Abstract
Osteosarcoma (OS) is the eighth most common form of childhood and adolescence cancer. Approximately 10%–20% of patients present metastatic disease at diagnosis and the 5-year overall survival remains around 70% for nonmetastatic patients and around 30% for metastatic patients. Metastatic disease at diagnosis and the necrosis grade induced by preoperative treatment are the only well-established prognostic factors for osteosarcoma. The DNA aberrant methylation is a frequent epigenetic alteration in humans and has been described as a molecular marker in different tumor types. This study evaluated the DNA aberrant methylation status of 18 genes in 34 OS samples without previous chemotherapy treatment and in four normal bone specimens and compared the methylation profile with clinicopathological characteristics of the patients. We were able to define a three-gene panel (AIM1, p14ARF, and ESR1) in which methylation was correlated with OS cases. The hypermethylation of p14ARF showed a significant association with the absence of metastases at diagnoses, while ESR1 hypermethylation was marginally associated with worse overall survival. This study demonstrated that aberrant promoter methylation is a common event in OS and provides evidence that p14ARF and ESR1 hypermethylation could be useful as a prognostic indicator for this disease.
Introduction
Osteosarcoma (OS) is the eighth most common form of childhood and adolescence cancer, comprising 2.4% of all malignancies in pediatric patients, and approximately 20% of all primary bone cancers.Citation1 Characteristically, OS is found in the metaphyseal regions of long bones in the appendicular skeleton, with the majority of patients developing the disease during the period of active bone growth in early adolescence. More than 15% of patients present clinically detectable pulmonary metastases.Citation2
The implementation of combined treatments (neoadjuvant chemotherapy, surgery, and adjuvant chemotherapy) and the use of multi-agent chemotherapy have improved prognosis over the last several decades, reaching 5-year survival rates of up to 70% for patients without metastatic disease at diagnosis.Citation3,Citation4 Despite these advances, for patients that present with metastases at diagnosis or those with tumors showing a poor response to chemotherapy, the prognosis is still unsatisfactory (5-year survival rates, 20%–40%), even with dose-intensive or high-dose chemotherapy.Citation5–Citation9 This suggests that even in tumors with the same histologic type, different genetic and epigenetic mechanisms may be operating, altering the response to chemotherapy and the metastatic capability.
The best prognostic factor for OS is the presence of metastatic disease at diagnosis and the necrosis grade induced by chemotherapy.Citation10 At the molecular level, OS is characterized by frequent genomic instability, highly heterogeneous karyotypes, gross changes in gene expression, and recurrent epigenetic alterations.Citation11–Citation16
Genetic alterations caused either by loss of heterozygosity or by mutations have been reported in OS. Besides this, most of the available data suggests that this bone tumor arises as a result of the inactivation of different tumor suppressor genes.Citation17 However, none of these alterations can characteristically reflect the biologic nature or clinical features of all OS cases. Therefore, assessment of more genetic and epigenetic data from OS tumors could provide important insights concerning both OS oncogenesis and molecular alterations governing differential clinical response to treatment.
Aberrant DNA methylation (hypermethylation) of gene promoter regions is the most widely epigenetic abnormality studied in human malignancies and is an important epigenetic mechanism of gene transcription regulation. This process is catalyzed by DNA methyltransferases and involves the addition of a methyl group to the carbon 5 position of the cytosine ring in CpG dinucleotides.Citation18 It is associated with several changes in chromatin structure, including the regulation of histone methylation and acetylation and the recruitment of proteins to the methylated sites. The methylation usually leads to the obstruction of the promoter region, hindering gene transcription and subsequently causing gene silencing.Citation19 In addition to genetic aberrations, there is increasing evidence that epigenetic processes also play a major role in carcinogenesis. Hypermethylation of the gene promoter region acts as an alternative to mutations in disrupting tumor suppressor gene function.Citation20
Aberrant promoter methylation has been found in several genes in various malignant diseases, and each tumor type may have its own distinct pattern of methylation.Citation21 It was reported previously that hypermethylation of different genes may occur in OS cases, among them are CDKN2A, CDKN2B, RASSF1A, DAPK, MGMT, TIMP3, and RB1.Citation22–Citation26 All of these studies focused on a few genes and most of them were conducted using conventional methylation-specific (MSP) polymerase chain reaction (PCR) technique.
