Abstract
Breast cancer includes a body of molecularly distinct subgroups, characterized by different presentation, prognosis, and sensitivity to treatments. Significant advances in our understanding of the complex architecture of this pathology have been achieved in the last few decades, thanks to new biotechnologies that have recently come into the research field and the clinical practice, giving oncologists new instruments that are based on biomarkers and allowing them to set up a personalized approach for each individual patient. Here we review the main treatments available or in preclinical development, the biomolecular diagnostic and prognostic approaches that changed our perspective about breast cancer, giving an overview of targeted therapies that represent the current standard of care for these patients. Finally, we report some examples of how new technologies in clinical practice can set in motion the development of new drugs.
Introduction
In the last decade, impressive steps toward understanding the biology of breast cancer have been accomplished, thanks to the use of biotechnologies. At present a window of opportunity exists to identify and use these biomarkers, to develop new therapies in a mechanistic-based rational approach, and to assist in the identification of patients requiring a treatment from those who do not, in a very early phase of the disease. According to the literature, a biomarker is:
[…] a characteristic that is objectively measured and evaluated as an indicator of normal biologic or pathogenic processes, or pharmacologic responses to a therapeutic intervention.Citation1
The first identification of breast cancer biomarkers dates back to the 1970s, with the discovery of the estrogen receptor (ER) and the progesterone receptor (PgR) by immunohistochemistry (IHC). Twenty years later, the second generation of breast cancer biomarkers was found with the use of gene amplification detection by in situ hybridization and their clinical impact has been dramatic in patients with the human epidermal growth factor 2 (HER2) overexpressing tumors.Citation2,Citation3 More recently, the turning point that led to the acceleration of breast cancer research has been represented by the use of microarrays for gene and microRNA expression profiling.Citation4 Afterwards, the acquisition of next-generation sequencing techniques for genetic mapping, mutational analysis, and genome-wide monitoring of the gene expression permitted the investigation of thousands of transcripts simultaneously. This review aims to explore the main clinical effects of new technologies in the diagnostic, prognostic, and treatment course of breast cancer patients. For this purpose, a search of the online PubMed database (all years) was undertaken to identify relevant previous and current clinical studies using the search terms “breast cancer gene expression profile,” “next generation sequencing,” and “personalized medicine.”
Current and future diagnostic technologies used in personalized medicine
Gene expression profile as a prognostic tool
The first pivotal study that paved the way for a new breast cancer classification and for the molecular taxonomy of subsequent investigations came from the laboratories of Perou and Sørlie more than 10 years ago.Citation5,Citation6 Using DNA microarrays, these authors identified five distinct molecular subgroups of breast cancer with a different prognosis, namely luminal A, luminal B, HER2-enriched, basal-like and normal-like. That was the first demonstration that breast cancer is not a single disease with different morphologic patterns but rather a heterogeneous group of diseases defined by the differential intrinsic gene signature. The main differentially expressed genes, which distinguished the five molecular intrinsic subtypes, were the ER and ER-related genes, proliferation-related genes, HER2, and the genes mapping to the region of the HER2 amplicon on chromosome 17.Citation7 After this forerunner study, additional simplified gene signatures with prognostic value were published with the aim of identifying a minimal gene set. Among these, the 70-gene prognosis signature (MammaPrint®; Agendia, Irvine, CA, USA),Citation8 the 97-gene histologic grade predictor (MapQuant Dx™ Genomic Grade; Ipsogen, Marseilles, France, and New Haven, CT, USA),Citation9 the 21-gene recurrence score (Oncotype Dx®; Genomic Health Inc., Redwood City, CA, USA),Citation10 and the 14-gene distant metastasis signature (BreastOncPx™; Integrated Oncology, Irvine, CA, USA),Citation11 Theros H/ISM and Theros MGISM Breast Cancer Index (bioMérieux, Marcy-l’Etoile, France)Citation12,Citation13 have been extensively evaluated in tumor specimens from patients with early breast cancer to establish different prognostic scores based on the gene expression profile and, therefore, to assign – or not – adjuvant treatment. Two large prospective trials – the EORTC (European Organization for Research and Treatment of Cancer) 10041/BIG (Breast International Group) 03-04 MINDACT (Microarray In Node-negative and 1–3 node positive Disease may Avoid Chemo-Therapy), and the TAILORx (Trial Assigning IndividuaLized Options for Treatment Trial) – are evaluating the MammaPrint (MammaPrint; Symphony Suite, Agendia, Irvine, CA, USA, and Amsterdam, the Netherlands) and the Oncotype DX® Recurrence Score (Genomic Health, Inc., Redwood City, CA, USA), respectively, with the aim to validate the clinical utility of these signatures as a prognostic tool for the decision-making process in early breast cancer.Citation14,Citation15 The results of these studies are awaited with great expectation, as they would optimize and overcome the conventional algorithms used for the decision on adjuvant systemic therapy, based on menopausal status, tumor size, nodal involvement, ER and HER2 status, and tumor grade.Citation16 In the meanwhile, data from a recent meta-analysis of the published gene signatures provided the evidence that most breast cancer patients can be stratified in the same risk group, according to the expression of genes that compose the proliferation, ER, and HER2 signatures.Citation17 It is important to note that these signatures displayed a decrease in the prediction accuracy from 5–10 years after the diagnosis.Citation18,Citation19 Furthermore, the application of gene expression in each different subgroup defined by the intrinsic subtype was a further implementation in molecular characterization of breast cancer. It became immediately evident that the same biological markers are not associated to all the molecular subtypes of breast cancer.Citation20–Citation23 In particular, a crucial role in the ER-positive patients is played by genes related to cell cycle progression and proliferation, while in ER-negative patients, especially in the HER2-positive and triple negative ones, a nodal point is represented by the involvement of the immune system.Citation24–Citation27
Gene expression profile as a predictive tool
Gene expression profiling has been studied not only as a prognostic tool, but also as a predictor of chemo-and hormone-sensitivity. Indeed, a plethora of studies have been conducted to verify whether the sensitivity to anticancer agents can be ascribed to a specific intrinsic molecular subtype rather than to the clinical/pathological presentation of the disease.Citation28–Citation37 In addition, these studies aimed to identify new targetable pathways in chemotherapy-refractory cases. Unfortunately, none of these trials reported data of general clinical interest. This is likely due to the simplification of the complexity of tumor heterogeneity that is an intrinsic limitation of gene profiling. Therefore, despite the initial enthusiasm regarding the molecular profiling of breast cancer, its role in clinical practice is still controversial. Another possible explanation is that the aforementioned studies were conducted in specific patient populations. For example, the analysis performed on women enrolled in the ATAC (Arimidex, Tamoxifen, Alone or in Combination) trialCitation38 and treated without chemotherapy, revealed that Oncotype DX® is substantially equal in terms of predicting metastatic recurrence to accurate quantitative IHC measures of ER, PgR, HER2, and Ki-67.Citation39 This information has been subsequently confirmed in a cohort of 786 patients. It is important to note that this study was conducted in a very restricted population, ie, ER-positive and/or PgR-positive postmenopausal women who were not treated with chemotherapy, which cannot be assumed as valid for the general breast cancer population.
Beyond gene expression profile: mutational analysis
In the very recent years, research has moved from gene expression profiling into a more detailed overview through biological mechanisms of carcinogenesis and tumor progression by mutational profiling. The first approach to sequencing of the genome has been Sanger sequencing, which was extremely sensitive but, in the meantime, hugely expensive in terms of time and resources – a burden with very low throughput.Citation40 Indeed, the Sanger instrument could only support 96 parallel reactions, and the cost per each genome analysis was in the order of 1 million USD. That incited academies and companies in the research of new technologies, passing from the first-generation sequencing to the most cutting-edge one, represented to date by next generation sequencing (NGS). The main characteristic of this procedure – known as massive-parallel sequencing – is its high sensitivity, high throughput, and reduced cost; about 1,000 USD per genome. The NGS can be applied to study the whole genome (exons, introns, and intergenic regions for about 22,000 genes), more specifically to the whole exome (about 1% of the genome) or to the 200–400 potentially targetable exons (about 0.003% of the genome). The very high sensitivity of this technique allows the evaluation of single nucleotide variants (SNVs), small insertions or deletions, copy number alternations (CNAs, gain or losses) and structural variations (translocations, inversions). The clinical translation of these investigations results in the discovery of actionable mutations. Furthermore, the NGS can be applied to the RNA, with the whole transcriptome approach (RNA-sequencing) for expression level analysis and to alternative splicing, RNA editing, and fusion transcripts.Citation41 It is remarkable to highlight that the NGS can be applied to tumor tissues compared with its normal counterparts, with the acquisition of information about somatic mutations or to the peripheral blood samples – with the aim to investigate germline alterations. The study of germline aberrations could open new key insights into germline actionable mutations, toxicity susceptibility, drug metabolism, and familial disease susceptibility. A more extensive description of the molecular architecture of cancer cells must include the epigenome, that can be investigated by several new-generation technologies (bisulfite sequencing [Bisulfite-Seq] and chromatin immunoprecipitation sequencing [ChIP-seq]).Citation41
The application of NGS to breast cancer research has led to the publication of several studies, from a comprehensive examination of the genome/transcriptomeCitation42 to whole exome sequences of DNA,Citation43 to studies in specific breast cancer subtypes,Citation44,Citation45 catalogs of somatic mutations,Citation46 and exploration of rearrangement patterns.Citation47 Furthermore, NGS has been applied to search for predictive biomarkers.Citation48 The Cancer Genome Atlas Network performed one of the widest analyses of breast cancer biology, using and integrating all the cutting-edge technologies available and investigating more than 800 patients.Citation42 Authors confirmed the well-known classification in four breast cancer subgroups characterized by substantial differences in their molecular complexity. Only three genes, TP53, PIK3CA, and GATA3, revealed somatic mutations in more than 10% through the different subgroups, and most of the genetic/epigenetic alterations were found to be subgroup-restricted, ie, specific mutations in GATA3, PIK3CA, and MAP3K1 were associated with luminal A breast cancer.
