Abstract
Background/objective
MicroRNAs (miRNAs) are small noncoding RNAs (ribonucleic acids), approximately 22 nucleotides in length, that function as regulators of gene expression. Dysregulation of miRNAs has been associated with the initiation and progression of oncogenesis in humans. The cell division cycle (CDC)25 phosphatases are important regulators of the cell cycle. Their abnormal expression detected in a number of tumors implies that their dysregulation is involved in malignant transformation.
Methods
Using miRNA target prediction software, we found that miR-141 could target the 3′ untranslated region (3′UTR) sequence of CDC25B. To shed light on the role of miR-141 in renal cell carcinogenesis, the expression of miR-141 was examined by real-time polymerase chain reaction (RT-PCR) in renal cell carcinoma and normal tissues. The impact of miR-141 re-expression on 769-P cells was analyzed using 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and colony-forming assay. A luciferase reporter assay was applied to prove the functionality of the miR-141 binding site.
Results
miR-141 is significantly downregulated in renal cell carcinoma. miR-141 re-expression suppressed cell growth in 769-P cells. Luciferase expression from a reporter vector containing the CDC25B-3′UTR was decreased when this construct was transfected with miR-141 in 769-P cells. The overexpression of miR-141 suppressed the endogenous CDC25B protein level in 769-P cells.
Conclusion
For the first time, we demonstrated that CDC25B is a direct target of miR-141 in renal cell carcinoma. The transcriptional loss of miR-141 and the resultant increase in CDC25B expression facilitates increased genomic instability at an early stage of renal cell carcinoma development.
Introduction
Renal cell carcinoma (RCC) is the third most common urological cancer after prostate and bladder cancer, but it has the highest mortality rate, at over 40%. RCC of the clear cell type is the most common malignant tumor of the kidney and has a poor prognosis.Citation1,Citation2 Approximately one-third of patients with RCC develop metastasis, and systemic treatment, such as chemotherapy and cytokines, has not proven to be effective. Some studies have identified some putative oncogenes involved in the carcinogenesis of RCC, but the molecular mechanisms regulating the aggressive properties of RCC are still poorly understood.Citation3,Citation4
The cell division cycle (CDC)25 phosphatases are dual-specificity phosphatases that regulate the cell cycle progression of eukaryotes in response to DNA damage.Citation5 There are three CDC25 isoforms in humans: CDC25A, CDC25B, and CDC25C. CDC25A has a role in promoting the entry into S phase, and the CDC25A, -B, and -C isoforms cooperate to drive entry into mitosis.Citation6 CDC25B is activated in G2 phase and is responsible for the activation of cyclin A/Cdk2 in G2 phaseCitation7 and the initiation of cyclin B/Cdk1 activation and mitotic entry.Citation8–Citation10 It also has a unique function in recovery from a G2-phase checkpoint arrest.Citation11 CDC25B can be regulated by a lot of mechanisms, including its expression, stability, localization, and so on. CDC25B protein expression increases in G2 phase and peaks in metaphase, prior to its degradation upon mitotic exit.Citation8
MicroRNAs are small noncoding RNAs that participate in diverse biological processes, including tumorigenesis by sequence-specific targeting of particular messenger RNAs (mRNAs), primarily in the 3′ untranslated region (3′ UTR).Citation12 MicroRNAs negatively regulate protein production by translational repression, mRNA destabilization, or a combination of both.Citation13 In mammals, miRNAs were recently shown to act predominantly through mRNA degradation.Citation14
Using miRNA target prediction software, we found several miRNAs that could target the 3′UTR sequence of CDC25B, including miR-141, miR-200a, and miR-148ab. In addition, the score of miR-141 is higher than that of the other miRNAs, so we paid more attention to the role of miR-141. We focused on the impact of miR-141 re-expression in 769-P cells and its potential role in proliferation and anchorage-independent growth. We also investigated whether miR-141 directly targeted the 3′UTR of CDC25B and whether overexpression of miR-141 impacted the expression of CDC25B at the protein level. Our data demonstrate that miR-141 was able to modulate the expression of CDC25B in renal cell carcinoma.
