Abstract
Background
Epigenetic modulations, including changes in DNA cytosine methylation, are implicated in the pathogenesis and progression of acute myeloid leukemia (AML). Azacitidine is a hypomethylating agent that is incorporated into RNA as well as DNA. Thus, there is a rationale to its use in patients with AML. We determined whether baseline and/or early changes in the methylation of long interspersed element (LINE)-1 or CDH13 correlate with bone marrow blast clearance, hematological response, or survival in patients with AML treated with azacitidine.
Methods
An open label, phase I/II trial was performed in 40 AML patients (median bone marrow blast count was 42%) unfit for intensive chemotherapy treated with azacitidine 75 mg/m2/day subcutaneously for 5 days every 4 weeks. Bone marrow mononuclear cell samples were taken on day 0 (pretreatment) and day 15 during the first treatment cycle; LINE-1 and CDH13 methylation levels were quantified by methylation-specific, semiquantitative, real-time polymerase chain reaction.
Results
Treatment with azacitidine significantly reduced LINE-1 but not CDH13 methylation levels over the first cycle (P < 0.0001). Absolute LINE-1 methylation levels tended to be lower on day 0 (P = 0.06) and day 15 of cycle 1 (P = 0.03) in patients who went on to achieve subsequent complete remission, partial remission or hematological improvement versus patients with stable disease. However, the decrease in LINE-1 methylation over the first treatment cycle did not correlate with subsequent response (P = 0.31). Baseline methylation levels of LINE-1 or CDH13 did not correlate with disease-related prognostic factors, including cytogenetic risk, relapsed/refractory AML, or presence of NPM1 or FLT3 mutations. No correlation was observed between LINE-1 or CDH13 methylation levels and overall survival.
Conclusion
Analysis of baseline LINE-1 methylation levels may help identify elderly AML patients who are most likely to respond to azacitidine therapy.
Introduction
Epigenetic modulations, including changes in DNA cytosine methylation, are strongly implicated in the pathogenesis and progression of acute myeloid leukemia (AML), primarily via the hypermethylation and associated downregulation of candidate tumor-suppressor genes such as p15CDKN2B or CDH13.Citation1,Citation2 More recently, genome-wide analyses have begun to identify characteristic patterns involving both hypermethylation of tumor-suppressor genes and hypomethylation of intergenic repeat sequences, such as long interspersed nuclear elements (LINEs), that define known and novel subtypes of AML.Citation3,Citation4 These studies suggest that DNA methylation profiling could be utilized to determine subgroups with specific therapeutic requirements in a clinical setting. Furthermore, they provide a rationale for the use of hypomethylating therapies in AML.
AML is particularly prevalent in older patients, the median age at onset being ~70 years.Citation5,Citation6 With advancing age, AML is also increasingly associated with unfavorable cytogenetic abnormalities, poor performance status, and comorbid disease. Consequently, patients are often considered unfit for intensive induction chemotherapy and generally have a very poor prognosis.Citation7–Citation12 Elderly patients with contraindications to intensive chemotherapy are usually offered best supportive care or low-dose chemotherapy, although outcome is generally poor in these cases.Citation13,Citation14 These factors, coupled with an aging population, identify novel therapeutic strategies for elderly patients with AML as an urgent unmet medical need.