Interestingly, the authors compared basal-like breast cancer with high-grade serous ovarian cancer, observing many similarities and thus suggesting a possible common therapeutic approach. It is important to underline that NGS is able to create a massive amount of information; it is intuitive that not each mutation/alteration found can become a target for specific therapy. Therefore, a priority scale of prognostic and predictive value should be applied. An example is offered by the METABRIC (Molecular Taxonomy of Breast Cancer International Consortium) study, where NGS was used to create CNAs, copy number variations (CNVs), and a single-nucleotide polymorphism (SNP) map, singling out somatic and germline abnormalities.Citation49 The authors identified 10 different subtypes with prognostic impact and found common, potentially targetable alterations, such as PPR2A, MAP2K4, and MTAP deletions. Alterations in the gene expression landscape can also be useful to guide treatment with conventional or experimental therapy. In the study by Bose et al, seven activating HER2 mutations were found in about 2% of HER2 nonamplified breast cancer patients.Citation50 Interestingly, HER2 mutant cells were demonstrated to be sensitive to neratinib but not to lapatinib, paving the way to Phase II clinical trials for the administration of neratinib in HER2 nonamplified mutant patients. More recently, the prospective multicentric molecular screening trial SAFIR 01 analyzed 423 patients with metastatic breast cancer, with no progressive disease at study entry.Citation51 Metastatic sites were biopsied and profiled using the copy number changes array and the Sanger sequencing on PIK3CA (exon 10/21) and AKT1 (exon 3). At the time of the progression, the patients were treated with a targeted therapy, matched with biopsy results. A total of 408 patients successfully underwent metastatic biopsy. The genome analysis was feasible in 71% of cases and informative in 67% of cases. The most frequent genomic alterations were the PIK3CA mutations, CCND1, FGF4, and FGFR1 amplifications. One quarter of the patients with targetable genomic alterations, representing 12% of the patients who had undergone biopsy, were treated with matched therapies.
Overall, 12 of 408 patients (3%) obtained a clinical benefit from the procedure. The first important conclusion from this study is that biopsies of metastatic sites are feasible and safe, with only nine cases of serious adverse events, and informative, with the highest rate of success reported for liver and nodal lesions. The innovative information derived from this study is that molecular-based personalized medicine is feasible, even with many challenges and limitations, which are now being addressed in ongoing studies. In the SAFIR 02 trial, NGS of metastatic lesions will be performed. Patients with HER2-positive breast cancer will be randomly assigned to receive targeted therapies versus standard therapy. In the NCI-MATCH trial, molecular profiling of 3,000 patients presenting progressive disease after systemic therapy will be performed with the aim to select 1,000 patients with molecular abnormalities who can be treated with targeted therapies already available. The results of these studies will be of great value to address the limitations of NGS.
In fact, despite the enthusiastic welcome given to NGS by scientists, many difficulties in its clinical application are still unresolved. The first is purely theoretic. Is it correct to search for every single gene alteration, or is it much more important to define pathway abnormalities? Second, there are biological issues due to tumor heterogeneity, clonal evolution, and the difficulty of discriminating between driver and passenger mutations. Third, there are some technical problems in terms of tumor tissue availability, stromal interferences, laboratory reproducibility of results, and the limited access to new bioactive drugs.
MicroRNAs and breast cancer
MicroRNAs (miRNAs) are a class of small (19–25 nucleotides) noncoding RNAs that are able to downregulate the expression of specific genes through the direct binding of the 3′ untranslated regions of their target messenger (m)RNAs, resulting in mRNA degradation or the inhibition of protein translation.Citation52 Several studies demonstrated that the microRNA-dependent regulation of gene expression modulates the various cellular processes, such as proliferation, differentiation, and apoptosis.Citation53 Moreover, the miRNA aberrant expression or mutation was described in a plethora of diseases, including cancer.Citation53,Citation54
In the last decade, different technologies, including miRNA microarrays, deep sequencing, and NanoString (NanoString® Technologies, Inc., Seattle, WA, USA), have been used to identify cancer-specific miRNA signatures. These studies allowed the identification of miRNAs specifically altered in their expression for any kind of human neoplasia, including breast cancer.Citation54–Citation56 Furthermore, the identification of target genes for these miRNAs led to the discovery of the new molecular players involved in tumor formation, progression, metastasis, and resistance to anticancer therapies.Citation57
In a first study, Iorio et al identified 29 miRNAs whose expression was significantly deregulated in breast cancer, with a smaller set of 15 miRNAs able to predict the nature of the sample analyzed (tumor or normal breast tissue) with 100% accuracy.Citation55 Differentially expressed miRNAs included, among others, miR-10b, miR-125b, miR-145, miR-21, and miR-155, suggesting their potential role as tumor suppressor genes or oncogenes. Other miRNAs were also found differentially expressed in breast tumors with distinct biopatho-logical features. Both ER-and PgR-negative breast tumors displayed reduced expression of the miR-30 family, while the let-7 miRNA was downregulated in those breast cancer patients with lymph node metastasis or a higher proliferation index. The miR-21 upregulation was observed in cancers with a high tumor stage, and a miR-9-3 downmodulation was associated with either a high vascular invasion or the presence of lymph node metastasis.
Further analysis also identified miRNAs differentially expressed in ductal carcinoma in situ (DCIS) or in invasive ductal carcinoma (IDC).Citation58 Based on deep-sequencing data sets, Volinia et al described a signature of 66 miRNAs whose expression levels were altered in DCIS when compared to the normal breast.Citation58 Moreover, comparing miRNA levels in DCIS versus IDC, an miRNA invasiveness-microsig-nature (including miR-210, let-7d, miR-181a, miR-221 as upregulated and miR-10b, miR-126, miR-218, miR-335-5p, and miR-143 as downregulated miRNAs) was also defined by this study.
The miRNAs identified were also correlated with clinical parameters, such as the time to metastasis and overall survival. Time to metastasis was significantly associated with miR-127-3p, miR-210, miR-185, miR-143*, and let-7b expression levels, while miR-210, miR-21, miR-221, and miR-652 were correlated with overall survival.
A recent report from Cascione et al also analyzed the miRNA expression levels in triple negative breast cancer and their metastasis, identifying 13 miRNAs differentially expressed in the normal versus the tumor comparison, and six miRNAs deregulated in tumor versus metastasis and a normal versus metastasis comparison.Citation59 Using univariate and multivariate Cox regression analysis, this group also generated two miRNA signatures prognostic for overall survival (OS) and distant disease-free survival (DDFS), consisting of four and seven miRNAs, respectively, with protective miR-16 and miR-374a and risk-associated miR-125b present in both signatures.
Along with their role as diagnostic and prognostic markers for breast cancer, the miRNAs can also confer antineo-plastic drug resistance through the modulation of specific cellular networks, such as the apoptotic pathway, the HER family driven or the ER-mediated signaling.Citation56
In fact, it has been demonstrated that the overexpression of the miRNA-221/222 cluster, whose expression is negatively regulated by ERα,Citation60,Citation61 confers tamoxifen resistance by targeting p27Kip1.Citation62 The upregulation of miR-125b, through the suppression of the proapoptotic B-cell lymphoma-2 (Bcl-2) antagonist killer 1 (Bak1) expression, induces breast cancer resistance to paclitaxel.Citation63 Epithelial cadherin (E-cadherin) downregulation by the miR-200 family alterations is related to the drug-resistant phenotype in breast cancer cells.Citation64 Anti-neoplastic effects of trastuzumab are negatively affected by the miR-21 overexpression.Citation65
Interestingly, circulating miR-221 levels were found to be a predictive biomarker for sensitivity to neoadjuvant chemotherapy in breast cancer patients.Citation66 These examples strongly indicate that the miRNA expression levels might also represent potential predictive markers of response to conventional and targeted antineoplastic treatments.
Taken together, these studies indicate that the miRNA signatures can represent a valid approach for the correct diagnosis and classification of the various subtypes of breast cancer, also providing the clinicians with new prognostic markers for overall survival and disease-free survival, along with predictive indicators of treatment responses and be potentially useful for the tailoring of patient-specific anticancer therapies.