Materials and methods
Cell culture
The human renal carcinoma cell line 769-P was obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, People’s Republic of China). The 769-P cells were maintained in Roswell Park Memorial Institute (RPMI) (Buffalo, NY, USA) 1640 medium supplemented with 10% fetal bovine serum (FBS), 100 units/mL penicillin, and 100 μg/mL streptomycin, in a humidified atmosphere of 5% CO2 at 37°C (all from Sigma-Aldrich, St Louis, MO, USA).
Transient transfection of miR-141 mimics
The 769-P cells were seeded in six-well plates. The cells were transfected in solution with 10 nM miR-141 mimics (Shanghai GenePharma Co, Ltd, Shanghai, People’s Republic of China), and 10 nM AllStars (Qiagen, Venlo, The Netherlands) served as control. The transfections of miRNA mimics were carried out using Lipofectamine™ 2000 (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions.
After 24 hours, the medium was replaced. Cells were harvested in RIPA buffer 96 hours posttransfection and subjected to Western blot analysis using CDC25B antibody.
Real-time polymerase chain reaction (RT-PCR)
The 769-P cells were seeded in six-well plates and transfected with miRNAs, and harvested 48 hours after transfection, before total RNA was extracted and DNase-treated using the miRNeasy Mini Kit (Qiagen). Mature miRNA expression of RCC, paired normal tissues, and 769-P cells were analyzed by TaqMan® miRNA-assays (Life Technologies). For cDNA synthesis, 50 ng of total RNA were reverse transcribed using the specific looped primer.
Western blot
Denatured protein was separated on a sodium dodecyl sulfate (SDS)-polyacrylamide gel and transferred to an Amersham™ Hybond™ membrane (GE Healthcare, Little Chalfont, UK), which was then blocked overnight in 5% skim milk in 2-Amino-2-hydroxymethyl-propane-1,3- diol (Tris)-buffered saline with Tween® 20 (TBST). For immunoblotting, the membrane was incubated overnight at 4°C with the rabbit antibody against CDC25B (1:100) (ab70927; Abcam, Cambridge, UK). Then, it was rinsed with TBST and incubated with immunoglobulin G (IgG) conjugated to horseradish peroxidase (1:1000) (Dako, Carpinteria, CA, USA) for 2 hours. Bands were visualized by ECL-Plus Western Blotting Detection Reagents (GE Healthcare). After that, the membrane was washed with Restore™ Western Blotting Stripping Solution (Thermo Fisher Scientific, Waltham, MA, USA) for 15 minutes and treated as described above, except that mouse anti-β-actin antibody (1:1000) (Santa Cruz Biotech, Dallas, Texas, USA) was used as an internal control.
Subjects
Surgical specimens were obtained between 1996 and 2010 from patients with newly diagnosed primary RCC in the First Hospital of China Medical University, Shenyang, People’s Republic of China. None of the patients underwent chemotherapy or radiotherapy treatment before surgery. Patient consent for the research use of tumor tissue was obtained, and the research protocol was approved by the Ethical Committee at China Medical University.
Proliferation assay
Cell viability was measured by a 3-(4,5-dimethyl thiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay. MTT solution was added to the cells in 96-well plates to the final concentration of 0.5 mg/mL, and the cells were incubated at 37°C for 4 hours. After removing the culture medium, 150 μL of dimethyl sulfoxide (DMSO) was added, and the optical density of each well was read at 590 nm.
Colony-forming assay
For anchorage-independent growth, 3 × 105 769-P cells were seeded per well on 0.7%. agarose (Life Technologies) in a 0.3% agarose solution in a six-well plate. The cells were fed every third day and cultivated over 4 weeks. Then, the cells were stained with iodonitrotetrazolium chloride solution overnight. The stained cells were scanned on an Epson GT-8000 (Longbeach, CA, USA) scanner.