Decitabine (deoxyazacytidine) is a hypomethylating agent that is incorporated into DNA and inhibits DNA methyltransferase. This agent was recently approved in Europe for the treatment of AML in patients older than 65 years with contraindications for standard chemotherapy. This approval was based on an unplanned survival analysis, which showed a benefit for decitabine compared with supportive care/low-dose cytarabine in a multicenter randomized trial.Citation15 Azacitidine is a related agent that is incorporated into RNA as well as DNA, and has a distinct methyltransferase-inhibitor activity.Citation16 Azacitidine can reactivate tumor-suppressor gene expression and has a direct cytotoxic effect on leukemic cells.Citation17,Citation18 It has been demonstrated to prolong overall survival (OS) versus conventional care regimens in patients with higher-risk myelodysplastic syndromes and World Health Organization (WHO)-defined AML (20%–30% bone marrow blasts).Citation12,Citation19 Furthermore, a recent phase I/II study reported that azacitidine is active and well tolerated in older patients with newly diagnosed and relapsed/refractory AML who are medically unfit for intensive chemotherapy, including those with >30% blasts.Citation20 These findings are supported by a recent “compassionate use” program.Citation21 In these studies, adverse events were generally predictable, manageable and resolved on treatment. Moreover, azacitidine has been reported to significantly improve patient quality of life, which is a key consideration in elderly patients in a palliative care setting.Citation22
The precise mechanism of action of azacitidine in AML and myelodysplastic syndromes (MDS) remains unclear, but is probably distinct from the direct demethylation of tumor-suppressor genes mediated by decitabine. Nonetheless, it will be important to identify biomarkers for the prediction and monitoring of clinical response. Here, we have examined the methylation state of the LINE-1 repetitive sequence as an indicator of general methylation before and during azacitidine therapy in older patients with AML enrolled in a phase I/II study.Citation20 As a comparison, we have also analyzed the methylation state of CDH13 as one of the most commonly methylated tumor-suppressor loci in AML. Specifically, we have investigated whether there is an association between baseline and/or early treatment-related changes in the methylation of LINE-1 and CDH13 and response to azacitidine.
Patients and methods
Study design and patients
An open label, phase I/II investigator-initiated trial was performed in three centers in Germany (OSHO #075, EU Clinical Trials Register number 2007-001194-29). The main details of the trial have been reported elsewhere.Citation20 The study was compliant with International Conference on Harmonisation – Good Clinical Practice guidelines, the Declaration of Helsinki and national regulations. Approval from local ethics committees was gained at every participating center, and all patients provided written informed consent. Patient eligibility and baseline characteristics were as detailed in the main report of this trial, with the main criteria including newly diagnosed (n = 20), or relapsed/refractory AML (n = 20) patients naive to hypomethylating agents who were ineligible for intensive chemotherapy and had been classified according to WHO criteria. All patients received azacitidine 75 mg/m2/day subcutaneously for 5 days every 4 weeks, plus supportive care, until progressive disease or relapse occurred.Citation20
High-risk cytogenetics were defined as –5/5q–, –7/7q–, abn(11q23), or complex karyotype with three or more cytogenetic abnormalities. Normal and all other karyotypes, other than core-binding factor leukemias, were defined as intermediate-risk. Mutational status of FLT3 and NPM1 were assessed in all patients by polymerase chain reaction.
Clinical response
Assessment of clinical response according to the International Working Group criteria for AML was performed after every two cycles for the first eight cycles, and every 3 months thereafter.Citation23 Patients demonstrating complete remission (CR), partial response (PR), hematological improvement (HI), or stable disease (SD) were eligible to continue receiving further cycles. Following any instances of progressive disease or relapse, patients were discontinued from treatment.
Bone marrow aspirations were undertaken on day 0 (pretreatment) and during the first treatment cycle (day 15 with a time window of up to 3 extra days) in order to assess early changes in bone marrow blast count and methylation pattern in response to azacitidine. A separate written informed consent was required to conduct the extra bone marrow aspiration and assessment following cycle 1.
Methylation-specific quantitative real-time polymerase chain reaction
The CpG methylation levels for LINE-1 and CDH13 were determined pre and during cycle 1 using semiquantitative methylation-specific real-time PCR (MSqPCR).Citation24 Genomic DNA was prepared from 5 × 106 bone marrow mononuclear cells using a DNeasy Tissue Kit (Qiagen, Hilden, Germany). Subsequently, 2 μg of genomic DNA was subjected to bisulfite treatment using an Epitect Kit (Qiagen), leading to conversion of nonmethylated cytosine to uracil. The relative amounts of methylated and unmethylated target in each sample were then assessed in parallel PCR reactions using primers specific for unmethylated or methylated CpG rich sequences of LINE-1 and for the upstream region of CDH13.Citation24,Citation25 Primers to a region of the myogenic regulatory factor MyoD locus devoid of CpG sequences served to control for successful bisulfite conversion.