Selected examples of personalized medicine available today for breast cancer patients
Treatment options and matched diagnostic/exploitable predictive markers, according to different breast cancer subtypes, are reported in . It is clearly evident that most of the markers of response to chemo-and/or targeted-therapy refer to ER and to HER2 breast cancer; triple negative is still a targetless population.Citation67
Table 1 Treatment options, current, and future biomarkers in different subgroups of breast cancer
Therapeutic agents targeting ER and PgR-positive breast cancer
The first targeted therapy that demonstrated a substantial benefit in terms of progression free survival (PFS) and OS in women with ER-positive breast cancer was represented by the selective ER modulator tamoxifen. Its development passed through the US Food and Drug Administration (FDA) approval: first, it passed for the treatment of postmenopausal patients with advanced breast cancer; second, it passed for the adjuvant therapy but only for cases with nodal involvement, independent from the ER status and subsequently for premenopausal patients with advanced breast cancer; and, third, for all women with hormone-receptor positive breast cancer, independent from the menopausal status and nodal involvement, as adjuvant therapy. Among the milestones that built the history of this drug, the NSABP (National Surgical Adjuvant Breast and Bowel Project) trial demonstrated a significant increase in terms of PFS with the administration of tamoxifen 10 mg twice a day for 5 years as adjuvant treatment for pre-or postmenopausal women with node-negative, ER-positive breast cancer, compared to the placebo (PFS 83% versus 77%, P<0.00001).Citation68
Another class of endocrine treatment is represented by the aromatase inhibitors (AIs), which prevent the conversion of androgens to estrogens in peripheral tissues, ie, the main estrogen production mechanism in postmenopausal women. After two generations of AIs characterized by low specificity and poor handling, the third generation deposed the use of tamoxifen as an adjuvant treatment and first-line therapy for hormone receptor (HR)-positive breast cancer in postmenopausal patients. Anastrozole and letrozole were the first registered nonsteroidal agents noncovalently and reversibly binding the aromatase enzyme. Following the registration for patients progressing to tamoxifen,Citation69–Citation71 the demonstration of the superiority of anastrozole or letrozole versus tamoxifen in terms of time to progression and overall response rate led to their registration as first-line therapies.Citation72–Citation74 The third AI that has been developed is exemestane, a steroidal agent that covalently and irreversibly binds the target enzyme. Like the other AIs, it was first approved in the metastatic setting, then in the adjuvant one.Citation75,Citation76 Two large Phase III trials, the ATAC trial and the BIG 1–98 trial, showed a greater benefit in terms of disease-free survival with anastrozole and letrozole, respectively, compared to tamoxifen as an adjuvant treatment for HR-positive early breast cancer in postmenopausal patients (hazard ratio 0.83 in the first analysis; 0.87 at the 5-year follow-up; 0.91 at the 10-year follow-up in favor of anastrozole; hazard ratio 0.81 in favor of letrozole).Citation77–Citation80
A subsequent issue has been the role of AIs as the continuation of adjuvant therapy after the initial treatment with tamoxifen. A big meta-analysis of three Phase III trials showed an improvement in disease-free survival, event-free survival, and overall survival in patients switching to anastrozole after 2–3 years of tamoxifen for the subsequent 2–3 years (hazard ratio 0.59, 0.55, and 0.71, respectively).Citation81
A still controversial topic is whether to continue the adjuvant treatment beyond 5 years. While the extended adjuvant therapy with AIs after 5 years of tamoxifen showed an improvement in disease-free survival and overall survival,Citation82–Citation84 the continuation of tamoxifen after 5 years of treatment had discordant results.Citation85 Interestingly, tamoxifen metabolites have recently been demonstrated to inhibit aromatase enzyme in vitro.Citation86,Citation87 These data could open new perspectives in the identification of novel AIs with a better tolerability profile.
The last endocrine treatment registered has been fulvestrant, a pure ER antagonist. It was first approved for the treatment of postmenopausal women with metastatic breast cancer after progression on tamoxifen, at a dose of 250 mg, based on two Phase III trials that demonstrated no difference in time to progression between fulvestrant and anastrozole.Citation88,Citation89 Later, a Phase III trial showed a benefit in time to progression when a 500 mg dose of fulvestrant was administered; thus the scheduled dose was amended to 500 mg.Citation90 The only Phase II study evaluating the higher dose regimen of fulvestrant compared to AI anastrozole as a first-line therapy in postmenopausal patients proved a benefit in terms of time to progression in favor of the antiestrogen drug (median time to progression 23.4 months for fulvestrant versus 13.1 months for anastrozole), with a 34% reduction in risk of progression (P=0.01).Citation91
Biomarkers and endocrine therapy
Two isoforms of ER exist – ERα and ERβ – which are encoded by two different genes (ESR1 and ESR2, respectively). Different studies have evaluated the correlation between ERα, ERβ, response to endocrine therapies, and prognosis, but with discordant results. Even if the ERα expression is – most of the time – associated with hormonal therapy sensitivity, and its expression level is considered as the main predictive factor to tamoxifen sensitivity,Citation92 many pre-or posttranslational alterations of the receptor could negatively influence the response to targeted treatments. In particular, the ERα-36 variant correlates with a lower tamoxifen response and worse outcome.Citation93 The ERα phosphorylation also seems to be associated with a resistance to antiestrogen therapies.Citation94–Citation96 These data suggest that a better understanding of ERα presentations could open new perspectives on both the selection of which patients would probably have a greater benefit from its inhibition and new combination treatments.
While the role of ERα is well-established in breast cancer tumorigenesis and progression, the same cannot be said for ERβ. There are many isoforms of this nuclear receptor and ERβ1, ERβ2, and ERβ5, which are the most involved in breast cancer.Citation97 ERβ is mainly expressed in ERα-positive tumors, even if fewer of the ERβ-positive cases are ERα-negative.Citation98,Citation99 Different isoforms of ERβ probably play different roles in breast cancer, and this behavior correlates with their intracellular localization. In fact, there is evidence that the nuclear expression of ERβ1 correlates with a better outcome, while the cytoplasmic expression of ERβ2 seems to be a poor prognosis marker.Citation100–Citation102 Several studies have evaluated the correlation between ERα, ERβ, a response to endocrine therapies, and a prognosis, but with discordant results, and – to the best of our knowledge – there is not a consensus about the clinical utility of testing ERβ.
ER and PgR assays are currently performed by IHC and the hormone receptor-positive status has been historically defined as 10% or more positive cancer cells to nuclear staining.Citation103 However, in very recent years, this threshold has been reduced to more than 1%, as recommended by the American Society of Clinical Oncology and the American College of Pathologists.Citation104 There is still not a collegial agreement about this new subgroup of weakly ER-positive breast cancer, that should therefore be treated with endocrine therapy. In a study published last year, only 24% of the borderline ER-positive cancer evaluated showed the ESR1 mRNA expression. Furthermore, the average ER gene signature scores of these tumors were more similar to ER-negative than ER-positive cases with more than 10% staining.Citation105
ER-positive breast cancer heterogeneity
In a meta-analysis that included 10,645 ER positive patients, treatment with 5 years of adjuvant tamoxifen reduced the risk of breast cancer death by one-third after 15 years of followup.Citation106 For postmenopausal patients with early breast cancer, a superior benefit was reported with the use of aromatase inhibitors.Citation76–Citation80 In the metastatic setting, another therapeutic option is offered by the pure ER antagonist fulvestrant, which is now approved for postmenopausal patients in progression after antiestrogen therapy.Citation90 Since the publication of the intrinsic gene signature, the existence of at least two subtypes of ER-positive breast cancers have been unanimously acknowledged. Luminal A and luminal B breast cancer cases are characterized not only by distinctive expression levels of ER, PgR, tumor grade, proliferation-related genes, and pathways activation, but also by a very different prognostic and predictive impact.Citation5,Citation6 In particular, the low expression of ER, found in luminal B tumors, correlates with poorer sensitivity to antiestrogen therapies as compared to luminal A cancer; whereas, the high tumor grade proliferation index that is characteristic of the luminal B subtype may justify at least in part the greater benefit from cytotoxic treatments compared with luminal A, as reported in the Spanish Breast Cancer Research Group (GEICAM)/2006-03 neoadjuvant trial.Citation107 On the other hand, luminal B tumors demonstrated fewer benefits from chemotherapy when compared to HER2-enriched and basal-like breast cancer cases.Citation108 As many endocrine therapies are now available for the oncologist and therapeutic decisions are still based on menopausal status, it is intuitive that new predictive and targetable markers are urgently needed for ER-positive and, particularly, in luminal B breast cancer patients.
Overcoming hormonal resistance by new targeted treatment
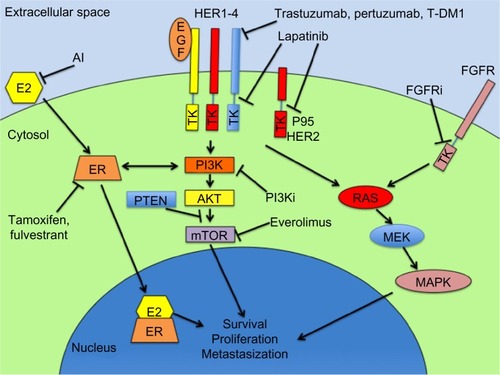
Presuming that breast cancer can acquire resistance to endocrine therapies through pathways that are alternative to ER activation, and since the phosphatidylinositol 3-kinase (PI3K)-serine/threonine-specific protein kinase (AKT)-mammalian target of rapamycin (mTOR) cascade is one of the main downstream nongenomic signals of the ER (), it is intuitive to hypothesize that the mTOR blockade can restore hormone sensitivity.Citation109
Figure 1 Schematic representation of the main targeted pathways and their inhibitory drugs in breast cancer treatment.
Abbreviations: E2, estradiol; AI, aromatase inhibitors; EGF, epidermal growth factor; HER, human epidermal growth factor; ER, estrogen receptor; PTEN, phosphatase and tensin homolog; PI3Ki, PI3 kinase inhibitors (ie, BKM120, GDC0941, XL147, BYL719, BEZ235); TK, tyrosine kinase; T-DM1, trastuzumab emtansine; AKT, serine/threonine-specific protein kinase; mTOR, mammalian target of rapamycin; RAS, reticular activating system; MEK, mitogen-activated extracellular signal-regulated kinase; MAPK, mitogen-activated protein kinase; FGFRi, fibroblast growth factor receptor inhibitors (ie, dovitinib, intedanib, brivanib).