Luciferase reporter assay
Luciferase constructs were made by ligating the 210 base pair (bp) 3′ of the 3′UTR of CDC25B as well as the mutated version after the luciferase open reading frame in the pGL3. The cells were triple-transfected using Lipofectamine-2000 (Life Technologies) with pGL3 containing either the wild type or mutant CDC25B sequence, in combination with 8 nM miR-141 mimics, and 20 ng pGL-Renilla luciferase control reporter vector (Promega Corp, Fitchburg, WI, USA) served as a transfection control. As a negative control, 8 nM AllStars siRNA oligos (Qiagen) was used. The luciferase was measured 48 hours after transfection, using the Dual-Luciferase® Reporter Assay System kit (Promega) according to the manufacturer’s instructions. Firefly luciferase was normalized against Renilla luciferase. All experiments were performed at least three independent times, with two parallels.
Results
miR-141 is significantly downregulated in renal cell carcinoma
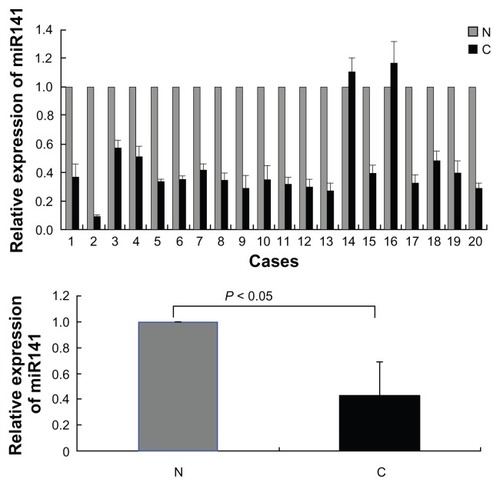
To confirm and validate the predicted result by software on miR-141, we first compared the transcription level of miR-141 in samples of 20 patients with RCC with 20 samples of matched adjacent normal tissues. The mean transcription level of miR-141 was significantly lower in RCC compared with the nontumor samples (P < 0.05) ().
Figure 1 The miR-141 expression is significantly downregulated in renal cell carcinoma.
Abbreviations: N, normal tissues; C, cancer; RT-PCR, real-time polymerase chain reaction; SE, standard error.
miR-141 re-expression suppressed cell growth in 769-P cells
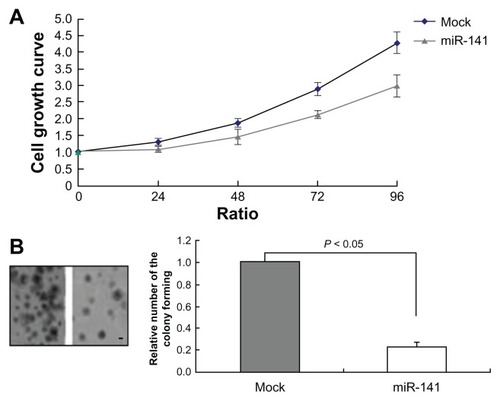
As miR-141-expression was silenced in RCC, we analyzed the functional impact of miR-141 in the RCC cell line 769-P. We generated miR-141-overexpressing 769-P cells by transient transfection of miR-141 mimics. The validity of miR-141 expression was confirmed by RT-PCR. We observed 80-fold increased miR-141 expression in comparison with the respective mock transduced cells (). We analyzed the impact of miR-141 re-expression on 769-P cells using MTT and colony-forming assay in this system (). The MTT assay revealed that there was significant change in the cell growth. At the end of the assay, we observed an obvious reduction in cell growth about the miR-141-overexpressing cells compared with the mock transduced ones (P < 0.05). This led to the assumption that miR-141 overexpression significantly reduces the tumor cell viability (). The colony-forming assay showed that foci from the miR-141-overexpressing 769-P cells were reduced in comparison with the mock transduced cells after 4 weeks ().
Figure 2 Average miR-141 relative expression levels were determined using RT-PCR in 769-P cells overexpressing miR-141.
Abbreviations: RT-PCR, real-time polymerase chain reaction; SE, standard error.