Therefore, we sought to evaluate the presence of aberrant DNA methylation in promoter regions of 18 candidate genes in 34 OS specimens as well as four normal bone samples by a real-time quantitative MSP (QMSP) PCR approach. Thus, the aims of our study were (1) to determine the methylation profile of a panel of genes in OS; (2) to correlate the molecular data with the clinicopathological characteristics of the patients; and (3) to identify epigenetic biomarkers that may be useful for diagnosis and/or as prognostic factors for OS cases.
Materials and methods
Patients, sample collection, and DNA preparation
The study involved 34 patients with OS treated between 1996 and 2004 at A C Camargo Hospital, São Paulo, Brazil and at the Pediatric Oncology Institute, GRAACC/Federal University of São Paulo, São Paulo, Brazil. Fresh OS biopsies were collected at diagnosis, without previous chemotherapy treatment. Additionally, four fresh normal bone specimens (distal femurs) from patients without bone related sarcomas who underwent inferior member amputation during radical surgery treatment at Barretos Cancer Hospital, Barretos, Brazil were included as normal controls.
Tissue sections of all samples were stained with hematoxylin and eosin according to standard procedures and reviewed by a senior pathologist in order to confirm the diagnosis of OS. Clinical information was collected from the patients’ medical records. The study was approved by the Ethics Committees of all the institutions.
DNA was isolated from bone specimens using the TRIzol reagent (Life Technologies, Carlsbad CA, USA) following the manufacturer’s recommendations.
Bisulfite treatment
Bisulfite treatment of DNA converts unmethylated cytosines to uracil, while the methylated ones remain as cytosines. Sodium-bisulfite conversion of 2 μg of DNA was performed according to a previously described method with modifications.Citation27 In brief, 2 μg of DNA from each sample was denatured in 0.2 M of NaOH for 20 minutes at 50°C (in a total volume of 20 μL). The denatured DNA was diluted in 500 μL of bisulfite solution (2.5 M of sodium metabisulfite, 125 mM of hydroquinone, 350 mM of sodium chloride, pH 5.0) and incubated for 3 hours at 70°C in the dark. Bisulfite-modified DNA was purified using the Wizard DNA Clean-Up System (Promega, Madison, WI, USA) according to the manufacturer’s instructions and eluted in 45 μL of 80°C water. After treatment with NaOH (final concentration 0.3 M) for 10 minutes at room temperature, the treated DNA was precipitated by the addition of 75 μL of ammonium acetate, 2.5 volumes of ethanol, and 2 μL of glycogen (5 mg/mL). Each resulting DNA pellet was washed with 70% ethanol, dried dissolved in 110 μL of water, and stored at −80°C.
Gene selection
After a literature examination and mining in different public databases, 18 genes were selected for the evaluation of methylation abnormalities. All genes analyzed in this study were previously reported as targets for epigenetic silencing in different human cancers. The majority of these genes present tumor suppressor activities and their silencing could contribute to the tumorigenesis process. Among these genes are CCNA1, CDKN2A, HIC1, p14ARF, RB1, and SOCS1 which are involved in cell cycle control; CDH1 in cell adhesion; ESR1, APC, DAPK, RASSF1A, RARβ, and THBS1 in signal transduction processes; GSTP1 in cell detoxification; MLH1 in DNA repair; CALCA in cell-cell signaling processes; and SFRP1 in cell differentiation and proliferation. The methylation pattern of AIM1 was also examined but its function is not yet well understood. It has been previously shown that these genes are affected by aberrant promoter methylation in association with transcription silencing in different types of human malignancies.Citation28–Citation34
QMSP PCR analyses
The QMSP PCR analyses were conducted as previously described.Citation35 Basically bisulfite-modified DNA was used as a template in fluorogenic QMSP assays carried out in a final volume of 20 μL in a 7500 Real Time PCR System (Life Technologies). PCR was done in separate wells for each primer/probe set and each sample was run in triplicate. The final reaction mixture contained 3 μL of bisulfite-modified DNA, 1.2 μM of forward and reverse primers, 200 nM of probe, 0.6 U of platinum Taq polymerase (Life Technologies), 200 μM of dNTPs, 16.6 mM of ammonium sulfate, 67 mM of Tris-HCl pH 8.0, 6.7 mM of magnesium chloride (2.5 mM for CDKN2A), 10 mM of mercaptoethanol, 0.1% DMSO and 1× ROX dye (Life Technologies). PCR was conducted with the following conditions: 95°C for 2 minutes, followed by 45 cycles at 95°C for 15 seconds and 60°C for 1 minute.