The most currently developed mTOR inhibitor in the clinical phase is everolimus, and the Phase III study that led its registration in the metastatic setting is the Breast cancer trials of OraL EveROlimus-2 (BOLERO) trial.Citation110 In this study, 724 women with advanced breast cancer were randomized to receive exemestane (25 mg daily) plus everolimus (10 mg daily) versus exemestane plus placebo. This study proved that the addition of everolimus to hormonotherapy prolongs PFS from 2.8 months to 6.9 months, according to the local investigators, and from 4.1 to 10.6 months, according to the central reviewer, at the preplanned interim analysis (P<0.001). At two later follow-ups, PFS was confirmed as statistically longer in the exemestane plus everolimus arm (7.4 versus 3.2 months and 7.8 versus 3.2 months, respectively, at the local assessment and 11.0 versus 4.1 months in both cases as per central assessment).Citation111,Citation112 On the basis of this study, both the FDA and European Medicines Agency (EMA) approved everolimus in combination with exemestane for the treatment of postmenopausal patients with advanced hormone-receptor positive, HER2 negative breast cancer, after recurrence or progression to letrozole or anastrozole.Citation113,Citation114 A recent exploratory study on 227 patients treated in the BOLERO 2 trial – 157 in the everolimus plus the exemestane arm and 70 in the placebo plus the exemestane-alone arm – investigated the possibility of discovering the gene alterations predictive of the response to everolimus.Citation115 The analysis by NGS of 3,230 exons of 182 oncogenes and tumor suppressor genes revealed – among the most common alterations – the PIK3CA (43%, most frequently missense) and TP53 (23%) mutations and FGFR1 (18%) and CCND1 amplifications (31%). Considering these genes one by one, wild-type (WT) and altered patients benefited equally from the combination therapy with everolimus, except for the cases of fibroblast growth factor receptors (FGFR) amplifications. Indeed, it seems that there is a reduced effect of mTOR inhibition in FGFR1/FGFR2 amplified cases. This data is only apparently in discord with the PIK3CA mutational substudy of the Phase II clinical trial that compared neoadjuvant letrozole plus everolimus versus letrozole plus placebo, where the mutations in the PIK3CA exon 9 helical domain were associated with a better response in terms of the proliferation index Ki67 reduction with the combination therapy compared to letrozole alone.Citation116 In fact, the PI3KCA mutations in that study were not associated with a specific benefit from everolimus, but rather to a reduced benefit from hormonotherapy. Interestingly, considering the combination of the different gene statuses, patients with no or only one genetic alteration in PI3K/phosphatase and tensin homolog (PTEN)/cyclin D1 (CCND1) or FGFR1/FGFR2 had the greatest benefit adding everolimus to hormonal treatment (hazard ratio 0.27 versus 0.40 of the full population). Even though preliminary, and with the limitations of an analysis performed mostly on the primary tumor rather than the metastatic sites, the BOLERO 2 results suggest that it is extremely improbable that a single biomarker could be responsible for everolimus efficacy, while a simultaneous analysis of the genes involved in the mTOR cascade is exploitable for future studies.
HER2-positive breast cancer
HER2 is a tyrosine-kinase transmembrane receptor of the HER family that is amplified in about 20% of breast cancer and that confers an aggressive phenotype and poor prognosis profile.Citation117 The humanized monoclonal antibody trastuzumab was the first therapy against the extracellular domain of the HER2 and revolutionized the clinical outcome of the HER2-positive breast cancer patient, both in the early and metastatic setting.Citation2,Citation3,Citation118,Citation119 The mechanism of the action of trastuzumab includes the inhibition of ligand-independent HER2 activation, the activation of antibody-dependent cellular toxicity, and the HER2 extracellular domain cleavage.Citation120 However, trastuzumab does not inhibit the heterodimerization of HER2 with other members of the HER family, especially HER3.Citation121 This is probably one of the main mechanisms of resistance to this drug. Consequently, many efforts have been made to develop alternative anti-HER2 treatments acting at different levels, such as the small-molecule tyrosine kinase inhibitor (TKI) directed both to HER2 and HER1, lapatinib, which has been already registered for the treatment of metastatic breast cancer in association with capecitabine or hormonotherapy.Citation122,Citation123 Another new anti-HER2 agent is pertuzumab, a humanized monoclonal antibody that binds the HER2 dimerization domain, impairing its dimerization with other HER2 proteins or HER2-family members. This mechanism of action induced researchers to suppose its possible synergic effect in association with trastuzumab. This hypothesis has been largely demonstrated in both the metastatic and in the early setting, in the CLEOPATRA (CLinical Evaluation Of Pertuzumab And TRAstuzumab) study and in the NeoSPHERE (Neoadjuvant Study of Pertuzumab and Herceptin in an Early Regimen Evaluation) trial, respectively.Citation124,Citation125 A subsequent pharmacological development of trastuzumab is the antibody conjugated to a derivative of maytansine trastuzumab emtansine (T-DM1), which demonstrated a high antitumoral effect and a very low toxicity profile.Citation126
HER2 breast cancer heterogeneity
HER2 status can be determined at protein, DNA, and RNA level. Current assays to evaluate the HER2 status in breast cancer include IHC and in situ hybridization. In clinical practice, a tumor is defined as HER2-positive if 3+ at IHC on a scale of 0–3, uniform intense membrane staining of >30% of invasive tumor cells, or fluorescence in situ hybridization (FISH) amplified, ie, ratio of HER2 to centromeric region of chromosome 17 (CEP17) of >2.2 or average HER2 gene copy number >6 signals/nucleus for those test systems without an internal control probe.Citation127 The degree of HER2 staining intensity is very variable among HER2-positive cases, but it did not show a prognostic or predictive value.Citation128–Citation130 Another intriguing way to investigate the HER2 status is the recently released HERmark™ (Monogram Biosciences, San Francisco, CA, USA) breast cancer assay.Citation131 This technique allows measurement of both the total HER2 protein and the functional HER2 homodimer level on the breast cancer cells’ surface. If validated in prospective trials, HERmark™ could be a useful, predictive marker of trastuzumab sensitivity.
Increasing evidence demonstrates that aberrations of the HER2 protein can affect tumor sensitivity to targeted therapies. The mainly studied HER2 alteration is the p95-HER2 truncated form. This isoform is the result of a 95-kDa or 100-kDa break of the carboxy terminal fragment of the HER2 that is lacking the binding epitope of trastuzumab and that is able to constitutively form homodimers, which activate not only the HER2 classical downstream pathway, but also other molecular effectors involved in the metastasization process.Citation132,Citation133 As a consequence, the p95-HER2 positive tumors have proved to be a highly aggressive subgroup of HER2-positive breast cancer characterized by a poor prognosis.Citation134 Due to its conformation, it is intuitive that p95-HER2 is not inhibited by trastuzumab, which binds the extracellular domain of HER2. Preliminary data in the metastatic setting, using immunofluorescence assays, proved that the p95-HER2 positive patients are resistant to treatment with trastuzumab and sensitive to lapatinib as p95-negative patients.Citation135–Citation137 The p95-HER2 is, therefore, not only a poor prognosis marker, but it is also a possible predictive biomarker of response to biological treatments. However, recent neoadjuvant studies, which analyzed p95 by IHC, did not replicate the findings obtained in patients with metastatic disease. This controversial data can be ascribed to the poor specificity of the anti-p95 antibody used and – secondarily – to the coexpression of p95 with the full-length HER2.
Therefore, no definite conclusion on the value of p95 in clinical practice can be drawn until the use of a more specific antibody and a simultaneous analysis of the levels of HER2 in the samples with truncated forms. In this sense, the upcoming results of the analysis of the Neo ALTTO (Neoadjuvant Lapatinib and/or Trastuzumab Treatment Optimisation) study, which treated patients with neoadjuvant trastuzumab, lapatinib, or their combination, are awaited with great expectation. Ongoing studies are also evaluating another alteration of the HER2 protein represented by a splice variant lacking exon 16, which is found in breast cancer patients, and is able to confer trastuzumab resistance in preclinical models.Citation138
Among HER2-positive breast cancer patients, those with ER-positive tumors are emerging as a different subgroup with a distinct prognosis and therapeutic outcome. ER is present in about 50% of the HER2-positive tumors, albeit with a lower rate in comparison with HER2-negative cases.Citation139 The formal molecular definition of HER2 and ER positive breast cancer as a distinct subtype came from molecular profiling. Indeed, both the PAM50 gene signature and the aforementioned ATLAS (ATLAS.ti Scientific Software Development GmbH, Berlin, Germany) analysis identified this good prognosis subgroup as luminal-mRNA subtype/HER2-positive, whose main characteristic is the overexpression of luminal genes.Citation42,Citation140 Preclinical models have explored in depth the crosstalk between ER and HER2, revealing a bidirectional scenario, in which ER mediates anti-HER2 resistance and vice versa.Citation141–Citation144 The ER expression in HER2-positive breast cancer has been shown to be not only a prognostic marker, but it also predicts benefit from chemotherapy and trastuzumab.Citation145 In addition, the difference in response rates to the HER2-targeted therapy between HER2-positive breast cancer patients with positive or negative expression of ER emerged dramatically in the neoadjuvant setting. Of note, the low rate of response to the HER2-targeted agents of the HER2 and ER positive breast cancer triples with the combination of hormonotherapy. Therefore, there is a growing need for additional markers of tumor response to hormone-and HER2-targeted therapy to further advance the field for women diagnosed with HER-positive and ER-positive tumors and to spare cytotoxic treatment when unnecessary.
As far as predictive biomarkers for trastuzumab sensitivity are concerned, it is important to mention the role of the immune system. In fact, the inhibition of ligand-independent HER2 activation is not the only mechanism of action for trastuzumab, which is also able to activate both the innate and adaptive immune response through antibody-dependent cellular toxicity. There is emerging evidence about how the immune system plays a major role in the clinical effectiveness of anti-HER2-directed therapies analyzed in depth by Andre et al.Citation146 However, no immune marker is currently available in clinical practice.
Overcoming anti-HER2 resistance by new targeted treatments
One of the main trastuzumab-resistance mechanisms is the activation of the downstream pathways, potentially due to a number of factors, including loss of PTEN, PI3K mutations, PI3K and Src activation by other receptors, such as insulin-like growth factor 1 (IGF-1R), MET, erythropoietin receptor (Epo-R), and ephrin type-A receptor 2 (EPHA2).Citation147 Because mTOR is the ultimate player of this pathway, its inhibition may overcome all these anti-HER2 escapes. In particular, the BOLERO 3 trial evaluated the clinical benefit of everolimus when combined to trastuzumab and vinorelbine in the metastatic HER2-positive and trastuzumab-resistant breast cancer patients pretreated with taxanes.