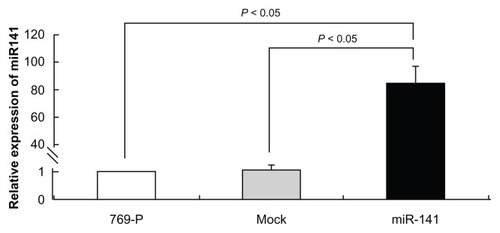
Figure 3 miR-141 reduced cell growth and colony formation. (A) Growth kinetics of mock 769-P cells and miR-141-overexpressing 769-P cells, using MTT assay (P < 0.05). (B) Colony-forming assay of mock 769-P cells and miR-141- overexpressing 769-P cells.
Abbreviations: MTT, 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; SE, standard error.
CDC25B is a candidate target of miR-141
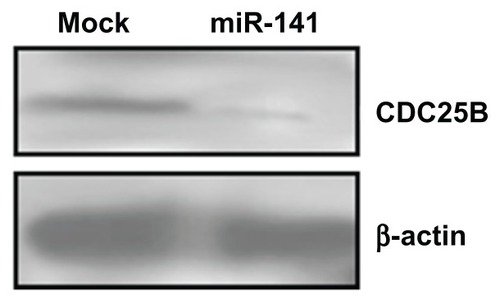
TargetScan v5.1 (Whitehead Institute for Biomedical Research, Cambridge, MA, USA) target prediction software was used to identify miRNAs that could potentially target the human CDC25B-3′UTR. CDC25B was the most promising candidate target for miR-141, which also contained a putative miR-141 binding site at the end of its 3′UTR (). A luciferase reporter assay was performed to prove the functionality of the miR-141 binding site. Therefore, we cloned the 210 bp 3′ of the CDC25B-3′UTR containing the predicted seed sequence downstream of the luciferase reporter gene. At 32 hours posttransfection, the luciferase activity was monitored in the group of miR-141 mimics and the negative control group in the human osteosarcoma cell line U2-OS, respectively. The luciferase reporter activity decreased approximately 55% in the U2-OS cells containing the CDC25B wild-type 3′UTR fragment. However, no significant change was found in the luciferase activity in cells expressing the mutated 3′UTR (). These data demonstrate the potential direct interaction between miR-141 and the 3′UTR of CDC25B mRNA. Next, we wanted to know the impact of miR-141 on the CDC25B protein expression in miR-141-overexpressing 769-P cells. We found that the overexpression of miR-141 suppressed the endogenous CDC25B protein level in 769-P cells ().
Figure 4 Suppression of CDC25B by miR-141. (A) Sequence complementarity between hsa-miR-141 and the 576–583 bp of the CDC25B 3′UTR. (B) Interaction between miR-141 and 3′ UTR of CDC25B. The U2-OS cells were transfected with WT and Mut 3′UTR of CDC25B containing luciferase reporter vector together, miR-141, and the control vector, respectively, as indicated.
Abbreviations: 3′ UTR, 3′ untranslated region; bp, base pair; CDC, cell division cycle [phosphatase]; hsa, Homo sapiens; Mut, mutant; SE, standard error; WT, wild-type.
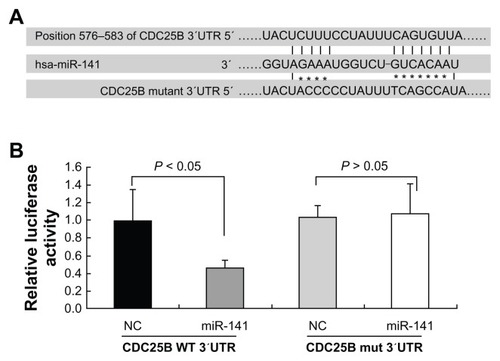
Figure 5 Alteration of CDC25B expression, by miR-141 Western blot analysis, demonstrating a reduction in CDC25B protein in 769-P cells transfected with miR-141 mimics.
Abbreviations: CDC, cell division cycle [phosphatase]; SE, standard error.