Primers are shown below, with methylation-sensitive positions underlined.
| Unmethylated LINE-1 forward: | 5′ GTTGAATAGGAATAGTTTTG GTTT 3′ |
| Unmethylated LINE-1 reverse: | 5′ ACTCCCTAACCCCTTACACTT 3′ |
| Methylated LINE-1 forward: | 5′ GTCGAATAGGAATAGTTTCGG 3′ |
| Methylated LINE-1 reverse: | 5′ ACTCCCTAACCCCTTACGCT 3′ |
| Unmethylated CDH13 forward: | 5′ TTGTGGGGTTTGTTTTTTGT 3′ |
| Unmethylated CDH13 reverse: | 5′ AACATTTTCATTCATACACACA 3′ |
| Methylated CDH13 forward: | 5′ TCGCGGGGTTCGTTTTTCGC 3′ |
| Methylated CDH13 reverse: | 5′ GACGTTTTCATTCATACACGCG 3′ |
| MyoD forward: | 5′ TAGGTGAGATGAGGTTAGGTTTTTAG 3′ |
| MyoD reverse: | 5′ TTTCCACAACAAAAATATCCAATAC 3′. |
MSqPCR reactions were performed in a final volume of 25 μL, containing 2 μL (5 ng) bisulfite-converted DNA, primers to a final concentration of 0.2 μM, and a 1× dilution of Quantitect SYBR Green PCR master mix (Qiagen). Amplification was performed in an AB 7300 Real-Time PCR System (Applied Biosystems, Darmstadt, Germany), using the following conditions: denaturation at 95°C for 15 minutes; amplification through 45 cycles of 94°C for 15 seconds, 55°C for 30 seconds, and 72°C for 35 seconds; a melting curve analysis of PCR products with dissociation at 95°C for 15 seconds, annealing at 60°C for 30 seconds, and melting by stepwise increases of 1°C every 15 seconds to 95°C.
The ratio of methylated to unmethylated target (methylation ratio) was calculated as 2ΔCt, where ΔCt is the difference between the cycle threshold (Ct) values for the methylated and unmethylated PCR reactions.
Statistical analyses
Demographic characteristics and responses to azacitidine were summarized using descriptive statistics, as previously detailed.Citation20 All analyses were performed on the intention-to-treat principle. Descriptive statistics and the Wilcoxon test were used to compare calculated ratios of methylated/unmethylated DNA for both LINE-1 and CDH13 methylation. Changes in methylation before and during the first treatment cycle were reported as a percentage of the initial ratio.
Pretreatment and cycle 1 methylation ratios and changes in methylation were correlated with disease status (newly diagnosed versus relapsed or refractory), pretreatment bone marrow blast count (continuous variable), bone marrow blast count after the first cycle (>45% versus ≤45%), cytogenetics (high-risk versus intermediate-risk), FLT3 and NPM1 (wild-type versus mutated), as well as with survival and response.
Univariate analysis was performed using the Chi-squared test, Fisher’s exact test for categorical data, and the Mann-Whitney test for continuous variables. The Kruskal-Wallis test was used for the correlation between methylation and cytogenetics. For nonparametric correlations, Spearman’s rho test and Pearson correlation were used. Data were analyzed using SPSS 15 software (IBM, Armonk, NY, USA). P-values of <0.05 were considered statistically significant.