The preliminary results of this randomized Phase III trial were presented at the 2013 American Society of Clinical Oncology annual meeting.Citation148 Patients were randomized to receive weekly vinorelbine 25 mg/m2 intravenously, plus weekly trastuzumab 2 mg/kg, plus either daily everolimus 5 mg by mouth or placebo. The primary endpoint was PFS. The addition of everolimus significantly improved PFS from 5.78 to 7.00 months (P=0.0067), while the OS data are not available yet. What is really interesting is the subgroup analysis. Indeed, the greatest benefit from the mTOR inhibition was obtained in a very clear subpopulation of patients younger than 65 years old without liver involvement, and – even more relevant – the patients who received trastuzumab in the early stage of disease (adjuvant or neoadjuvant setting) and who did not express hormone receptors. This last observation entails many questions about the use of mTOR inhibitors in the HER2-positive patients: should this therapy be restricted to ER-negative disease or should the additional combination of everolimus plus anti-HER2 therapy plus antiestrogen-targeted treatment be hypothesized? Further studies are essential to address these questions. Another fundamental study whose results are still awaited is the BOLERO 1 trial, a randomized, Phase III study of everolimus in combination with trastuzumab and paclitaxel as first-line treatment in the HER2-positive metastatic breast cancer patients.Citation149
Another druggable target to overcome the anti-HER2 resistance is represented by the heat shock protein 90 (Hsp90). Hsp90 is the ubiquitous well-conserved adenosine 5′-triphosphatase that fulfills a crucial role in the protein synthesis processes, found overexpressed in many types of tumors, and involved in a variety of oncogenic pathways. It allows cancer cells to survive despite exogenous and endogenous injuries.Citation150 As HER2 is an Hsp90 client, a synergistic activity of their inhibitors has been hypothesized and demonstrated in preclinical models.Citation151,Citation152 At least 13 Hsp90 inhibitors have entered clinical development in a variety of tumors, including breast cancer, and have already shown their potential, even in the very early clinical study phase and despite the difficulties due to the low pharmacokinetic and the high toxic profile of their predecessors.Citation153 First, tanespimycin (17-AAG) showed promising activity in combination with trastuzumab in pretrastuzumab-treated metastatic HER2-positive breast cancer patients.Citation154,Citation155 Indeed, in a Phase II trial, the overall response rate was 22%, with a clinical benefit rate of 59%. These encouraging results stress the biological rationale and the clinical utility of combining the Hsp90 inhibition to the anti-HER2 treatment. It is not our objective to discuss every Hsp90 inhibitor that is under clinical development in breast cancer. A very detailed review about this topic is in press.Citation156 It is very interesting to note that the p95-HER2 showed to be Hsp90-dependent, both in vitro and in vivo. Preclinical models demonstrated that the Hsp90 inhibition can suppress the p95-HER2 pathway and the tumor cells’ proliferation, and that the trastuzumab-resistant p95-HER2-positive cancer cells are Hsp90-inhibitor sensitive.Citation157 As we have discussed above, the p95-HER2 is a poor prognosis marker and is a predictive factor for trastuzumab resistance. These very early results opened a window for this poor prognosis subgroup.
Selected examples of novel clinical molecular diagnostics and cancer therapeutics
PI3K pathway dysregulation and resistance to breast cancer treatment
The PI3K-AKT-mTOR pathway plays a pivotal role in breast cancer oncogenesis, progression, and resistance to both the ER and the HER2-targeted therapies.Citation158 The complexity of this axis allows the possibility of accumulating alterations in many of its steps, making it a very ambitious target. Indeed, there are several inhibitors in clinical development that act at different levels of this cascade: pan-PI3K inhibitors, isoform-specific PI3K inhibitors, dual PI3K/mammalian target of rapamycin complex (mTORC)1/2 inhibitors, mTORC1/2 inhibitors, and pan-AKT inhibitors. Furthermore, emerging evidence indicates that different subtypes of breast cancer present distinct alterations in the PI3K-signaling cascade, making a focused diagnostic and therapeutic approach essential, case by case.Citation159 Among the number of alterations that occur to the PI3K gene, mutations within exon 9 of the helical domain and exon 20 of the catalytic domain are the most common.Citation160 Other mechanisms by which the PI3K-AKT-mTOR pathway is abnormally activated are: the PI3K and AKT2 gene amplification, AKT1 mutations, and the loss of PTEN, its physiological inhibitor by loss of heterozygosity or hypermethylation of its promoter.Citation161–Citation163 The PI3K-AKT-mTOR pathway abnormal activation has been related to trastuzumab and lapatinib resistance and poor outcome.Citation164–Citation166
One of the main mechanisms by which PI3K-AKT-mTOR pathway is constitutively active in cancer is the loss of PTEN. Thus, it is not surprising that the loss of PTEN has been associated with a worse prognosis and trastuzumab resistance.Citation167
We have already mentioned the solid connection between the PI3K-AKT-mTOR pathway and the ER signaling that lead to the registration of the mTOR inhibitor everolimus in ER-positive patients. From a predictive point of view, in the preclinical models PI3K-AKT-mTOR activation has been related with resistance to all the hormonal therapies available, making it a very promising target for the combination strategies.Citation168–Citation170
Currently available therapies for PI3K-activated breast cancer
The first generation of PI3K inhibitors did not go beyond the preclinical phase because of their poor pharmacokinetic profile and their high toxic effects. Many of the second-generation PI3K inhibitors are in clinical development. One of the most advanced is BKM120, a pan-PI3K inhibitor that is now in a Phase III clinical stage in two different ongoing protocols.Citation171 The Buparlisib brEast cancer cLinicaL Evaluation (BELLE) 2 trial evaluates the association of BKM120 to fulvestrant in postmenopausal patients with HR-positive/HER2-negative locally advanced or metastatic breast cancer refractory to AIs (NCT01610284).Citation172 The BELLE 3 trial is studying the same regimen in the same subgroup of patients but who progressed on or after mTOR inhibitors (NCT01633060).Citation173 BKM120 is also under investigation in the HER2-positive patients, following the Phase I trial of combination with trastuzumab in the trastuzumab-resistant patients.Citation174 This early study demonstrated that the PI3K inhibition could restore the sensitivity to the anti-HER2 targeted therapies. Other promising PI3K inhibitors include GDC 0941, XL 147, BYL 719, an isoform-specific inhibitor, and BEZ235, a dual PI3K-mTOR inhibitor.Citation175–Citation177
Currently, no exhaustive clinical data are available about the effect of PI3K mutations on the sensitivity to PI3K inhibitors. In the context of the Phase I program at the MD Anderson Institute at The University of Texas (Austin, TX, USA), the mutational status of PIK3CA, along with K-RAS, N-RAS, and BRAF, has been evaluated in patients with several types of tumors, including breast cancer, treated with mTOR inhibitors.Citation178 In this study, authors reported a higher response rate in patients harboring PIK3CA mutations compared to the WT ones (30% versus 10%). However, this data contain many issues, as there is no preclinical definitive evidence of the correlation between the PIK3CA mutational status and the benefit from the PI3K inhibitors, even taking into account the many differences in isoform-specific drugs.Citation179 Furthermore, due to the complexity of the PI3K-AKT-mTOR pathway, several other steps, including crosstalk with the other signaling cascade, may affect tumor susceptibility. As an example, in preclinical models, the inhibition of the PI3K-AKT-mTOR signal resulted in a negative feedback loop with the drawback activation of the RAS-RAF-MEK-ERK pathway.Citation180
FGFR amplification
The FGFR family includes four tyrosine-kinase receptors (FGFR1, FGFR2, FGFR3, and FGFR4) that have been deeply involved in tumorigenesis.Citation181 Only sporadic examples of FGFRs’ mutations have been identified in breast cancer patients, while amplifications appear to be prevalent. The different receptors are not crosswise represented, but they are associated to particular biological subtypes, making FGFRs excellent candidates for the single-patient therapeutic choice. Even if the FGFR1 amplification range in the general breast cancer population varies from 7%–17%, in luminal B, it reaches 27%.Citation182,Citation183 The FGFR2 amplification has been reported in 4% of triple negative breast cancer.Citation184
Relationship between FGFR activation and response therapy
The possible prognostic and predictive impact of the FGFRs has been hypothesized, especially for FGFR1, which has been related to chemotherapy sensitivity, resistance to hormone treatments, and to poor prognosis.Citation183,Citation185,Citation186
Whether this behavior depends on FGFR1 amplification itself or on its association with the luminal B subtype is still unknown. Single observations suggested there was a correlation between the FGFR2 protein levels and a poor prognosis as well as between FGFR3 and tamoxifen resistance, and between FGFR4, tamoxifen sensitivity, and prognosis.Citation187–Citation189
Currently available therapies for FGFR-activated breast cancer
Despite the relatively young age of FGFR as a potential target in cancer treatment, several therapeutic approaches have been already attempted. The most advanced in clinical development are the tyrosine kinase inhibitors. Two subsequent generations of FGFR-directed TKIs are already in Phase II studies. The first generation is represented by multitargeting adenosine triphosphate competitive inhibitors, whereas the second generation targets selectively FGFR and is characterized by a higher potency. The most advanced first-generation small molecules that inhibit FGFR are TKI258 (dovitinib), BIBF 1120 (intedanib), and BMS540215 (brivanib). Dovitinib targets FGFR, platelet-derived growth factor receptor (PDGFR), and vascular endothelial growth factor receptor (VEGFR). In a Phase II trial, treatment with dovitinib induced an unconfirmed response or stable disease for more than 6 months in 25% of patients with FGFR1-amplified ER-positive and HER2-negative metastatic breast cancer, but only in the 3% of the FGFR1 not-amplified cases.Citation190 Another possible way to target the FGFR pathway is with monoclonal antibodies binding the FGFR, ligand traps, or downstream blockage, but they are still in a very premature development phase. Taken together, these results suggest that the FGFRs’ amplification status could be not only a predictive and prognostic marker, but it could also be a potential antitumor target and that the FGFR inhibition could be a valid approach for a selected subpopulation of breast cancer patients, probably in association to conventional therapies.
