versus C (odds ratio [OR] 1.09, 95% confidence interval [CI] 1.01–1.18), the homozygote codominant model TT versus CC (OR 1.47, 95% CI 1.12–1.92), and the recessive model TT versus CC/CT (OR 1.45, 95% CI 1.10–1.89).
Conclusion:
Our study indicates that minor allele T of prohibitin 1630 C>T polymorphism is associated with increased susceptibility to breast cancer.
Introduction
Prohibitin is a candidate tumor suppressor gene encoding a 30 kDa intracellular protein which regulates cell cycle progression in multiple cell types. It interacts with the retinoblastoma tumor suppressor protein and its family members to suppress E2F-mediated transcription, and binds to p53 protein, increasing p53 transcriptional activity via increased DNA binding.Citation1,Citation2 The human prohibitin gene is located on chromosome 17q21, a region of frequent loss of heterozygosity in breast cancers, spanning approximately 11 kb and consisting of seven exons.Citation3 In total, 217 single nucleotide polymorphisms have been identified in the prohibitin gene region, and 38 nucleotide polymorphisms in the coding region (http://www.ncbi.nlm.nih.gov/SNP/). Of these, 14 nucleotide polymorphisms have been reported in the 3′-untranslated region, as shown in , only five nucleotide polymorphisms (rs6917, rs9893420, rs111398671, rs112294663, rs73324369) have minor allele frequencies available, and the potential microRNA binding sites are summarized in . The most extensively studied nucleotide polymorphism of prohibitin is a C-to-T transition at position 1630 in the 3′-untranslated region, that creates a variant with hsa-miR-1292 and hsa-miR-886-5p as potential binding sites (http://snpinfo.niehs.nih.gov/cgi-bin/snpinfo/snpfunc.cgi). This variant lacks antiproliferative activity and significantly reduces cell motility.Citation4–Citation6
Table 1 The SNPs of prohibitin 3′UTR and MicroRNA binding sites
Recent studies have evaluated the potential role of prohibitin in development of breast cancer and risk modification associated with prohibitin 1630 C>T polymorphism, but there are still no consistent data to indicate the molecular mechanism of 1630 C>T polymorphism in the regulation of prohibitin mRNA expression and its role in carcinogenesis. Although the T allele has been associated with an increased risk of breast cancer in women aged younger than 50 years who have a first-degree relative with breast cancer,Citation7 there are other studies that have not found an association between this polymorphism and breast cancer. In order to evaluate this potential association more precisely, we identified all published case-control studies, amounting to 5072 cases and 4796 controls, and undertook a quantitative analysis to identify evidence of an association between prohibitin 1630 C>T polymorphism and breast cancer risk.
Materials and methods
Genotype and mRNA expression data in lymphoblastoid cell lines
We used additional data on prohibitin genotypes and mRNA levels available online (http://app3.titan.uio.no/biotools/help.php?app=snpexp) for analysis of the genotype-phenotype relationship.Citation8 We analyzed the variation in gene expression using genome-wide expression arrays (47,294 transcripts) from Epstein-Barr virus-transformed lymphoblastoid cell lines from the same 270 HapMap individuals.Citation9 The genotyping data were from the HapMap Phase II release 23 data set consisting of 3.96 million single nucleotide polymorphism genotypes from 270 individuals in four populations.Citation10
Publication search and data extraction
Eligible studies were identified by searching in the PubMed, ISI Web of Knowledge, and Embase databases for relevant reports (last search update, August 2012), using the search terms “PHB” or “prohibitin”, “polymorphism”, and “breast cancer”. We did not define any minimum number of patients to be included for meta-analysis. When multiple studies of the same patient population were identified, we included the published report with the largest sample size.
Inclusion criteria were: evaluation of prohibitin 1630 C>T polymorphism and breast cancer risk, case-control study design, and sufficient published data for estimating an odds ratio (OR) with a 95% confidence interval (CI). Only the most recent or complete study was used if the same study subjects were included in more than one publication. The main exclusion criteria were: no control population, no available genotype frequency, and overlapping data.
Two authors reviewed the articles separately and extracted the data from all eligible publications according to the criteria listed above. Any discrepancies between investigators were resolved by discussion and consultation with a third reviewer. The first author’s surname, year of publication, country of original ethnicity, study design, genotyping method, and numbers of genotyped cases and controls (CC, CT, and TT genotypes) were recorded for each study.