Results
The median age was 72 (range 32–84) years, with 48% of patients being male. A total of 203 treatment cycles were administered (range 1–16), and the median follow-up duration was 13 (range 9–16) months.Citation20
Overall clinical efficacy
Clinical efficacy has been previously reported.Citation20 In summary, 68% of patients experienced SD or better, and the median duration of response was 5.5 months. CR, PR, or HI was reported in 50% of patients with newly diagnosed AML and 10% of patients with relapsed or refractory AML. Median OS for patients with SD was 3.8 months, whereas the median OS for patients with CR, PR, or HI was not reached at 13 months (P = 0.045).Citation20
Early changes in methylation over time
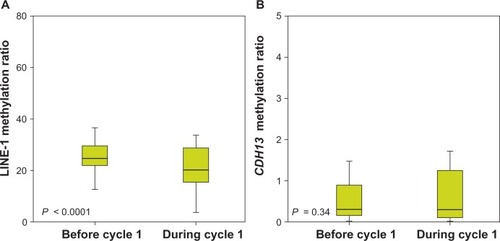
Assessable samples were available in 33 patients both pretreatment and at day 15 of cycle 1. Azacitidine induced early hypomethylation of the LINE-1 repetitive sequences. Median LINE-1 methylation ratio decreased from 25 (range 13–64) at day 0 to 20 (range 4–54) following treatment cycle 1 (P < 0.0001); decreased LINE-1 methylation was observed in 27/33 (82%) of patients (). Median methylation ratio of CDH13 was 0.29 (range 0–119) at day 0 and remained unchanged after the first cycle with a median of 0.29 (range 0–50, P = 0.34), with 16 (48%) patients showing a decrease and 14 (42%) an increase ().
Figure 1 Long interspersed element (LINE)-1 (A) and CDH13 (B) DNA methylation before (day 0) and during (day 15) the first cycle of azacitidine treatment. The ratio of methylated to unmethylated target (methylation ratio) was calculated as 2ΔCt, where ΔCt is the difference between the cycle threshold (Ct) values for the methylated and unmethylated polymerase chain reactions.
Relationship between methylation status and disease-related factors
LINE-1 and CDH13 methylation status before and after the first treatment cycle were assessed with respect to a number of disease-related prognostic factors. These analyses demonstrated that there was no significant difference in methylation status of either LINE-1 or CDH13: (1) between newly diagnosed versus relapsed/refractory patients (P = 0.8); (2) in patients with intermediate- versus high-risk cytogenetics (P = 0.4); (3) in patients with varying baseline blast count levels; (4) in patients with wild-type versus mutated NPM1 (P = 0.5); or (5) between patients with wild-type versus mutated FLT3 (P = 0.8). In none of these cases did methylation status correlate with a known disease-related prognostic factor.
Relationship between pretreatment methylation status and early marrow blast clearance under azacitidine
As reported previously for this study, the marrow blast count on day 15 during the first treatment cycle was highly predictive of subsequent treatment response, irrespective of the pretreatment blast count or the extent of blast clearance up to day 15. No patient with marrow blasts <45% on day 15 of cycle 1 achieved a subsequent response. As per protocol, the earliest assessment of response was performed after two cycles on day 56.
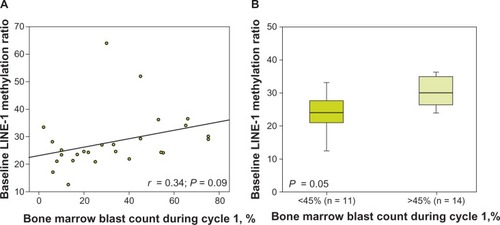
In the current analysis, there was no significant correlation between the methylation status of either LINE-1 or CDH13 (initial or during cycle 1) and blast count at the equivalent time point. However, linear analysis indicated that there was a possible relationship between baseline LINE-1 methylation levels and blast count on day 15 of cycle 1; initial LINE-1 methylation levels appeared to be lower in patients with low blast counts during the first cycle of azacitidine, though this correlation did not achieve statistical significance (r = 0,34, P = 0.09; ). Having previously observed that a bone marrow blast count of <45% during cycle 1 was predictive of subsequent response,Citation20 we compared baseline LINE-1 methylation levels in patients with bone marrow blast counts of <45% versus >45% during cycle 1. The initial LINE-1 methylation ratio on day 15 was lower in the group with <45% blasts versus the group with >45% blasts (24 vs 30, P = 0.05; ). Notably, changes in methylation ratios for either LINE-1 or CDH13 occurring over the first treatment cycle showed no correlation to bone marrow blast percentage on day 15 of cycle 1.