Future directions of diagnostics and therapeutics in breast cancer: the HER2-positive lesson
Recent neoadjuvant studies in the early HER2-positive disease represent the ideal model of how new targeted therapies can be tested in parallel with correlative studies on biomarkers. In the Neo ALTTO study, the combination of trastuzumab plus lapatinib to standard chemotherapy resulted in a pathological complete response (pCR) rate of 51% versus 24%–29% of patients treated with chemotherapy, plus a single HER2 blockade.Citation191
Similarly, in the NeoSPHERE trial, the therapeutic scheme including both trastuzumab and pertuzumab plus chemotherapy resulted in a 46% pCR rate.Citation125 It is very interesting to note that in this trial a treatment arm was planned to receive only the targeted combined therapies before the surgery, postponing chemotherapy to the adjuvant setting. In this subgroup, a 17% pCR rate was obtained, pointing out the existence of a minority of patients who could be theoretically cured without the use of cytotoxic regimens. Unfortunately, no markers are available for the prediction of which population would not need chemotherapy, that therefore remains not excludable from a therapeutic plan so far. An interesting substudy of the NeoSPHERE trial identified the high programmed cell death-1 ligand-1 expression as a poor predictive marker for the pCR in all the chemotherapy-containing arms. (The subgroup treated with only targeted therapies in the neoadjuvant setting showed a similar trend). A good predictive value was associated to high interferon gamma and/or the signal transducers and activators of transcription 1 expression. These preliminary results highlight the role of the immune system in response to the anti-HER2 treatments and paves the way to new therapeutic combinations (anti-programmed cell death-1 ligand-1).Citation192
In the metastatic setting, there are many anti-HER2 therapies, but disappointingly, no marker is still available to define the best anti-HER2 agent or combined therapy and the best order of treatment for breast cancer patients. A critical comparison between pertuzumab, T-DM1 and lapatinib derived from three randomized clinical trials (CLEOPATRA, EMILIA and EGF 104900) allows us to assume that a possible sequence for the anti-HER2 treatments still strictly depends on the level of sensitivity displayed by the disease to trastuzumab. In patients not treated with trastuzumab or showing a recurrence after more than 1 year from the adjuvant therapy, the first-line treatment of choice seems to be a combination of chemotherapy, trastuzumab, and pertuzumab, followed by T-DM1, capecitabine, and lapatinib and – finally – trastuzumab and lapatinib combinations.Citation124,Citation126,Citation193 On the other hand, for patients with unknown or limited responsiveness to trastuzumab (less than 1 year before the recurrence of the disease), there is no preferred first-line therapy, and if an experimental treatment is not available, the T-DM1 is a reasonable option. In fact, clinical trials for patients recurring early after the adjuvant trastuzumab, are missing, whereas this patient population is increasing and urgently deserves dedicated therapies. As far as biomarkers for the outcome prediction and the prognosis are concerned, the substudy from EMILIA indicates that the HER2 mRNA levels are associated with a better outcome, and patients displaying high HER2 mRNA levels showed an enhanced survival benefit from T-DM1 treatment. Both the EMILIA and the CLEOPATRA studies analyzed the mutational status of PIK3CA, demonstrating that the mutational status of this gene is associated to poor prognosis. These studies reported a higher beneficial effect of combined HER2 double blockade in WT patients, while patients carrying a mutant allele of PIK3CA displayed a higher sensitivity to the T-DM1 treatment.
Conclusion
In conclusion, new technologies are significantly improving our knowledge about the prognostic and predictive biomarkers. Many new targeted therapies will soon be available for experimentation, but the large studies are required to identify specific subsets of patients who will take advantage of these treatments. Moreover, these investigations will also provide us with data sets that could allow the clinician to predict the possibility to safely avoid standard chemotherapy for specific patients, preventing them from undergoing all the toxic side effects associated with conventional anticancer treatments.
Disclosure
The authors report no conflicts of interest in this work.
References
- Biomarkers Definition Working GroupBiomarkers and surrogate endpoints: preferred definitions and conceptual frameworkClin Pharmacol Ther2001693899511240971
- SlamonDJLeyland-JonesBShakSUse of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2N Engl J Med20013441178379211248153
- Piccart-GebhartMJProcterMLeyland-JonesBHerceptin Adjuvant (HERA) Trial Study TeamTrastuzumab after adjuvant chemotherapy in HER2-positive breast cancerN Engl J Med2005353161659167216236737
- EisenMBBrownPODNA arrays for analysis of gene expressionMethods Enzymol199930317920510349646
- PerouCMSørlieTEisenMBMolecular portraits of human breast tumoursNature2000406679774775210963602
- SørlieTPerouCMTibshiraniRGene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implicationsProc Natl Acad Sci U S A20019819108691087411553815
- SørlieTTibshiraniRParkerJRepeated observation of breast tumor subtypes in independent gene expression data setsProc Natl Acad Sci U S A2003100188418842312829800
- van ‘t VeerLJDaiHvan de VijverMJGene expression profiling predicts clinical outcome of breast cancerNature2002415687153053611823860
- SotiriouCWirapatiPLoiSGene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosisJ Natl Cancer Inst200698426227216478745
- PaikSShakSTangGA multi-gene assay to predict recurrence of tamoxifen-treated, node-negative breast cancerN Engl J Med2004351272817282615591335
- TuttAWangARowlandCRisk estimation of distant metastasis in node-negative, estrogen receptor-positive breast cancer patients using an RT-PCR based prognostic expression signatureBMC Cancer2008833919025599
- MaXJWangZRyanPDA two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifenCancer Cell20045660761615193263
- JansenMPSieuwertsAMLookMPHOXB13-to-IL17BR expression ratio is related with tumor aggressiveness and response to tamoxifen of recurrent breast cancer: a retrospective studyJ Clin Oncol200725666266817308270
- BogaertsJCardosoFBuyseMTRANSBIG consortiumGene signature evaluation as a prognostic tool: challenges in the design of the MINDACT trialNat Clin Pract Oncol200631054055117019432
- SparanoJATAILORx: trial assigning individualized options for treatment (Rx)Clin Breast Cancer20067434735017092406
- RavdinPMSiminoffLADavisGJComputer program to assist in making decisions about adjuvant therapy for women with early breast cancerJ Clin Oncol200119498099111181660
- WirapatiPSotiriouCKunkelSMeta-analysis of gene-expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signaturesBreast Cancer Res2008104R6518662380
- BuyseMLoiSvan’t VeerLTRANSBIG ConsortiumValidation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancerJ Natl Cancer Inst200698171183119216954471
- DesmedtCPietteFLoiSTRANSBIG ConsortiumStrong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation seriesClin Cancer Res200713113207321417545524
- DaiHvan’t VeerLLambJA cell proliferation signature is a marker of extremely poor outcome in a subpopulation of breast cancer patientsCancer Res200565104059406615899795
- YuJXSieuwertsAMZhangYPathway analysis of gene signatures predicting metastasis of node-negative primary breast cancerBMC Cancer2007718217894856
- TordaiAWangJAndreFEvaluation of biological pathways involved in chemotherapy response in breast cancerBreast Cancer Res2008102R3718445275
- IwamotoTBianchiniGBooserDGene pathways associated with prognosis and chemotherapy sensitivity in molecular subtypes of breast cancerJ Natl Cancer Inst2011103326427221191116
- AlexeGDalginGSScanfeldDHigh expression of lymphocyte-associated genes in node-negative HER2+ breast cancers correlates with lower recurrence ratesCancer Res20076722106691067618006808
- TeschendorffAEMiremadiAPinderSEEllisIOCaldasCAn immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancerGenome Biol200788R15717683518
- SchmidtMBohmDvon TorneCThe humoral immune system has a key prognostic impact in node-negative breast cancerCancer Res200868135405541318593943
- RodyAHoltrichUPusztaiLT-cell metagene predicts a favorable prognosis in estrogen receptor-negative and HER2-positive breast cancersBreast Cancer Res2009112R1519272155
- ChangJCWootenECTsimelzonAGene expression profiling for the prediction of therapeutic response to docetaxel in patients with breast cancerLancet2003362938136236912907009
- AyersMSymmansWFStecJGene expression profiles predict complete pathologic response to neoadjuvant paclitaxel and fluorou-racil, doxorubicin, and cyclophosphamide chemotherapy in breast cancerJ Clin Oncol200422122284229315136595
- GianniLZambettiMClarkKGene expression profiles in paraffin-embedded core biopsy tissue predict response to chemotherapy in women with locally advanced breast cancerJ Clin Oncol200523297265727716145055
- Iwao-KoizumiKMatobaRUenoNPrediction of docetaxel response in human breast cancer by gene expression profilingJ Clin Oncol200523342243115659489
- HessKRAndersonKSymmansWFPharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancerJ Clin Oncol200624264236424416896004
- ThuerigenOSchneeweissAToedtGGene expression signature predicting pathologic complete response with gemcitabine, epirubicin, and docetaxel in primary breast cancerJ Clin Oncol200624121839184516622258
- FarmerPBonnefoiHAnderlePA stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancerNat Med2009151687419122658
- KordeLALusaLMcShaneLGene expression pathway analysis to predict response to neoadjuvant docetaxel and capecitabine for breast cancerBreast Cancer Res Treat2010119368569920012355
