Abstract
Objective
Single nucleotide polymorphisms (SNPs) accumulated frequently in the mitochondrial displacement loop (D-loop) in many cancers. We had identified cancer risk-associated SNPs in the D-loop of non-Hodgkin lymphoma (NHL) patients previously, in this study, we investigated the association of age at onset and D-loop SNPs in NHL patients.
Materials and methods
The D-loop region of mtDNA was sequenced for 133 NHL patients recorded at the Fourth Hospital of Hebei Medical University. The Kaplan–Meier method was used to identify age at onset-associated SNPs in the D-loop of NHL patients. The Cox proportional hazards model was used to identify independent risk factors for age at onset.
Results
The SNP sites of nucleotides 146C/T, 151T/C, 194T/C, 315C/C insert, 523Del/A, and 525Del/C were identified for their association with age at onset, by the logrank test. In an overall multivariate analysis, allele 146 (relative risk, 0.403; 95% confidence interval [CI]: 0.182–0.895) (P = 0.026), allele 151 (relative risk, 0.378; 95% CI: 0.165–0.868) (P = 0.022), and allele 315 (relative risk, 3.554; 95% CI: 1.344–9.400) (P = 0.011) were identified as independent predictors for age at onset in NHL patients.
Conclusion
SNPs in the D-loop can predict age at onset in NHL patients. Analysis of the D-loop SNPs can help identify NHL patient subgroups at high risk of early onset.
Introduction
Non-Hodgkin lymphoma (NHL) is the fifth most frequent form of cancer among men and women, accounting for 2.15% of all cancer-related deaths in the People’s Republic of China over the period 2003–2007.Citation1,Citation2 Although the etiology of NHL is poorly understood, the immune-related conditions, family histology of hematopoietic malignancy, and body mass index have been shown to have an association with the cancer risk.Citation3,Citation4 Some studies have also shown that genetic factors are important to development of this cancer.Citation5–Citation7 The polymorphism of genes in the oxidative stress pathway also was identified for their association with a risk of NHL. However, the true mechanism of this cancer remains unknown.Citation8–Citation10 Although some studies have focused on genetics associated with the age at onset of NHL, they have yet to demonstrate a genetic basis of this disease.Citation11
The human mitochondrial genome is 16 kilobase (kb) in length and is formed as a closed-circular duplex molecule.Citation12 Mitochondrial deoxyribonucleic acid (mtDNA) is believed to be more susceptible to DNA damage and so acquires mutations at a higher rate than nuclear DNA because of the high levels of reactive oxygen species (ROS) generation, lack of protective histones, and limited DNA repair capacity in the mitochondria.Citation13,Citation14
The mitochondrial displacement loop (D-loop) region is important for regulating both replication and expression of the mitochondrial genome because it contains the leading-strand origin of replication and the main promoter for transcription. In cancer patients, sequence changes accumulated extensively in this region. Citation15–Citation18 Previously, we sequenced the D-loop region in a case-control study of NHL patients and identified 140 single nucleotide polymorphisms (SNPs), including 26 SNPs with frequency distribution of minor allele greater than 5%, and we also identified NHL risk-associated SNPs.Citation19 In this study, we investigated the association of age at onset and SNPs of the D-loop, in NHL patients.
Materials and methods
Tissue specimens and DNA extraction
Diagnosis of lymphoma was made according to the WHO Classification of Tumors.Citation20 Blood samples were collected from 133 NHL patients who received treatment in the Respiratory Department at the Fourth Hospital of Hebei University, Shijiazhuang, People’s Republic of China, between 1990 and 2011. The genomic deoxyribonucleic acid (DNA) was immediately extracted using the Wizard® Genomic DNA Purification Kit (Promega Corporation, Fitchburg, WI, USA) and stored at −20°C. All procedures were supervised and approved by the Human Tissue Research Committee at the hospital. Informed consent was obtained from all participants before enrollment.