Each plate included DNA samples, multiple water blanks, a negative control (normal leukocyte DNA), and serial dilutions (90–0.009 ng) of a positive control for constructing the calibration curves. Leukocyte DNA from a healthy individual was methylated in vitro with SssI methyltransferase (New England Biolabs Inc, Ipswich, MA, USA) to generate completely methylated DNA at all CpG and used as a positive control.
Primers and probes were designed to specifically amplify the promoter regions of the 18 genes of interest and the internal reference gene, ACTB (Supplementary Table S1). The relative level of methylated DNA of each gene in each sample was determined as a ratio of MSP PCR-amplified gene to ACTB and then multiplied by 100 for easier tabulation (average value of triplicates of specific gene divided by the average value of triplicates of ACTB × 100). Cases were scored as positive if a percentage value of ≥0.1% was obtained. This cutoff was chosen for being clinically relevant and also to exclude very low-level background readings that can occur in certain individual for certain genes.Citation36
Statistical analysis
SPSS 13.0 (IBM Corporation, Armonk, NY, USA) for Windows was used for all statistical analyses. Descriptive statistics were used to summarize study data. For all analysis we considered statistical significance when P-value <0.05. Comparisons between clinical-demographic variables and methylation patterns were performed using the chi-square test or Fisher’s exact test. Survival curves were estimated using the Kaplan–Meier method. Survival data were censored for patients alive at the last observation. The log-rank test was used to compare survival outcomes. The univariate Cox regression model was used to evaluate the methylation level influence in the overall survival.
Results
Patient characteristics and clinical predictors
Clinical characteristics of the patients are summarized in . The age at diagnosis ranged from 7 to 29 years (median of 14 years). The most frequent site of the primary tumor was the femur (52.9%), followed by tibia (26.5%), humerus (8.8%), and fibula (5.9%). Metastasis at diagnosis was detectable in 29.4% of the cases. Osteoblastic (61.8%) was the most common histological subtype, followed by chondroblastic (17.6%) and telangiectasic (5.9%).
Table 1 Clinical characteristics of the osteosarcoma patients included in this study
The necrosis grade (Huvos Grade) was evaluated in the surgical specimens after neoadjuvant chemotherapy and 29.4% of patients were classified as Huvos I (less than 50% of tumor necrosis), 29.4% as Huvos II (51% to 90% of necrosis), 20.6% as Huvos III (90% to 99% of necrosis), and 20.6% as Huvos IV (100% of necrosis). Recurrent disease was observed in 52.9% of patients with 17.7% showing combined local and lung relapses, 8.8% local recurrence alone, 20.6% isolated pulmonary relapses, and 5.8% bone relapse at a different site from the primary one (data not shown).
QMSP PCR in osteosarcoma
In the first series, the promoter methylation status of 18 genes was evaluated in DNA from 13 OS biopsy samples, collected at diagnosis, without any previous treatment. This analysis showed that CDKN2A, CCNA1, GSTP1, THBS1, RB1, and DAPK were unmethylated in the tested samples and the hypermethylation of RASSF1A (7.6%), GSTP1 (10.0%), RARβ (15.3%), APC (23.0%), SOCS1 (23.0%), and MLH1 (23.0%) were rare events. Conversely, ESR1 (30.7%), AIM1 (30.7%), p14ARF (61.5%), SFRP1 (61.5%), CALCA (76.0%), CDH1 (76.0%), and HIC1 (92.0%) were found to be frequently methylated (). Representative examples of QMSP results are shown in Supplementary Figure S1. The most appropriate genes for the additional analyses were those that were shown to be frequently methylated in the OS samples evaluated in the first series. Thus, in the second series, the presence of promoter methylation of seven genes, namely, AIM1, CALCA, CDH1, ESR1, HIC1, p14ARF, and SFRP1, was tested in 21 additional cases and four normal bone samples.