- KnauerMMookSRutgersEJThe predictive value of the 70-gene signature for adjuvant chemotherapy in early breast cancerBreast Cancer Res Treat2010120365566120204499
- MillerTWHennessyBTGonzalez-AnguloAMHyperactivation of phosphatidylinositol-3 kinase promotes escape from hormone dependence in estrogen receptor-positive human breast cancerJ Clin Invest201012072406241320530877
- DowsettMCuzickJWaleCPrediction of risk of distant recurrence using the 21-gene recurrence score in node-negative and node-positive postmenopausal patients with breast cancer treated with anastrozole or tamoxifen: a TransATAC studyJ Clin Oncol201028111829183420212256
- CuzickJDowsettMPinedaSPrognostic value of a combined estrogen receptor, progesterone receptor, Ki-67, and human epidermal growth factor receptor 2 immunohistochemical score and comparison with the Genomic Health recurrence score in early breast cancerJ Clin Oncol201129324273427821990413
- SangerFCoulsonARA rapid method for determining sequences in DNA by primed synthesis with DNA polymeraseJ Mol Biol19759434414481100841
- ShyrDLiuQNext generation sequencing in cancer research and clinical applicationBiol Proced Online2013151423406336
- Cancer Genome Atlas NetworkComprehensive molecular portraits of human breast tumoursNature20124907418617023000897
- BanerjiSCibulskisKRangel-EscarenoCSequence analysis of mutations and translocations across breast cancer subtypesNature2012486740340540922722202
- ShahSPMorinRDKhattraJMutational evolution in a lobular breast tumour profiled at single nucleotide resolutionNature2009461726580981319812674
- ShahSPRothAGoyaRThe clonal and mutational evolution spectrum of primary triple-negative breast cancersNature2012486740339539922495314
- Nik-ZainalSVan LooPWedgeDCBreast Cancer Working Group of the International Cancer Genome ConsortiumThe life history of 21 breast cancersCell20121495994100722608083
- StephensPJTarpeyPSDaviesHOslo Breast Cancer Consortium (OSBREAC)The landscape of cancer genes and mutational processes in breast cancerNature2012486740340040422722201
- EllisMJDingLShenDWhole-genome analysis informs breast cancer response to aromatase inhibitionNature2012486740335336022722193
- CurtisCShahSPChinSFThe genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroupsNature2012486740334635222522925
- BoseRKavuriSMSearlemanACActivating HER2 mutations in HER2 gene amplification negative breast cancerCancer Discov20133222423723220880
- AndreFBachelotThomas DenisCamponeMarioArray CGH and DNA sequencing to personalize targeted treatment of metastatic breast cancer (MBC) patients (pts): A prospective multicentric trial (SAFIR01)J Clin Oncol312013 (suppl; abstr 511)
- KimVNHanJSiomiMCBiogenesis of small RNAs in animalsNat Rev Mol Cell Biol200910212613919165215
- AmbrosVThe functions of animal microRNAsNature2004431700635035515372042
- IorioMVCroceCMCauses and consequences of microRNA dysregulationCancer J201218321522222647357
- IorioMVFerracinMLiuCGMicroRNA gene expression deregulation in human breast cancerCancer Res200565167065707016103053
- IorioMVCasaliniPTagliabueEMénardSCroceCMMicroRNA profiling as a tool to understand prognosis, therapy response and resistance in breast cancerEur J Cancer200844182753275919022662
- Nana-SinkamSPCroceCMMicroRNAs as therapeutic targets in cancerTransl Res2011157421622521420032
- VoliniaSGalassoMSanaMEBreast cancer signatures for invasiveness and prognosis defined by deep sequencing of microRNAProc Natl Acad Sci U S A201210983024302922315424
- CascioneLGaspariniPLovatFIntegrated microRNA and mRNA signatures associated with survival in triple negative breast cancerPLoS One201382e5591023405235
- ZhaoJJLinJYangHMicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancerJ Biol Chem200828345310793108618790736
- Di LevaGGaspariniPPiovanCMicroRNA cluster 221–222 and estrogen receptor alpha interactions in breast cancerJ Natl Cancer Inst20101021070672120388878
- MillerTEGhoshalKRamaswamyBMicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1J Biol Chem200828344298972990318708351
- ZhouMLiuZZhaoYMicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of proapoptotic Bcl-2 antagonist killer 1 (Bak1) expressionJ Biol Chem201028528214962150720460378
- ZhangHFXuLYLiEMA Family of Pleiotropically Acting MicroRNAs in Cancer Progression, miR-200: Potential Cancer Therapeutic TargetsCurr Pharm Des EpubJuly192013
- GongCYaoYWangYUpregulation of miR-21 mediates resistance to trastuzumab therapy for breast cancerJ Biol Chem201128621191271913721471222
- ZhaoRWuJJiaWPlasma miR-221 as a predictive biomarker for chemoresistance in breast cancer patients who previously received neoadjuvant chemotherapyOnkologie2011341267568022156446
- O’TooleSABeithJMMillarEKTherapeutic targets in triple negative breast cancerJ Clin Pathol201366653054223436929
- FisherBCostantinoJRedmondCA randomized clinical trial evaluating tamoxifen in the treatment of patients with node-negative breast cancer who have estrogen-receptor-positive tumorsN Engl J Med198932084794842644532
- BuzdarAJonatWHowellAAnastrozole, a potent and selective aromatase inhibitor, versus megestrol acetate in postmenopausal women with advanced breast cancer: results of overview analysis of two phase III trials. Arimidex Study GroupJ Clin Oncol1996147200020118683230
- GershanovichMChaudriHACamposDLetrozole, a new oral aromatase inhibitor: randomised trial comparing 2.5 mg daily, 0.5 mg daily and aminoglutethimide in postmenopausal women with advanced breast cancer. Letrozole International Trial Group (AR/BC3)Ann Oncol1998966396459681078
- DombernowskyPSmithIFalksonGLetrozole, a new oral aromatase inhibitor for advanced breast cancer: double-blind randomized trial showing a dose effect and improved efficacy and tolerability compared with megestrol acetateJ Clin Oncol19981624534619469328
- NabholtzJMBuzdarAPollakMAnastrozole is superior to tamoxifen as first-line therapy for advanced breast cancer in postmenopausal women: results of a North American multicenter randomized trial. Arimidex Study GroupJ Clin Oncol200018223758376711078488
- BonneterreJThürlimannBRobertsonJFAnastrozole versus tamoxifen as first-line therapy for advanced breast cancer in 668 postmenopausal women: results of the Tamoxifen or Arimidex Randomized Group Efficacy and Tolerability studyJ Clin Oncol200018223748375711078487
- MouridsenHGershanovichMSunYSuperior efficacy of letrozole versus tamoxifen as first-line therapy for postmenopausal women with advanced breast cancer: results of a phase III study of the International Letrozole Breast Cancer GroupJ Clin Oncol200119102596260611352951
- ParidaensRJDirixLYBeexLVPhase III study comparing exemestane with tamoxifen as first-line hormonal treatment of metastatic breast cancer in postmenopausal women: the European Organisation for Research and Treatment of Cancer Breast Cancer Cooperative GroupJ Clin Oncol200826304883489018794551
- CoombesRCHallEGibsonLJIntergroup Exemestane StudyA randomized trial of exemestane after two to three years of tamoxifen therapy in postmenopausal women with primary breast cancerN Engl J Med2004350111081109215014181
- BaumMBudzarAUCuzickJATAC Trialists’ GroupAnastrozole alone or in combination with tamoxifen versus tamoxifen alone for adjuvant treatment of postmenopausal women with early breast cancer: first results of the ATAC randomised trialLancet200235993242131213912090977
- HowellACuzickJBaumMATAC Trialists’ GroupResults of the ATAC (Arimidex, Tamoxifen, Alone or in Combination) trial after completion of 5 years’ adjuvant treatment for breast cancerLancet20053659453606215639680
- CuzickJSestakIBaumMATAC/LATTE investigatorsEffect of anastrozole and tamoxifen as adjuvant treatment for early-stage breast cancer: 10-year analysis of the ATAC trialLancet Oncol201011121135114121087898
- The Breast International Group (BIG) 1–98 Collaborative Group; ThürlimannBKeshaviahACoatesASA comparison of letro-zole and tamoxifen in postmenopausal women with early breast cancerN Engl J Med2005353262747275716382061
- JonatWGnantMBoccardoFEffectiveness of switching from adjuvant tamoxifen to anastrozole in postmenopausal women with hormone-sensitive early-stage breast cancer: a meta-analysisLancet Oncol200671299199617138220
- GossPEIngleJNMartinoSRandomized trial of letrozole following tamoxifen as extended adjuvant therapy in receptor-positive breast cancer: updated findings from NCIC CTG MA.17J Natl Cancer Inst200597171262127116145047
- JakeszRGreilRGnantMAustrian Breast and Colorectal Cancer Study GroupExtended adjuvant therapy with anastrozole among postmenopausal breast cancer patients: results from the randomized Austrian Breast Cancer and Colorectal Cancer Study Group Trial 6aJ Natl Cancer Inst200799241845185318073378
- MamounasEPJeongJHWickerhamDLBenefit From exemestane as extended adjuvant therapy after 5 years of adjuvant tamoxifen: intention-to-treat analysis of the National Surgical Adjuvant Breast and Bowel Project B-33 trialJ Clin Oncol200826121965197118332472
- DaviesCPanHGodwinJAdjuvant Tamoxifen: Longer Against Shorter (ATLAS) Collaborative GroupLong-term effects of continuing adjuvant tamoxifen to 10 years versus stopping at 5 years after diagnosis of oestrogen receptor-positive breast cancer: ATLAS, a randomised trialLancet2013381986980581623219286
- LuWJXuCPeiZMayhoubASCushmanMFlockhartDAThe tamoxifen metabolite norendoxifen is a potent and selective inhibitor of aromatase (CYP19) and a potential lead compound for novel therapeutic agentsBreast Cancer Res Treat201213319910921814747
- LuWJDestaZFlockhartDATamoxifen metabolites as active inhibitors of aromatase in the treatment of breast cancerBreast Cancer Res Treat2012131247348121390495
- HowellARobertsonJFQuaresmaAJFulvestrant, formerly ICI 182,780, is as effective as anastrozole in postmenopausal women with advanced breast cancer progressing after prior endocrine treatmentJ Clin Oncol200220163396340312177099
- OsborneCKPippenJJonesSEDouble-blind, randomized trial comparing the efficacy and tolerability of fulvestrant versus anastrozole in postmenopausal women with advanced breast cancer progressing on prior endocrine therapy: results of a North American trialJ Clin Oncol200220163386339512177098