Polymerase chain reaction (PCR) amplification and sequence analysis
A 982-base pair (bp) product of D-Loop was amplified with the forward primer 5′-CCCCATGCTTACAAGCAAGT-3′ (nucleotide 16190–16209) and reverse primer 5′-GCTTTGAGGAGGTAAGCTAC-3′ (nucleotide 602–583). PCR was run with a PCR Master Mix Kit (Promega Corporation) and purified prior to sequencing. Cycle sequencing was performed with the BigDye Terminator v3.1 Cycle Sequencing Kit (Life Technologies, Carlsbad CA, USA), and the products were then read with the ABI PRISM® 3100 Genetic Analyzer (Life Technologies). Polymorphisms were confirmed by repeated analyses from both strands.
Statistical analysis
The age at onset curve of NHL patients was calculated using the Kaplan–Meier method at each of the SNP sites, and compared with the logrank test. Multivariate analysis was performed with a Cox proportional hazards model. All of the statistical analysis was done with the SPSS Statistics for Windows, Version 17.0 software package (SPSS Inc, Chicago, IL, USA). A P-value of <0.05 was considered statistically significant.
Results
The age at onset distribution of the NHL patients was listed in . Those analyzed included: 25 patients at age less than 30 years old; 16 patients at age 30–40; 28 patients at age 41–50; 30 patients at age 51–60; 20 patients at age 61–70, and 14 patients at age older than 70. Age at onset was analyzed together with clinical characteristics, including gender and TNM (tumor node metastasis) classification, using the Kaplan–Meier method, and these were compared by the logrank test. TNM stage was associated with the age at onset, at statistically significant levels ( and ).
Figure 1 Comparison of age at onset for NHL patients according to (A) sex and (B) TNM classification with Kaplan–Meier methods.
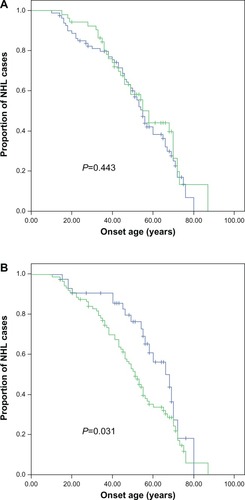
Table 1 Onset age distribution in NHL patients
Table 2 Clinical characteristics and their association with onset age in NHL patients
Twenty-six SNPs with minor allele frequency higher than 5% were subsequently evaluated for their association with age at onset. The NHL patients were divided into two groups on the basis of their genotype at each SNP site, and the association of SNPs in the D-loop and age at onset was calculated with Kaplan–Meier method. The SNP sites of nucleotides 146C/T, 151T/C, 194T/C, 315C/C insert, 523Del/A, and 525Del/C were identified for their association with age at onset by the logrank test ( and ); the SNP site of nucleotide 16261 was also identified for its association with age at onset but with borderline statistical significance (P = 0.083). We performed multivariate analysis for these predictors, including these SNPs and the clinical characteristics, with the Cox proportional hazards model. Allele 146 (relative risk, 0.403; 95% confidence interval [CI]: 0.182–0.895) (P = 0.026), allele 151 (relative risk, 0.378; 95% CI: 0.165–0.868) (P = 0.022), and allele 315 (relative risk, 3.554; 95% CI: 1.344–9.400) (P = 0.011) were identified as independent predictors for age at onset in the NHL patients ().
Figure 2 Comparison of age at onset for NHL patients, according to the genotype of nucleotides 146, 151, 194, 16261, 315, 523, and 525 in the mitochondrial D-loop, using the Kaplan–Meier method.
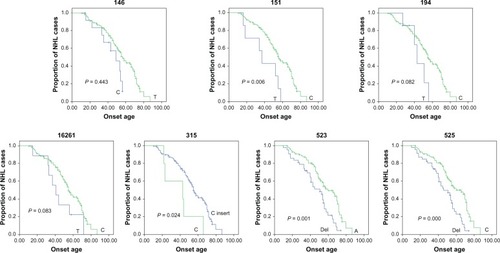
Table 3 Polymorphic sites of the D-loop and their association with onset age in NHL patients
Table 4 Multivariate analysis of predictors associated with age at onset of NHL patients
Discussion
Many SNPs in the D-loop region have been identified for their association with cancer risk and disease outcome in cancers.Citation21–Citation27 The present study has extended those analyses to determine the relationship between age at onset and germline SNPs, in NHL patients. The alleles 146, 151, and 315 were identified for their association with age at onset, at statistically significant levels, by multivariate analysis.