Figure 2 Long interspersed element (LINE)-1 DNA methylation at day 0 versus blast count during cycle 1 (A) and in patients who achieved or did not achieve a bone marrow blast threshold of <45% during cycle 1 (B). The ratio of methylated to unmethylated target (methylation ratio) was calculated as 2ΔCt, where ΔCt is the difference between the cycle threshold (Ct) values for the methylated and unmethylated polymerase chain reactions.
Relationship between methylation and subsequent response to azacitidine
For both LINE-1 and CDH13, neither methylation levels (initial or during cycle 1) nor changes in the extent of methylation during cycle 1 correlated with OS.
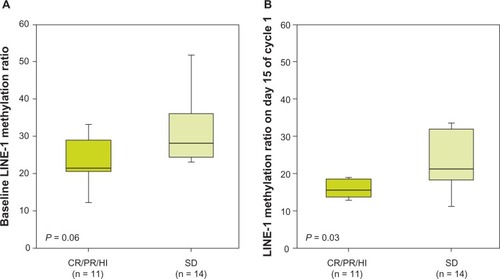
At both pretreatment and day 15 of cycle 1, assessable samples were available in eleven of twelve (92%) patients in whom CR, PR, or HI was documented later and 14 of 15 (93%) patients with SD only. No correlation was found between response (CR, PR, HI versus SD) and CDH13 DNA methylation either before or after treatment cycle 1. In contrast, LINE-1 methylation levels did appear to be predictive of clinical response. LINE-1 methylation ratios were lower in patients who subsequently achieved CR, PR, or HI versus those who achieved SD only, both prior to treatment (22 versus 26, P = 0.06; ) and during the first treatment cycle on day 15 (15.4 versus 20.9, P = 0.03; ). There was no association between the type of response and changes in methylation over the first cycle, either for LINE-1 (P = 0.31) or for CDH13 (P = 0.4).
Figure 3 Long interspersed element (LINE)-1 DNA methylation at day 0 (A) and during cycle 1 (day 15) of azacitidine treatment (B) and subsequent clinical response. The ratio of methylated to unmethylated target (methylation ratio) was calculated as 2ΔCt, where ΔCt is the difference between the cycle threshold (Ct) values for the methylated and unmethylated polymerase chain reactions.
Discussion
The purpose of the current analysis was to determine whether initial levels of or early changes in methylation of LINE-1 and CDH13 correlated with clinical outcomes including early bone marrow blast clearance after the first cycle of azacitidine, hematological response, or survival. Our analysis revealed that the initial degree of LINE-1 methylation but not subsequent treatment-associated changes were predictive for clinical response.