Table 2 Promoter methylation frequency for the 18 genes analyzed in osteosarcoma samples (pilot group n = 13 and total group n = 34) and in the normal control group (n = 4)
By the end, CALCA was methylated in 79.4% of all analyzed cases (27/34), SFRP1 and HIC1 in 76.5% (26/34), CDH1 in 61.8% (21/34), AIM1 in 38.2% (13/34), p14ARF in 23.5% (8/34), and ESR1 in 14.7% (5/34) (). Although CALCA, HIC1, CDH1, and SFRP1 were found to be methylated in OS samples, they were also methylated in the normal bone samples used as the control (75%, 75% 50%, and 50%, respectively) and, for this reason, they were not good candidates for tumor markers. On the other hand, the methylation of AIM1, ESR1, and p14ARF seems to be specific to tumor samples because no hypermethylation was detected in the control samples ().
Methylation levels and clinical-pathologic correlations
The methylation patterns of AIM1, ESR1, and p14ARF were analyzed for potential correlations with clinical characteristics of patients with OS, including age, gender, primary tumor site, histologic subtype, Huvos Grade, presence of metastasis at diagnosis, and recurrence. Hypermethylation of p14ARF was significantly associated with the absence of metastasis (P = 0.041), while unmethylation of AIM1 was associated with the chondroblastic OS histological subtype (P = 0.038) (). No significant correlation was observed between the other clinical features and methylation status of the remaining genes tested.
Table 3 Correlations between hypermethylation pattern and clinicopathological parameters of OS patients evaluated
The 5-year overall survival for all OS patients included in this study was 64%. There was no significant difference in overall survival by gender (P = 0.184), age (P = 0.690), primary tumor site (P = 0.096), histological subtype (P = 0.343), and Huvos Grade (P = 0.407). On the other hand, as expected, the presence of metastasis at diagnosis (16.7% metastatic at diagnosis versus 86.8% nonmetastatic; P = 0.001) and tumor recurrence after pre-adjuvant chemotherapeutic treatment (41.3% with recurrence versus 92.9% nonrecurrence; P = 0.024) influenced the overall survival of OS patients ().
Table 4 Univariate analysis of selected factors for overall survival
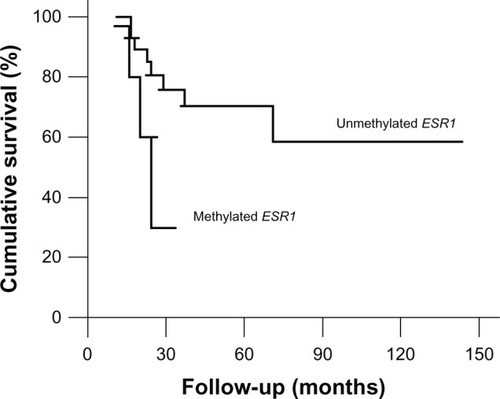
There was no significant association between the overall survival and the hypermethylation profile of the genes investigated. However, although the difference in overall survival between patients with and without ESR1 hypermethylation was not significant (70.4% hypermethylated versus 30% non-methylated; P = 0.059), the patients with methylated ESR1 seem to have had a worse prognosis when compared with OS patients with unmethylated ESR1 (hazard ratio = 3.554; confidence interval = 0.873–14.475; P = 0.077) ( and ).
Discussion
Many studies have demonstrated the importance of DNA hypermethylation in the extinction of tumor suppressor gene activity in different human cancers. Considerable variations exist in promoter methylation profiles of different cancers, such that, individual tumor types have characteristic methylation profiles.Citation21
To date, few studies have attempted to assess the OS methylation pattern. All but one of them relied on the conventional MSP PCR approach and evaluated a few genes in a small number of cases.Citation15,Citation16,Citation26 Thus, the present study is pioneering in its use of the QMSP approach to conduct an extensive analysis of the OS methylation profile. Given the sensitivity of the QMSP technique used to detect the presence of methylated alleles in a background of normal cells at a threshold of 1/1000 to 1/10,000, this strategy allowed us to define methylated genes that were highly specific for tumor, and rarely or never present in normal bone.Citation37
In the present study, we evaluated the hypermethylation pattern of 18 genes in 34 OS cases. Of note, this panel of 18 genes included genes already reported to be methylated in OS (CDKN2A, DAPK, RASSF1A, and p14ARF) and genes not yet evaluated in this neoplasia (AIM1, APC, CALCA, CCNA1, CDH1, ESR1, GSTP1, HIC1, MLH1, THBS1, RARβ, SFRP1, and SOCS1).