Statistical methods
The genotype and phenotype relationship analysis was performed using SAS software (version 9.1, SAS Institute, Cary, NC). A pooled OR and 95% CI were calculated to estimate the risk of breast cancer associated with prohibitin 1630 C>T. For all studies, we estimated the association under five different types of OR, namely the allele contrast model (T versus C), homozygote codominant model (TT versus CC), heterozygote codominant model (CT versus CC), dominant model (TT/CT versus CC), and recessive model (TT versus CC/CT). Hardy-Weinberg equilibrium was investigated using the χ2 test. The Q-statistic and I2 test were used to investigate the degree of heterogeneity between studies. When P ≥ 0.1 or I2 ≤ 50% indicated a lack of heterogeneity, the fixed-effects model (Mantel-Haenszel method) was used. Otherwise, the random-effects model (DerSimonian-Laird method) was chosen. Egger’s test and inverted Begg’s funnel plots were used to detect any publication bias. A sensitivity analysis was also performed by repeating the meta-analysis and omitting each study at each iteration.Citation11,Citation12 The data were analyzed using Revman 5.0 software (http://ims.cochrane.org/revman).
Results
Prohibitin mRNA expression by genotype in lymphoblastoid cell lines
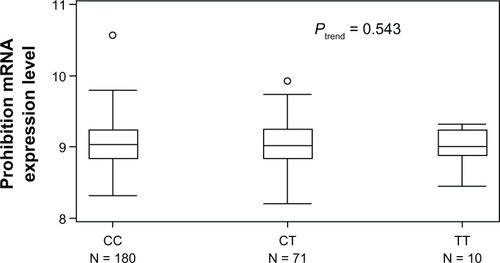
We used the available HapMap-cDNA expression database for correlation analysis of prohibitin genotype and mRNA expression in 270 HapMap lymphoblastoid cell lines. Except for nine cell lines with no available values, 180 (68.9%) cell lines had the CC genotype, 71 (27.3%) had the CT genotype, and 10 (3.8%) had the TT genotype. shows prohibitin mRNA expression according to 1630 C>T genotype for the lymphoblastoid cell lines. There was no significant difference in prohibitin mRNA expression level between cell lines carrying the TT genotype (9.05 ± 0.31), TC genotype (9.04 ± 0.33), or CC genotype (8.96 ± 0.29, Ptrend = 0.543, ).
Study characteristics
After careful examination according to the inclusion criteria, six publications on polymorphisms of prohibitin 1630 C>T and breast cancer risk were eligible,Citation4,Citation7,Citation13–Citation16 of which the study by Jakubowska et alCitation14 was reported twice. For the overlapping studies, only the one with the largest sample numbers was included. Jupe et alCitation7 only provided information on C/T or T/T versus C/C. Hence, a total of five publications including 5072 cases and 4796 controls were used in the present meta-analysis. lists the main characteristics of these studies. All cases were histologically confirmed as breast cancer, and controls were cancer-free and hospital-based populations matched for age and gender. The genotype distribution of the controls was in Hardy-Weinberg equilibrium, except for one study.Citation7
Table 2 Characteristics of the studies included in the meta-analysis
Meta-analysis results
The results of the meta-analysis are shown in . Because the between-study heterogeneity of each study included in our meta-analysis was not statistically significant, all pooled ORs were derived from fixed-effects models. We observed that the prohibitin 1630 C>T polymorphism was significantly correlated with risk of breast cancer in the allele contrast model T versus C (OR 1.09, 95% CI 1.01–1.18, ), the homozygote codominant model TT versus CC (OR 1.47, 95% CI 1.12–1.92, ), and the recessive model TT versus CC/CT (OR 1.45, 95% CI 1.10–1.89, ). However, no significant association was detected for the heterozygote codominant model CT versus CC (OR 1.04, 95% CI 0.95–1.14, ) or the dominant model TT/CT versus CC (OR 1.08, 95% CI 0.99–1.18, ).
Figure 2 Forest plot for the association between the prohibitin 1630 C>T polymorphism and breast cancer risk (for T versus C) in a fixed-effects model.