The impact of azacitidine therapy on DNA methylation levels in patients with MDS or AML has been investigated in several previous studies with somewhat conflicting results, which probably refects differences in azacitidine treatment schedules (5 versus 7 days), the loci assessed, the techniques used to quantify methylation, the target cells under investigation, and time points at which samples were taken in relation to treatment.Citation26–Citation29 The rationale behind the experimental 5-day schedule of azacitidine in this study was to provide a Monday–Friday outpatient-treatment option to patients with AML who were medically unfit for or resistant to chemotherapy. Notwithstanding these caveats, the overall evidence from our current analysis and from previous studies indicates that changes in methylation under azacitidine therapy do not correlate with clinical response. In a previous study in patients with AML, MDS, or chronic myelomonocytic leukemia, early changes in the methylation of four tumor-suppressor genes (p15CDKN2B, CDH-1, DAPK-1, and SOCS-1) or LINE-1 in bone marrow mononuclear cells or purified CD34+ cells did not predict response to azacitidine in combination with the histone deacetylase inhibitor entinostat.Citation28 In another study, reduction in the methylation at the p15CDKN2B locus after treatment with azacitidine did not correlate with treatment response in patients with MDS.Citation27
In this respect, the predictive factors for clinical response to azacitidine appear to differ from those that predict response to decitabine. In contrast to azacitidine, several studies in patients with AML have shown that response to decitabine clearly correlates with early epigenetic changes. For example, the methylation levels of Alu repeats, LINE-1, and total genomic DNA were found to decrease significantly within 5 days of treatment with decitabine in patients with AML, with reduced methylation correlating with response.Citation30 Similarly, in a more recent study, decitabine-mediated hypomethylation of several tumor-suppressor genes, including p15CDK2NB, within the first cycle of treatment correlated with subsequent clinical response.Citation31 Thus, the extent of demethylation under decitabine but not under azacitidine appears to be predictive of clinical response.
At the same time, several studies of patients with MDS have indicated that pretreatment methylation of tumor-suppressor loci is associated with subsequent response to azacitidine, with low methylation correlating with better prognosis.Citation27,Citation29 Consistent with these findings, the current study demonstrated that pretreatment LINE-1 methylation levels are indicative of subsequent response to azacitidine in AML patients. One possibility is that the clinical response to azacitidine depends on DNA methylation being reduced below a threshold value after cycle 1, and that this is more likely to occur in AML cells that start with lower levels of LINE-1 methylation. Although our data would be consistent with this explanation, they do not distinguish dynamic methylation changes at the cellular level from those occurring at the population level as a result of selective death of cells with a particularly high or low level of methylation. This selective cell death may contribute to the methylation changes observed. The relationship between methylation of repetitive sequences and that of tumor-suppressor genes appears to be complex. On one hand, methylation of tumor-suppressor genes and LINE-1 (this study) appears to show similar correlations with response.Citation27,Citation29 Conversely, other observations suggest that LINE-1 and p15CDK2B methylation are inversely correlated with response.Citation2 A better understanding of the relationship between the methylation states of tumor-suppressor genes and repetitive sequences, and their relevance to prognosis, can be expected to emerge from genome-wide analyses of CpG island methylation. A genome-wide analysis has already revealed clustered methylation patterns distinctive of both known and novel functional subgroups of AML.Citation3,Citation4 Interestingly, older individuals with AML who were comparable to the group of patients studied here were found to have a distinct methylation cluster, suggesting that they may have a distinct response to demethylating agents.Citation3
Notwithstanding the association between initial LINE-1 methylation and subsequent response, it is possible that the clinical effects of azacitidine involve activities other than hypomethylation of DNA, in which case the pretreatment level of LINE-1 methylation would be a surrogate marker of susceptibility. Up to 90% of azacitidine becomes incorporated into RNA, leading to reduced protein synthesis.Citation32
In this way, the downregulation of ribonucleotide reductase, a rate-limiting enzyme in DNA synthesis, has recently been suggested to mediate the activity of azacitidine in patients with AML.Citation33 Furthermore, experimental evidence indicates that azacitidine inhibits a transfer RNA methyltransferase (TRDMT1), raising the possibility that changes in the protein-translation machinery itself may also be involved in the clinical effects.Citation16
Finally, emerging data suggest that azacitidine may have immunomodulatory as well as cytotoxic effects.Citation18,Citation34–Citation36 Given the numerous effects of azacitidine that may contribute to clinical outcome, it is of interest to investigate the activity of azacitidine on subpopulations of cells, particularly leukemic stem cells that are important for response.