OS is the most commonly diagnosed primary malignancy of bone, particularly among children and adolescents; however, it is rare, representing less than 1% of all cancers. Due to restrictions in the available amount of DNA from some cases, we first investigated the panel of 18 genes in 13 tumor samples and then the seven most frequently methylated genes in a second series of cases (n = 21) and in four normal bone samples. We were able to define a three-gene panel (AIM1, p14ARF, and ESR1) for which methylation was correlated with OS cases. Worth mentioning, the clinical and pathological characteristics of the cohorts analyzed in the first and second series were similar.
AIM1 (absent in melanoma 1) is one of the newest cancer-associated genes discovered and its function is poorly understood. It was found to be involved in melanoma tumorigenesis and in calcium binding.Citation38 Ray et al described a possible role of AIM1 in processes of stress response, differentiation, and changes in cell morphology through interactions with the cytoskeleton.Citation38 This gene was found to be methylated in different tumors such as lung, bladder, and nasopharyngeal carcinoma,Citation39–Citation41 but there is no previous report of AIM1 methylation in OS. According to our results, the absence of methylation in the AIM1 promoter region is significantly correlated with the OS chondroblastic histological subtype. Worth mentioning, AIM1 is localized in chromosome 6 (6q21) and this locus is described as frequently deleted in OS.Citation42,Citation43 Thereby, not only hypermethylation, but also chromosomal deletion, is contributing to AIM1 silencing in OS, corroborating its role as a tumor suppressor gene.
The p14ARF gene is encoded in the INK4a/ARF locus, situated on chromosome 9p21, which also encodes the cyclin inhibitor CDKN2A. The p14ARF protein is translated from an alternative reading frame of CDKN2A.Citation44 The p14ARF gene induces growth arrest and acts as a negative regulator of cell proliferation.Citation45 It has been demonstrated that p14ARF binds to MDM2 and inhibits the ubiquitination of p53, thereby stabilizing p53.Citation46 p14ARF is regulated mainly at the transcriptional level by DNA hypermethylation of its promoter region, which has come to the forefront of many studies.Citation47 Oh et al evaluated the hypermethylation profile of the p14ARF promoter region in 32 OS samples using conventional MSP.Citation16 They were able to detect p14ARF methylation in 47% of the samples and this alteration was correlated to worse overall survival (79% of survival for unmethylated versus 31% of survival for methylated, P = 0.03). Controversially, in our study, p14ARF was found to be methylated in 23.5% of the OS cases and this methylation was associated with the absence of metastases at diagnosis (P = 0.04), a clinical indicator for favorable outcome. So the hypermethylation of p14ARF seems to be correlated with a favorable prognosis for OS patients. Other studies with different tumor types have also identified the aberrant methylation of p14ARF as a favorable prognostic factor. Sailasree et al analyzed 116 oral tumor samples and concluded that methylation of p14ARF is related to a low rate of local recurrence and a better prognosis compared with patients who present this gene unmethylated.Citation48
ESR1 encodes the estrogen receptor 1, a protein that can initiate or enhance gene transcription in response to estrogen stimulation. Estrogen has a multifunctional role, influencing the growth, differentiation, and function of different tissues. In bone tissue, estrogen has an important role in regulating bone growth during puberty and bone remodeling in adults.Citation49 Some authors have suggested that the estrogen effect mediated by ESR1 is involved with bone mineralization and the homozygous deletion of this gene could be associated to severe osteoporosis and increased bone turnover.Citation50 In addition, several studies have shown the presence of these receptors in osteoblasts and osteoclasts, but with different activities.Citation51 This gene was found to be methylated in 14.7% of the samples and this group of patients showed a worse 5-year overall survival (non-methylated: 60%; methylated: 30%, P = 0.059). Despite the fact that this difference was not statistically significant, the survival curves show a clear trend to separate the two groups. We do not exclude the possibility that the trend to significant association observed between aberrant methylation of ESR1 and poor prognosis could be due to the small sample size. Susman analyzed the expression of ESR1 in tumor samples from 110 OS patients by immunohistochemistry and found that ESR1 expression was associated with localized disease at presentation and improved outcomes.Citation52 According to Susman, high levels of ESR1 inhibited tumor proliferation and ESR1 expression could be associated with event-free survival. Issa et al have already demonstrated that hypermethylation is an important mechanism of transcriptional repression and gene silencing for the ESR1 gene.Citation53 So, taking that into account, our findings are in complete concordance with Susman’s because both studies found that the OS patients expressing ESR1 (unmethylated cases) presented better overall survival rates.