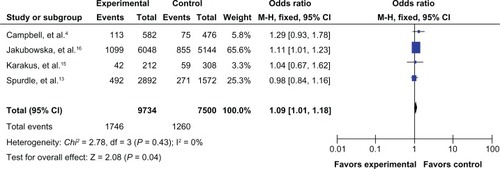
Figure 3 Forest plot for the association between the prohibitin 1630 C>T polymorphism and breast cancer risk (for TT versus CC) in a fixed-effects model.
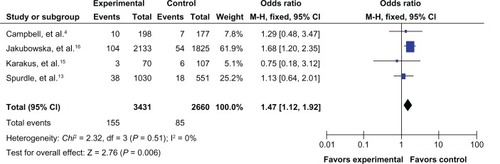
Figure 4 Forest plot for the association between the prohibitin 1630 C>T polymorphism and breast cancer risk (for TT versus CC/CT) in a fixed-effects model.
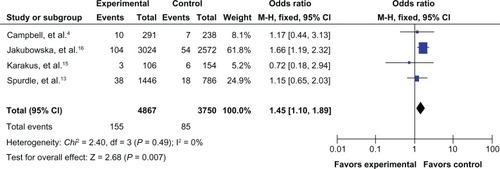
Figure 5 Forest plot for the association between the prohibitin 1630 C>T polymorphism and breast cancer risk (for CT versus CC) in a fixed-effects model.
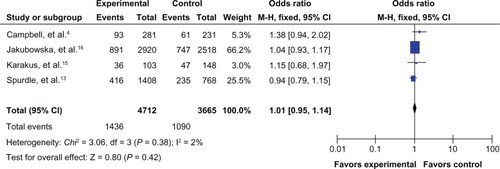
Figure 6 Forest plot for the association between the prohibitin 1630 C>T polymorphism and breast cancer risk (for TT/CT versus CC) in a fixed-effects model.
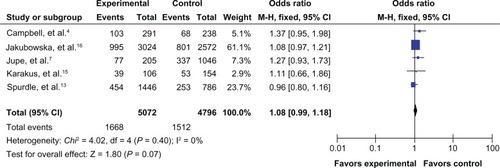
Table 3 Result of meta-analysis for prohibitin 3′UTR 1630 C>T polymorphism and breast cancer risk
Publication bias
Funnel plots and Egger’s test were used to assess publication bias in the literature. There was no evidence of publication bias for prohibitin 1630 C>T polymorphism, and the results of the Egger’s test suggested no publication bias for the allele contrast model (P = 0.685), homozygote codominant model (P = 0.810), heterozygote codominant model (P = 0.926), dominant model (P = 0.639), or recessive model (P = 0.846).
Discussion
The 3′ untranslated region of the prohibitin gene which encodes a transacting regulatory RNA molecule arrests cell proliferation between the G1 and S phases of the cell cycle.Citation17 Jupe et al confirmed the antiproliferative activity of prohibitin by microinjection of prohibitin mRNA and protein into normal and immortalized cancer cell lines.Citation18 The protein-encoding region of the prohibitin gene was not found to be mutated, but the 3′-untranslated region of prohibitin mutations inhibited cell cycle progression in loss of human cancer cell lines.Citation5 The investigators confirmed that a single nucleotide polymorphism (C–T transition) in the prohibitin 3′-untranslated region creates a null (T) allele whereby the RNA product has lost its antiproliferative activity.Citation17 The results of our study are consistent with the functional prohibitin 3′-untranslated region 1630 C>T polymorphism resulting in increased risk of breast cancer, although there was no significant association between prohibitin 1630 C>T polymorphism and mRNA expression in lymphoblastoid cell lines from the HapMap database. Data being collected from different studies without stratification/adjustment for differences between studies and inconsistent use of selection criteria are possible explanations for this. Further investigations should be done in breast cancer tissue or cells to determine if a correlation exists between genotype and mRNA expression.
In this study, we investigated 5072 cases and 4796 controls, the allele contrast model, the homozygote codominant and the recessive model of prohibitin 1630 C>T polymorphism were found to be significantly associated with influencing the risk of breast cancer. Heterogeneity and publication bias were not observed in this study. Our findings suggest that prohibitin 1630 C>T polymorphism increases the risk of breast cancer.