In summary, recent studies have demonstrated that azacitidine has encouraging clinical activity and favorable tolerability in elderly patients with AML, including patients with >30% bone marrow blasts or relapsed/refractory disease, suggesting that this agent has considerable potential in a setting of significant unmet medical need.Citation19–Citation21 We report here that LINE-1 DNA methylation levels prior to treatment, but not treatment-induced changes in LINE-1 methylation, were indicative of clinical response to azacitidine. If these results are validated, pretreatment methylation analysis could potentially help to identify elderly AML patients most likely to benefit from azacitidine therapy.
Acknowledgments
Medical writing support was provided by Lindsay Armstrong and Kate Unsworth of the Investigator-Initiated Research Writing Group (part of the KnowledgePoint 360 Group).
Disclosure
This work was financed by a research grant funded by Celgene. HKA has received honoraria from Celgene. The authors report no other conflicts of interest in this work.
References
- CameronEEBaylinSBHermanJGp15(INK4B) CpG island methylation in primary acute leukemia is heterogeneous and suggests density as a critical factor for transcriptional silencingBlood19999472445245110498617
- KroegerHJelinekJEstécioMRHAberrant CpG island methylation in acute myeloid leukemia is accentuated at relapseBlood200811241366137318523155
- FigueroaMELugthartSLiYDNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemiaCancer Cell2010171132720060365
- SaiedMHMarzecJKhalidSGenome wide analysis of acute myeloid leukemia reveal leukemia specific methylome and subtype specific hypomethylation of repeatsPLoS One201273e3321322479372
- KnippSHildebrandBKündgenAIntensive chemotherapy is not recommended for patients aged >60 years who have myelodysplastic syndromes or acute myeloid leukemia with high-risk karyotypesCancer2007110234535217559141
- JuliussonGAntunovicPDerolfÅAge and acute myeloid leukemia: real world data on decision to treat and outcomes from the Swedish Acute Leukemia RegistryBlood2009113184179418719008455
- GrimwadeDWalkerHHarrisonGThe predictive value of hierarchical cytogenetic classification in older adults with acute myeloid leukemia (AML): analysis of 1065 patients entered into the United Kingdom Medical Research Council AML11 trialBlood20019851312132011520776
- StoneRMThe difficult problem of acute myeloid leukemia in the older adultCA Cancer J Clin200252636337112469764
- KantarjianHO’BrienSCortesJResults of intensive chemotherapy in 998 patients age 65 years or older with acute myeloid leukemia or high-risk myelodysplastic syndromeCancer200610651090109816435386
- EleniLDNicholasZCAlexandrosSChallenges in treating older patients with acute myeloid leukemiaJ Oncol2010201094382320628485
- Van Der HoltBBreemsDABerna BeverlooHVarious distinctive cytogenetic abnormalities in patients with acute myeloid leukaemia aged 60 years and older express adverse prognostic value: results from a prospective clinical trialBr J Haematol200713619610517129222
- FenauxPMuftiGJHellstrom-LindbergEEfficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III studyLancet Oncol200910322323219230772
- DeschlerBWitteT deMertelsmannRLuebbertMTreatment decision-making for older patients with high-risk myelodysplastic syndrome or acute myeloid leukemia: problems and approachesHaematologica200691111513152217082009
- BurnettAKMilliganDPrenticeAGA comparison of low-dose cytarabine and hydroxyurea with or without all-trans retinoic acid for acute myeloid leukemia and high-risk myelodysplastic syndrome in patients not considered fit for intensive treatmentCancer200710961114112417315155
- KantarjianHMThomasXGDmoszynskaAMulticenter, randomized, open-label, phase III trial of decitabine versus patient choice, with physician advice, of either supportive care or low-dose cytarabine for the treatment of older patients with newly diagnosed acute myeloid leukemiaJ Clin Oncol201230212670267722689805