The data generated in this study do not allow us to know if the aberrant methylation of ESR1 could be useful only as a marker of tumor progression or could serve to disrupt directly a gene that is critical to the biology of OS. But, for the first time, it was suggested that ESR1 aberrant methylation is a relevant molecular alteration related with poor prognosis of OS patients. We trust that this study shed some light on this field and more analyses are needed to dissect the association between the ESR1 pathway and OS. Besides, our results, together with other findings, provide some evidence that aberrant methylation is a common event in OS and suggest that patients with this bone neoplasia could also have benefited by demethylating drug treatments. However, this hypothesis needs to be proved by in vitro functional studies before using demethylating agents in OS clinical trials.
Histologic response to chemotherapy is currently the best prognostic parameter in OS, but it can be evaluated only after several weeks of chemotherapy. Thus, a prognostic parameter known at the time of diagnosis would be of great clinical benefit. We do not exclude the possibility that the limited number of cases available for analysis could bias our findings; however our data suggest that p14ARF and ESR1 methylation status may be useful as prognostic markers for OS. Of note, a larger patient cohort needs to be evaluated in support of our findings. In the future, preoperative tests could be performed and the methylation status of these genes could help in the choice of the best therapy scheme to be adopted.
Conclusion
This study represents the largest quantitative evaluation of the methylation profile of OS. We demonstrated that aberrant methylation is a frequent event in OS and our data provide further evidence that aberrant methylation of p14ARF was significantly associated with the absence of metastasis at diagnosis and ESR1 methylation could be correlated with poor prognosis.
Acknowledgments
This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico, Edital Universal (CNPq) grant 472193/2004-0 (to ALV). VS was the recipient of a fellowship from Fundação Antonio Prudente (FAP). ALV received a fellowship from CNPq, grant 472193/2004-0. The authors thank the tumor bank of A C Camargo Hospital for kindly providing tumor samples to be examined in this study.
Supplementary materials
Figure S1 Amplification plots representing the quantitative methylation-specific results.
Notes: (A) Calibration curves constructed by serial dilutions (90–0.009 ng) of a leukocyte DNA methylated in vitro. (B) Amplification of all samples by the reference gene ACTB. (C) A frequently unmethylated gene showing the amplification of the positive control only (leukocyte DNA methylated in vitro). (D) A frequently methylated gene.
Table S1 Primers and probes used in the quantitative methylation-specific assays
Disclosure
The authors report no conflicts of interest in this work.
References
- OttavianiGJaffeNThe epidemiology of osteosarcomaCancer Treat Res200915231320213383
- JaffeNAdjuvant chemotherapy in osteosarcoma: An odyssey of rejection and vindicationJaffeNBielackSSBrulandÃSPediatric and Adolescent Osteosarcoma, Cancer Treatment and ResearchNew YorkSpringer2009219238
- FerrariSPalmeriniEAdjuvant and neoadjuvant combination chemotherapy for osteogenic sarcomaCurr Opin Oncol200719434134617545797
- ChouAJGellerDSGorlickRTherapy for osteosarcoma: where do we go from here?Paediatr Drugs200810531532718754698
- BuddinghEPAnningaJKVersteeghMIPrognostic factors in pulmonary metastasized high-grade osteosarcomaPediatr Blood Cancer201054221622119890902
- LewisIJNooijMAWhelanJMRC BO06 and EORTC 80931 collaborators; European Osteosarcoma Intergroup. Improvement in histologic response but not survival in osteosarcoma patients treated with intensified chemotherapy: a randomized phase III trial of the European Osteosarcoma IntergroupJ Natl Cancer Inst200799211212817227995