Discussion
MicroRNAs posttranscriptionally regulate the expression of more than 30% of protein coding genes by translational repression, which also regulates the expression of several putative target genes by binding to a complementary sequence predominantly in their UTR.Citation15 This posttranscriptional regulation within the context of tumor development has been reported for the regulation of genes that have an impact on cell differentiation, apoptosis, and neoplastic transformation.Citation16,Citation17 The miR-200 family member miR-141 can differentiate papillary thyroid cancer cell lines from a control thyroid cell line. Hashimoto thyroiditis epithelium can be distinguished from classic papillary thyroid cancer epithelium and control epithelium based on the relative expression of miR-141.Citation18 The serum level of miR141 was significantly higher in lung cancer patients than that in patients with benign disease, and high serum miR141 values were associated with elevated levels of urokinase plasminogen activator.Citation19 However, as miRNAs are expected to have multiple targets, we sought to further investigate and expand the putative targets of miR-141 in RCC. Here we, for the first time, examined miR141 expression in RCC samples by RT-PCR. The mean level of miR-141 was significantly lower in RCC compared with nontumor samples. In the present study, the in vitro model was established: miR-141 overexpression based on the transient transfection of miR-141 mimics in the RCC cell line 769-P. We subsequently exploited this in vitro model to validate the tumor-suppressor role of miR-141 in RCC by demonstrating that miR-141 overexpression had an inhibitory influence on the growth potential of RCC cells. It was proposed that miR-141 might be an important negative regulator in RCC, in terms of growth and proliferation.
Using miRNA target prediction software, we found several miRNAs that could target the 3′UTR sequence of CDC25B, including miR-141, miR-200a, and miR-148ab. A body of evidence indicated that CDC25B abnormal expression detected in some tumors implies that its dysregulation is involved in malignant transformation. CDC25B plays a minor role in the pathogenesis and/or progression of vulvar carcinoma, which has been associated with malignant features and aggressive cancer phenotypes but not independently correlated to prognosis.Citation20 In contrast, we have identified that the expression of CDC25B was increased distinctively in carcinoma over that in the adjacent normal tissues, at both mRNA and protein levels, and there was a positive correlation between CDC25B expression and clinical stage and histopathologic grade of RCC.Citation21 However, the mechanism that leads to the dysregulation of CDC25B within the specific context of tumor proliferation and progression has largely remained elusive. The poor correlation between CDC25B mRNA and its corresponding protein abundance has often led to the speculation that the deregulation of CDC25B may occur at any stage between its transcription and translation.Citation6 It is reported that CDC25B was identified as a novel miRNA-148a target. CDC25B was downregulated, at the protein level, in miR-148a-overexpressing IMIM-PC2-cells and in transiently transfected pancreatic cell lines as well as in human pancreatic ductal adenocarcinoma.Citation22 The level of CDC25B was found to be lower in U2-OS cells compared with other cell lines, such as MiaPaCa or Hep2 cells,Citation23 so here, U2-OS cells were chosen to transfect the CDC25B wild-type or mutation-type 3′UTR fragment. Our results, for the first time, indicate a downregulation of CDC25B by miR-141 at the posttranscriptional level, by translational repression. This proposed interaction is evidenced from our reporter assay–based studies, where miR-141 was directly shown to influence the expression of the luciferase reporter by binding to the predicted target sequence in the CDC25B-3′UTR.
In summary, using an in vitro model based on miR-141 overexpression in the RCC cell line 769-P, we validated the tumor-suppressor role of miR-141 in RCC. Further, our results, for the first time, demonstrate that CDC25B is a direct target of miR-141 in RCC. The transcriptional loss of miR-141 and the resultant increase in CDC25B expression facilitate increased genomic instability at an early stage of RCC development.
Acknowledgments
This work was supported by National Natural Scientific Foundation of China (Grant No 81172438).
Disclosure
The authors report no conflicts of interest in this work.