We have previously identified age at onset-associated SNPs of the D-loop, in patients with hepatocellular carcinoma and esophageal squamous carcinoma.Citation28,Citation29 In this study, we also identified age at onset-associated SNPs in NHL patients.
All of the cancer risk-associated SNP sites are located in the hypervariable segment (HVII) region, mutational hotspots at which germline and tumor mtDNA mutations preferentially occur.Citation30 We have identified outcome-associated SNPs of these regions in other cancers.Citation21,Citation27 The allele 315 was identified as a cancer risk-associated SNP for NHL, and allele 151 was identified as a cancer risk-associated SNP for non-small cell lung cancer in previous study.Citation19,Citation31 Our data imply the important role of HVII region in modifying the onset age of cancer.
Since the mitochondrial D-loop is important for replication and expression of the mitochondrial genome, SNPs in this region might alter the function of electron transport chains by modifying mtDNA replication and transcription, which is responsible for the generation of ROS, and could contribute to nuclear genome damage as well as cancer initiation and promotion.Citation32–Citation35 These ROS-mediated mechanisms may accelerate earlier onset of this disease.
In conclusion, SNPs in the D-loop can predict age at onset in NHL patients, and analysis of D-loop SNPs can help to identify NHL patient subgroups at high risk of early onset.
Disclosure
The authors report no conflicts of interest in this work.
References
- GeyerSMMortonLMHabermannTMSmoking, alcohol use, obesity, and overall survival from non-Hodgkin lymphoma: a population-based studyCancer2010116122993300020564404
- ZhangMLiGCZhangYLZhangSWYangNN[An analysis of the incidence and mortality with malignant lymphoma in China during 2003–2007]China Cancer201221190196 Chinese
- MüllerAMIhorstGMertelsmannREngelhardtMEpidemiology of non-Hodgkin’s lymphoma (NHL): trends, geographic distribution, and etiologyAnn Hematol200584111215480663
- ChatterjeeNHartgePCerhanJRRisk of non-Hodgkin’s lymphoma and family history of lymphatic, hematologic, and other cancersCancer Epidemiol Biomarkers Prev20041391415142115342441
- LanQWangSSMenasheIGenetic variation in Th1/Th2 pathway genes and risk of non-Hodgkin lymphoma: a pooled analysis of three population-based case-control studiesBr J Haematol2011153334135021418175
- DiaoLPMaHWeiGCMatrix metalloproteinase-2 promoter and tissue inhibitor of metalloproteinase-2 gene polymorphisms in non-Hodgkin’s lymphomaInt J Cancer201213151095110322020421
- HosgoodHDPurdueMPWangSSA pooled analysis of three studies evaluating genetic variation in innate immunity genes and non-Hodgkin lymphoma riskBr J Haematol2011152672172621250972
- LanQZhengTShenMGenetic polymorphisms in the oxidative stress pathway and susceptibility to non-Hodgkin lymphomaHum Genet2007121216116817149600
- LightfootTJSkibolaCFSmithAGPolymorphisms in the oxidative stress genes, superoxide dismutase, glutathione peroxidase and catalase and risk of non-Hodgkin’s lymphomaHaematologica20069191222122716956821
- WangSSDavisSCerhanJRPolymorphisms in oxidative stress genes and risk for non-Hodgkin lymphomaCarcinogenesis20062791828183416543247
- WiernikPHWangSQHuXPMarinoPPaiettaEAge of onset evidence for anticipation in familial non-Hodgkin’s lymphomaBr J Haematol20001081727910651726
- ShadelGSClaytonDAMitochondrial DNA maintenance in vertebratesAnnu Rev Biochem1997664094359242913
- DiMauroSSchonEAMitochondrial DNA mutations in human diseaseAm J Med Genet20011061182611579421