- MankinHJHornicekFJRosenbergAEHarmonDCGebhardtMCSurvival data for 648 patients with osteosarcoma treated at one institutionClin Orthop Relat Res200442928629115577500
- BielackSCarrleDJostLESMO Guidelines Working GroupOsteosarcoma: ESMO clinical recommendations for diagnosis, treatment and follow-upAnn Oncol200819Suppl 2ii94ii96
- PetrilliASde CamargoBFilhoVOBrazilian Osteosarcoma Treatment Group Studies III and IVResults of the Brazilian Osteosarcoma Treatment Group Studies III and IV: prognostic factors and impact on survivalJ Clin Oncol20062471161116816505436
- SakamotoAIwamotoYCurrent status and perspectives regarding the treatment of osteo-sarcoma: chemotherapyRev Recent Clin Trials20083322823118782081
- BatanianJRCavalliLRAldosariNMEvaluation of paediatric osteosarcomas by classic cytogenetic and CGH analysesMol Pathol200255638939312456778
- GorlickRCurrent concepts on the molecular biology of osteosarcomaCancer Treat Res200915246747820213409
- LimGKaraskovaJBeheshtiBAn integrated mBAND and submegabase resolution tiling set (SMRT) CGH array analysis of focal amplification, microdeletions, and ladder structures consistent with breakage-fusion-bridge cycle events in osteosarcomaGenes Chromosomes Cancer200542439240315660435
- SelvarajahSYoshimotoMMaireGIdentification of cryptic microaberrations in osteosarcoma by high-definition oligonucleotide array comparative genomic hybridizationCancer Genet Cytogenet20071791526117981215
- CuiQJiangWGuoJRelationship between hypermethylated MGMT gene and osteosarcoma necrosis rate after chemotherapyPathol Oncol Res20111758759121424568
- OhJHKimHSKimHHKimWHLeeSHAberrant methylation of p14ARF gene correlates with poor survival in osteosarcomaClin Orthop Relat Res200644221622216394764
- GorlickRAndersonPAndrulisIBiology of childhood osteogenic sarcoma and potential targets for therapeutic development: meeting summaryClin Cancer Res20039155442545314654523
- HermanJGBaylinSBGene silencing in cancer in association with promoter hypermethylationN Engl J Med2003349212042205414627790
- GeimanTMRobertsonKDChromatin remodeling, histone modifications, and DNA methylation-how does it all fit together?J Cell Biochem200287211712512244565
- JonesPABaylinSBThe fundamental role of epigenetic events in cancerNat Rev Genet20023641542812042769
- EstellerMCornPGBaylinSBHermanJGA gene hypermethylation profile of human cancerCancer Res20016183225322911309270
- TsuchiyaTSekineKHinoharaSNamikiTNoboriTKanekoYAnalysis of the p16INK4, p14ARF, p15, TP53, and MDM2 genes and their prognostic implications in osteosarcoma and Ewing sarcomaCancer Genet Cytogenet20001202919810942797
- BenassiMSMolendiniLGamberiGInvolvement of INK4A gene products in the pathogenesis and development of human osteosarcomaCancer200192123062306711753985
- LimSYangMHParkJHInactivation of the RASSF1A in osteosarcomaOncol Rep200310489790112792742
- Patiño-GarciaAPiñeiroESDíezMZIturriagagoitiaLGKlüssmannFAAriznabarretaLSGenetic and epigenetic alterations of the cell cycle regulators and tumor suppressor genes in pediatric osteosarcomasJ Pediatr Hematol Oncol200325536236712759621
- HouPJiMYangBQuantitative analysis of promoter hypermethylation in multiple genes in osteosarcomaCancer200610671602160916502411
- VidalDOPaixãoVABraitMAberrant methylation in pediatric myelodysplastic syndromeLeuk Res200731217518116890288
- EadsCALordRVWickramasingheKEpigenetic patterns in the progression of esophageal adenocarcinomaCancer Res2001613410341811309301
- CarvalhoALJeronimoCKimMMEvaluation of promoter hypermethylation detection in body fluids as a screening/diagnosis tool for head and neck squamous cell carcinomaClin Cancer Res2008149710718172258
- HardenSVTokumaruYWestraWHGene promoter hypermethylation in tumors and lymph nodes of stage I lung cancer patientsClin Cancer Res200391370137512684406