References
- YoungANMasterVAPanerGPWangMDAminMBRenal epithelial neoplasms: diagnostic applications of gene expression profilingAdv Anat Pathol2008151283818156810
- LeroyXZiniLBuobDBallereauCVillersAAubertSRenal cell carcinoma with rhabdoid features: an aggressive neoplasm with overexpression of p53Arch Pathol Lab Med2007131110210617227108
- KardasIMrózekKBabinskaMCytogenetic and molecular findings in 75 clear cell renal cell carcinomasOncol Rep200513594995615809763
- GirolamiFPasseriniIGarganoDMicrosatellite analysis of chromosome 3p region in sporadic renal cell carcinomasPathol Oncol Res20028424124412579209
- BoutrosRDozierCDucommunBThe when and wheres of CDC25 phosphatasesCurr Opin Cell Biol200618218519116488126
- BoutrosRLobjoisVDucommunBCDC25 phosphatases in cancer cells: key players? Good targets?Nat Rev Cancer20077749550717568790
- GoldstoneSPaveySForrestASinnamonJGabrielliBCdc25-dependent activation of cyclin A/cdk2 is blocked in G2 phase arrested cells independently of ATM/ATROncogene200120892193211314027
- GabrielliBGDe SouzaCPTonksIDClarkJMHaywardNKEllemKACytoplasmic accumulation of cdc25B phosphatase in mitosis triggers centrosomal microtubule nucleation in HeLa cellsJ Cell Sci1996109Pt 5108110938743955
- KarlssonCKatichSHagtingAHoffmannIPinesJCdc25B and cdc25C differ markedly in their properties as initiators of mitosisJ Cell Biol1999146357358410444066
- LöfflerHRebaczBHoADLukasJBartekJKrämerAChk1-dependent regulation of Cdc25B functions to coordinate mitotic eventsCell Cycle20065212543254717106257
- van VugtMABrásAMedemaRHPolo-like kinase-1 controls recovery from a G2 DNA damage-induced arrest in mammalian cellsMol Cell200415579981115350223
- BartelDPMicroRNAs: target recognition and regulatory functionsCell2009136221523319167326
- FilipowiczWBhattacharyyaSNSonenbergNMechanisms of post-transcriptional regulation by microRNAs: are the answers in sight?Nat Rev Genet20089210211418197166
- GuoHIngoliaNTWeissmanJSBartelDPMammalian microRNAs predominantly act to decrease target mRNA levelsNature2010466730883584020703300
- CarthewRWGene regulation by microRNAsCurr Opin Genet Dev200616220320816503132
- DrakakiAIliopoulosDMicroRNA gene networks in oncogenesisCurr Genomics2009101354119721809
- HammondSMRNAi, microRNAs, and human diseaseCancer Chemother Pharmacol200658 Suppl 1S63S6817093929
- DorrisERSmythPO’LearyJJSheilsOMIR141 expression differentiates Hashimoto thyroiditis from PTC and benign thyrocytes in Irish archival thyroid tissuesFront Endocrinol (Lausanne)2012310222969748
- RothCKasimir-BauerSPantelKSchwarzenbachHScreening for circulating nucleic acids and caspase activity in the peripheral blood as potential diagnostic tools in lung cancerMol Oncol20115328129121398193
- WangZTropeCGFlørenesVASuoZNeslandJMHolmROverexpression of CDC25B, CDC25C and phospho-CDC25C (Ser216) in vulvar squamous cell carcinomas are associated with malignant features and aggressive cancer phenotypesBMC Cancer20101023320500813
- YuXYZhangZZhangGJGuoKFKongCZKnockdown of Cdc25B in renal cell carcinoma is associated with decreased malignant featuresAsian Pac J Cancer Prev201213393193522631674
- LiffersSTMundingJBVogtMMicroRNA-148a is down-regulated in human pancreatic ductal adenocarcinomas and regulates cell survival by targeting CDC25BLab Invest201191101472147921709669
- BuglerBQuarantaMAressyBBrezakMCPrevostGDucommunBGenotoxic-activated G2-M checkpoint exit is dependent on CDC25B phosphatase expressionMol Cancer Ther2006561446145116818502