- BealMFMitochondria, free radicals, and neurodegenerationCurr Opin Neurobiol1996656616668937831
- YoneyamaHHaraTKatoYYamoriTMatsuuraETKoikeKNucleotide sequence variation is frequent in the mitochondrial DNA displacement loop region of individual human tumor cellsMol Cancer Res200531142015671245
- NishikawaMNishiguchiSShiomiSSomatic mutation of mitochondrial DNA in cancerous and noncancerous liver tissue in individuals with hepatocellular carcinomaCancer Res20016151843184511280735
- Sanchez-CespedesMParrellaPNomotoSIdentification of a mononucleotide repeat as a major target for mitochondrial DNA alterations in human tumorsCancer Res200161197015701911585726
- TaanmanJWThe mitochondrial genome: structure, transcription, translation and replicationBiochim Biophys Acta19991410210312310076021
- GaoYZhaoGDiaoLGuoZIdentification of sequence polymorphisms in the D-loop region of mitochondrial DNA as a risk factor for non-Hodgkin lymphomaMitochondrial DNA2013
- SwerdlowSHCampoEHarrisNLWHO Classification of Tumours of Haematopoietic and Lymphoid Tissues4th edLyonInternational Agency for Research on Cancer (IARC)2008
- ZhangRWangRZhangFSingle nucleotide polymorphisms in the mitochondrial displacement loop and outcome of esophageal squamous cell carcinomaJ Exp Clin Cancer Res20102915521110870
- NavagliaFBassoDFogarPMitochondrial DNA D-loop in pancreatic cancer: somatic mutations are epiphenomena while the germline 16519 T variant worsens metabolism and outcomeAm J Clin Pathol2006126459360116938655
- WangLBamletWRde AndradeMMitochondrial genetic polymorphisms and pancreatic cancer riskCancer Epidemiol Biomarkers Prev20071671455145917627010
- WangLMcDonnellSKHebbringSJPolymorphisms in mitochondrial genes and prostate cancer riskCancer Epidemiol Biomarkers Prev200817123558356619064571
- BaiRKLealSMCovarrubiasDLiuAWongLJMitochondrial genetic background modifies breast cancer riskCancer Res200767104687469417510395
- ZhangRZhangFWangCWangSShiaoYHGuoZIdentification of sequence polymorphism in the D-Loop region of mitochondrial DNA as a risk factor for hepatocellular carcinoma with distinct etiologyJ Exp Clin Cancer Res20102913020849651
- WangCZhangFFanHSequence polymorphisms of mitochondrial D-loop and hepatocellular carcinoma outcomeBiochem Biophys Res Commun2011406349349621345333
- GuoZYangHWangCLiuSMitochondrial DNA haplogroup M is associated with late onset of hepatocellular carcinomaExp Ther Med20123349950222969918
- GuoZYangHZhangFZhangRWangCSingle nucleotide polymorphisms in the mitochondrial displacement loop and age-at-onset of esophageal squamous cell carcinomaOncol Lett20123248248422740936
- StonekingMHypervariable sites in the mtDNA control region are mutational hotspotsAm J Hum Genet20006741029103210968778
- DingCLiRWangPJinPLiSGuoZIdentification of sequence polymorphisms in the D-loop region of mitochondrial DNA as a risk factor for lung cancerMitochondrial DNA201223425125422708867
- BandyBDavisonAJMitochondrial mutations may increase oxidative stress: implications for carcinogenesis and aging?Free Radic Biol Med1990865235392193852
- GilleJJJoenjeHCell culture models for oxidative stress: superoxide and hydrogen peroxide versus normobaric hyperoxiaMutat Res19922753–64054141383781
- ShigenagaMKHagenTMAmesBNOxidative damage and mitochondrial decay in agingProc Natl Acad Sci U S A1994912310771107787971961
- DementGAMaloneySCReevesRNuclear HMGA1 nonhistone chromatin proteins directly influence mitochondrial transcription, maintenance, and functionExp Cell Res20073131778717045586