Some limitations of this meta-analysis need to be acknowledged when interpreting its findings. First, we presumed that ethnicity status and family history play diverse roles in the risk of breast cancer. In our study, we considered the possibility that the effect of prohibitin 1630 C>T polymorphism might be ethnicity-specific in mixed populations, but we did not perform subgroup analysis to detect an association between this polymorphism and ethnicity. Second, our results were based on unadjusted estimates, so a more precise analysis should be done when more detailed individual data become available. A recent study evaluated the association between genetic variants of prohibitin and breast cancer risk in BRCA1 or BRCA2 mutation carriers, and the findings showed that the prohibitin 1630TT genotype may modify breast cancer risk in these women.Citation16 Third, as we all know, cancer is a complicated disease, different genetic backgrounds may contribute to the discrepancy, and it is still necessary to conduct larger sample studies considering gene-gene and gene-environment interactions, which may be an important component of the association between prohibitin 1630 C>T polymorphism and risk of breast cancer. In conclusion, the results of this meta-analysis suggest that the prohibitin 1630 C>T variant was associated with a significant increase in the risk of breast cancer.
Acknowledgements
This project was supported by grants from the National Natural Science Foundation of China (81272252, 81141094 to XG) and the Natural Science Foundation of Jiangsu Province (BK2011656 to XG). The authors thank Hongliang Liu, Department of Epidemiology, The University of Texas MD Anderson Cancer Center, for assistance with data analysis.
Disclosure
The authors report no conflicts of interest in this work.
References
- WangSNathNAdlamMChellappanSProhibitin, a potential tumor suppressor, interacts with RB and regulates E2F functionOncogene199918233501351010376528
- FusaroGDasguptaPRastogiSProhibitin induces the transcriptional activity of p53 and is exported from the nucleus upon apoptotic signalingJ Biol Chem200327848478534786114500729
- WhiteJJLedbetterDHEddyRLAssignment of the human prohibitin gene (PHB) to chromosome-17q21Cytogenet Cell Genet1991583–420112011
- CampbellIGAllenJEcclesDMProhibitin 3′ untranslated region polymorphism and breast cancer riskCancer Epidemiol Biomarkers Prev200312111273127414652295
- JupeERLiuXTKiehlbauchJLProhibitin in breast cancer cell lines: loss of antiproliferative activity is linked to 3′ untranslated region mutationsCell Growth Differ1996778718788809404
- ManjeshwarSLernerMRZangXPExpression of prohibitin 3′ untranslated region suppressor RNA alters morphology and inhibits motility of breast cancer cellsJ Mol Histol200435663964615614618
- JupeERBadgettAANeasBRSingle nucleotide polymorphism in prohibitin 3′ untranslated region and breast-cancer susceptibilityLancet200135792681588158911377649
- HolmKMelumEFrankeAKarlsenTHSNPexp – a web tool for calculating and visualizing correlation between HapMap genotypes and gene expression levelsBMC Bioinformatics20101160021167019
- StrangerBEForrestMSDunningMRelative impact of nucleotide and copy number variation on gene expression phenotypesScience2007315581384885317289997
- GibbsRABelmontJWHardenbolPThe International HapMap ProjectNature2003426696878979614685227
- HigginsJPThompsonSGDeeksJJAltmanDGMeasuring inconsistency in meta-analysesBMJ2003327741455756012958120
- EggerMSmithGDSchneiderMMinderCBias in meta-analysis detected by a simple, graphical testBMJ199731571096296349310563
- SpurdleABHopperJLChenXQProhibitin 3′ untranslated region polymorphism and breast cancer risk in Australian womenLancet2002360933792592612354477
- JakubowskaAGronwaldJGorskiBThe 3′ untranslated region C>T polymorphism of prohibitin is a breast cancer risk modifier in Polish women carrying a BRCA1 mutationBreast Cancer Res Treat20071041677417004108
- KarakusNKaraNUlusoyANLack of association between prohibitin 3′ untranslated region C>T polymorphism and breast cancer in a Turkish populationDNA Cell Biol200827844945218494604
- JakubowskaARozkrutDAntoniouAAssociation of PHB 1630C>T and MTHFR 677C>T polymorphisms with breast and ovarian cancer risk in BRCA1/2 mutation carriers: results from a multicenter studyBr J Cancer2012106122016202422669161
- ManjeshwarSBranamDELernerMRTumor suppression by the prohibitin gene 3′ untranslated region RNA in human breast cancerCancer Res200363175251525614500355
- JupeERLiuXTKiehlbauchJLThe 3′ untranslated region of prohibitin and cellular immortalizationExp Cell Res199622411281358612677