- JeronimoCUsadelHHenriqueRQuantitation of GSTP1 methylation in non-neoplastic prostatic tissue and organ-confined prostate adenocarcinomaJ Natl Cancer Inst2001931747175211717336
- HoqueMOFengQTourePDetection of aberrant methylation of four genes in plasma DNA for the detection of breast cancerJ Clin Oncol2006244262426916908936
- LehmannULangerFFeistHQuantitative assessment of promoter hypermethylation during breast cancer developmentAm J Pathol200216060561211839581
- WeisenbergerDJSiegmundKDCampanMCpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancerNat Genet20063878779316804544
- de CarvalhoFColleoniGWAlmeidaMSCarvalhoALVettoreALTGFbetaR2 aberrant methylation is a potential prognostic marker and therapeutic target in multiple myelomaInt J Cancer200912581985199119548309
- BrabenderJUsadelHDanenbergKDAdenomatous polyposis coli gene promoter hypermethylation in non-small cell lung cancer is associated with survivalOncogene200120273528353211429699
- EadsCADanenbergKDKawakamiKMethyLight: a high-throughput assay to measure DNA methylationNucleic Acids Res2000288E3210734209
- RayMEWistowGSuYAMeltzerPSTrentJMAIM1, a novel non-lens member of the betagamma-crystallin superfamily, is associated with the control of tumorigenicity in human malignant melanomaProc Natl Acad Sci USA1997947322932349096375
- BegumSBraitMDasguptaSAn epigenetic marker panel for detection of lung cancer using cell-free serum DNAClin Cancer Res201117134494450321610147
- BraitMBegumSCarvalhoALAberrant promoter methylation of multiple genes during pathogenesis of bladder cancerCancer Epidemiol Biomarkers Prev200817102786279418843024
- LoyoMBraitMKimMSA survey of methylated candidate tumor suppressor genes in nasopharyngeal carcinomaInt J Cancer201112861393140320473931
- TarkkanenMElomaaIBlomqvistCDNA sequence copy number increase at 8q: a potential new prognostic marker in high-grade osteosarcomaInt J Cancer199984211412110096241
- FletcherJAGebhardtMCKozakewichHPCytogenetic aberrations in osteosarcomas. Nonrandom deletions, rings, and double-minute chromosomesCancer Genet Cytogenet199477181887923089
- QuelleDEZindyFAshmunRASherrCJAlternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrestCell199583699310008521522
- KamijoTZindyFRousselMFTumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARFCell19979156496599393858
- KamijoTWeberJDZambettiGZindyFRousselMFSherrCJFunctional and physical interactions of the ARF tumor suppressor with p53 and Mdm2Proc Natl Acad Sci USA19989514829282979653180
- RobertsonKDJonesPAThe human ARF cell cycle regulatory gene promoter is a CpG island which can be silenced by DNA methylation and down-regulated by wild-type p53Mol Cell Biol19981811645764739774662
- SailasreeRAbhilashASathyanKMNalinakumariKRThomasSKannanSDifferential roles of p16INK4A and p14ARF genes in prognosis of oral carcinomaCancer Epidemiol Biomarkers Prev200817241442018268126
- StossiFBarnettDHFrasorJKommBLyttleCRKatzenellenbogenBSTranscriptional profiling of estrogen-regulated gene expression via estrogen receptor (ER) alpha or ERbeta in human osteosarcoma cells: distinct and common target genes for these receptorsEndocrinology200414573473348615033914
- LimaFVicoLLafage-ProustMHvan der SaagPAlexandreCThomasTInteractions between estrogen and mechanical strain effects on U2OS human osteosarcoma cells are not influenced by estrogen receptor typeBone20043551127113515542038
- BordSHornerABeavanSCompstonJEstrogen receptors alpha and beta are differentially expressed in developing human boneJ Clin Endocrinol Metab20018652309231411344243
- SusmanEOsteosarcoma: Where Research Is Heading. Skeletal Complications of Malignancy SymposiumOncol Times200527183738
- IssaJPOttavianoYLCelanoPHamiltonSRDavidsonNEBaylinSBMethylation of the oestrogen receptor CpG island links ageing and neoplasia in human colonNat Genet1994745365407951326
- MüllerHMWidschwendterAFieglHDNA methylation in serum of breast cancer patients: an independent prognostic markerCancer Res2003637641764514633683