- SchaeferMHagemannSHannaKLykoFAzacytidine inhibits RNA methylation at DNMT2 target sites in human cancer cell linesCancer Res200969208127813219808971
- ChristmanJK5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapyOncogene200221355483549512154409
- HollenbachPWNguyenANBradyHA comparison of azacitidine and decitabine activities in acute myeloid leukemia cell linesPLoS One201052e900120126405
- FenauxPMuftiGJHellstroem-LindbergEAzacitidine prolongs overall survival compared with conventional care regimens in elderly patients with low bone marrow blast count acute myeloid leukemiaJ Clin Oncol201028456256920026804
- Al-AliHKJaekelNJunghanssCAzacitidine in patients with acute myeloid leukemia medically unfit for or resistant to chemotherapy: a multicenter phase I/II studyLeuk Lymphoma201253111011721767242
- MaurilloLVendittiASpagnoliAAzacitidine for the treatment of patients with acute myeloid leukemia: report of 82 patients enrolled in an Italian Compassionate ProgramCancer201211841014102221761399
- KornblithABHerndonJE2ndSilvermanLRImpact of azacytidine on the quality of life of patients with myelodysplastic syndrome treated in a randomized phase III Trial: A Cancer and Leukemia Group B StudyJ Clin Oncol200220102441245212011121
- ChesonBDBennettJMKopeckyKJRevised recommendations of the International Working Group for Diagnosis, Standardization of Response Criteria, Treatment Outcomes, and Reporting Standards for Therapeutic Trials in Acute Myeloid LeukemiaJ Clin Oncol200321244642464914673054
- Roman-GomezJJimenez-VelascoAAgirreXPromoter hypomethylation of the LINE-1 retrotransposable elements activates sense/antisense transcription and marks the progression of chronic myeloid leukemiaOncogene200524487213722316170379
- Roman-GomezJCastillejoJAJimenezACadherin-13, a mediator of calcium-dependent cell-cell adhesion, is silenced by methylation in chronic myeloid leukemia and correlates with pretreatment risk profile and cytogenetic response to interferon alfaJ Clin Oncol20032181472147912697869
- GoreSDBaylinSSugarECombined DNA methyltransferase and histone deacetylase inhibition in the treatment of myeloid neoplasmsCancer Res200666126361636916778214
- RajKJohnAHoACDKN2B methylation status and isolated chromosome 7 abnormalities predict responses to treatment with 5-azacytidineLeukemia20072191937194417611569
- FandyTEHermanJGKernsPEarly epigenetic changes and DNA damage do not predict clinical response in an overlapping schedule of 5-azacytidine and entinostat in patients with myeloid malignanciesBlood2009114132764277319546476
- TranHTKimHNLeeIKDNA methylation changes following 5-azacitidine treatment in patients with myelodysplastic syndromeJ Korean Med Sci201126220721321286011
- YangASDoshiKDChoiSWDNA methylation changes after 5-aza-2′-deoxycytidine therapy in patients with leukemiaCancer Res200666105495550316707479
- YanPFrankhouserDMurphyMGenome-wide methylation profiling in decitabine-treated patients with acute myeloid leukemiaBlood2012120122466247422786882
- LiLHOlinEJBuskirkHHReinekeLMCytotoxicity and mode of action of 5-azacytidine on L1210 leukemiaCancer Res19703011276027695487063
- AimiuwuJWangHChenPRNA-dependent inhibition of ribonucleotide reductase is a major pathway for 5-azacytidine activity in acute myeloid leukemiaBlood2012119225229523822517893
- IssaJPKantarjianHMTargeting DNA methylationClin Cancer Res200915123938394619509174
- Sánchez-AbarcaLIGutierrez-CosioSSantamaríaCImmunomodulatory effect of 5-azacytidine (5-azaC): potential role in the transplantation settingBlood2010115110712119887673
- XiaoWHodgeDRWangLYangXZhangXFarrarWLNF-kappaB activates IL-6 expression through cooperation with c-Jun and IL6-AP1 site, but is independent of its IL6-NFkappaB regulatory site in autocrine human multiple myeloma cellsCancer Biol Ther20043101007101715467434