- Di LeoAJerusalemGPetruzelkaLResults of the CONFIRM phase III trial comparing fulvestrant 250 mg with fulvestrant 500 mg in postmenopausal women with estrogen receptor-positive advanced breast cancerJ Clin Oncol201028304594460020855825
- RobertsonJFLindemannJPLlombart-CussacAFulvestrant 500 mg versus anastrozole 1 mg for the first-line treatment of advanced breast cancer: follow-up analysis from the randomized ‘FIRST’ studyBreast Cancer Res Treat2012136250351123065000
- Early Breast Cancer Trialists’ Collaborative Group (EBCTCG); DaviesCGodwinJGrayRRelevance of breast cancer hormone receptor and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trialsLancet2011378979377178421802721
- ShiLDongBLiZExpression of ER-{alpha}36, a novel variant of estrogen receptor {alpha}, and resistance to tamoxifen treatment in breast cancerJ Clin Oncol200927213423342919487384
- HolmCKokMMichalidesRPhosphorylation of the oestrogen receptor alpha at serine 305 and prediction of tamoxifen resistance in breast cancerJ Pathol2009217337237918991335
- SklirisGPNugentZJRowanBGPennerCRWatsonPHMurphyLCA phosphorylation code for oestrogen receptor-alpha predicts clinical outcome to endocrine therapy in breast cancerEndocr Relat Cancer201017358959720418363
- MurphyLCSeekalluSVWatsonPHClinical significance of estrogen receptor phosphorylationEndocr Relat Cancer2011181R1R1421149515
- SmithLColemanLJCummingsMExpression of oestrogen receptor beta isoforms is regulated by transcriptional and post-transcriptional mechanismsBiochem J2010429228329020462399
- JärvinenTAPelto-HuikkoMHolliKIsolaJEstrogen receptor beta is coexpressed with ERalpha and PR and associated with nodal status, grade, and proliferation rate in breast cancerAm J Pathol20001561293510623650
- SklirisGPLeygueEWatsonPHMurphyLCEstrogen receptor alpha negative breast cancer patients: estrogen receptor beta as a therapeutic targetJ Steroid Biochem Mol Biol20081091–211018243688
- LinCYStrömALi KongSInhibitory effects of estrogen receptor beta on specific hormone-responsive gene expression and association with disease outcome in primary breast cancerBreast Cancer Res200792R2517428314
- SugiuraHToyamaTHaraYExpression of estrogen receptor beta wild-type and its variant ERbetacx/beta2 is correlated with better prognosis in breast cancerJpn J Clin Oncol2007371182082817932113
- ShaabanAMGreenARKarthikSNuclear and cytoplasmic expression of ERbeta1, ERbeta2, and ERbeta5 identifies distinct prognostic outcome for breast cancer patientsClin Cancer Res200814165228523518698041
- ReganMMVialeGMastropasquaMGInternational Breast Cancer Study GroupRe-evaluating adjuvant breast cancer trials: assessing hormone receptor status by immunohistochemical versus extraction assaysJ Natl Cancer Inst200698211571158117077359
- HammondMEHayesDFDowsettMAmerican Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estro-gen and progesterone receptors in breast cancerJ Clin Oncol201028162784279520404251
- IwamotoTBooserDValeroVEstrogen receptor (ER) mRNA and ER-related gene expression in breast cancers that are 1% to 10% ER-positive by immunohistochemistryJ Clin Oncol201230772973422291085
- Early Breast Cancer Trialists’ Collaborative Group (EBCTCG); DaviesCGodwinJGrayRRelevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trialsLancet2011378979377178421802721
- AlbaEChaconJILluchAA randomized phase II trial of platinum salts in basal-like breast cancer patients in the neoadjuvant setting. Results from the GEICAM/2006-03, multicenter studyBreast Cancer Res Treat2012136248749323053638
- TranBBedardPLLuminal-B breast cancer and novel therapeutic targetsBreast Cancer Research201113622122217398
- RugoHSKeckSReversing hormone resistance: have we found the golden key?J Clin Oncol201230222707270922753913
- BaselgaJCamponeMPiccartMEverolimus in postmenopausal hormone-receptor-positive advanced breast cancerN Engl J Med2012366652052922149876
- HortobagyiGNPiccartMRugoHEverolimus for postmenopausal women with advanced breast cancer: updated results of the BOLERO-2 phase III trialCancer Res20117124 Suppl 3105s106s
- Piccart-GebhartMJNoguchiSPritchardKIEverolimus for postmenopausal women with advanced breast cancer: updated results of the BOLERO-2 phase III trialProgram and abstracts of the American Society of Clinical Oncology Annual MeetingJune 1–5, 2012Chicago, IL Abstract 559
- US Food Drug AdministrationFDA approves Afinitor for advanced breast cancer7202012 Available from: http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm312965.htm
- European Medicines AgencyAfinitor (Everolimus): Summary of Product CharacteristicsEuropean Medicines Agency2012 Available from: http://www.ema.europa.eu/ema/index.jsp?curl=pages/medicines/human/medicines/001038/human_med_000633.jsp&mid=WC0b01ac058001d124Accessed October 10, 2012
- HortobagyiGNPiccart-GebhartMRugoHCorrelation of molecular alterations with efficacy of everolimus in hormone receptor–positive, HER2-negative advanced breast cancer: Results from BOLERO-2J Clin Oncol312013 (suppl; abstr LBA509)
- BaselgaJSemiglazovVvan DamPPhase II randomized study of neoadjuvant everolimus plus letrozole compared with placebo plus letrozole in patients with estrogen receptor-positive breast cancerJ Clin Oncol200927162630263719380449
- SlamonDJClarkGMWongSGHuman breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogeneScience198723547851771823798106
- MartyMCognettiFMaraninchiDRandomized phase II trial of the efficacy and safety of trastuzumab combined with docetaxel in patients with human epidermal growth factor receptor 2-positive metastatic breast cancer administered as first-line treatment: the M77001 study groupJ Clin Oncol200523194265427415911866
- RomondEHPerezEABryantJTrastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancerN Engl J Med2005353161673168416236738
- HudisCATrastuzumab – mechanism of action and use in clinical practiceN Engl J Med20073571395117611206
- Lee-HoeflichSTCrockerLYaoEA central role for HER3 in HER2-amplified breast cancer: implications for targeted therapyCancer Res200868145878588718632642
- GeyerCEForsterJLindquistDLapatinib plus capecitabine for HER2-positive advanced breast cancerN Engl J Med2006355262733274317192538
- SchwartzbergLSFrancoSXFloranceAO’RourkeLMaltzmanJJohnstonSLapatinib plus letrozole as first-Line therapy for HER-2+ hormone receptor-positive metastatic breast cancerOncologist201015212212920156908
- BaselgaJCortésJKimSBCLEOPATRA Study GroupPertuzumab plus trastuzumab plus docetaxel for metastatic breast cancerN Engl J Med2012366210911922149875
- GianniLPienkowskiTImYHEfficacy and safety of neoad-juvant pertuzumab and trastuzumab in women with locally advanced, inflammatory, or early HER2-positive breast cancer (NeoSphere): a randomised multicentre, open-label, phase 2 trialLancet Oncol2012131253222153890
- VermaSMilesDGianniLEMILIA Study GroupTrastuzumab emtansine for HER2-positive advanced breast cancerN Engl J Med2012367191783179123020162
- WolffACHammondMESchwartzJNAmerican Society of Clinical OncologyCollege of American PathologistsAmerican Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancerJ Clin Oncol200725111814517159189
- DowsettMProcterMMcCaskill-StevensWDisease-free survival according to degree of HER2 amplification for patients treated with adjuvant chemotherapy with or without 1 year of trastuzumab: the HERA TrialJ Clin Oncol200927182962296919364966
- PerezEAReinholzMMHillmanDWHER2 and chromosome 17 effect on patient outcome in the N9831 adjuvant trastuzumab trialJ Clin Oncol201028284307431520697084
- ZabagloLStossORüschoffJHERA Trial Study TeamHER2 staining intensity in HER2-positive disease: relationship with FISH amplification and clinical outcome in the HERA trial of adjuvant trastuzumabAnn Oncol EpubJuly252013
- ShiYHuangWTanYA novel proximity assay for the detection of proteins and protein complexes: quantitation of HER1 and HER2 total protein expression and homodimerization in formalin-fixed, paraffin-embedded cell lines and breast cancer tissueDiagn Mol Pathol2009181112119214113
- ArribasJBaselgaJPedersenKParra-PalauJLp95HER2 and breast cancerCancer Res20117151515151921343397
- PedersenKAngeliniPDLaosSA naturally occurring HER2 carboxy-terminal fragment promotes mammary tumor growth and metastasisMol Cell Biol200929123319333119364815
- SáezRMolinaMARamseyEEp95HER-2 predicts worse outcome in patients with HER-2-positive breast cancerClin Cancer Res200612242443116428482
- ScaltritiMRojoFOcañaAExpression of p95HER2, a truncated form of the HER2 receptor, and response to anti-HER2 therapies in breast cancerJ Natl Cancer Inst200799862863817440164
- SperindeJJinXBanerjeeJQuantitation of p95HER2 in paraffin sections by using a p95-specific antibody and correlation with outcome in a cohort of trastuzumab-treated breast cancer patientsClin Cancer Res201016164226423520664024
- ScaltritiMChandarlapatySPrudkinLClinical benefit of lapatinib-based therapy in patients with human epidermal growth factor receptor 2-positive breast tumors coexpressing the truncated p95HER2 receptorClin Cancer Res20101692688269520406840
- CastiglioniFTagliabueECampiglioMPupaSMBalsariAMénardSRole of exon-16-deleted HER2 in breast carcinomasEndocr Relat Cancer200613122123216601290
- LalPTanLKChenBCorrelation of HER-2 status with estrogen and progesterone receptors and histologic features in 3,655 invasive breast carcinomasAm J Clin Pathol2005123454154615743737
- Gomez PardoPPratABianchiniGPAM50 intrinsic subtyping and pathologic responses to neoadjuvant trastuzumab-based chemotherapy in HER2-positive breast cancerJ Clin Oncol (Meeting Abstracts)201129 (Abstr 554)
- GuoSQSonensheinGEForkhead box transcription factor FOXO3a regulates estrogen receptor alpha expression and is repressed by the Her-2/neu/phosphatidylinositol 3-kinase/Akt signaling pathwayMol Cell Biology2004241986818690
- CreightonCJFuXHennessyBTProteomic and transcriptomic profiling reveals a link between the PI3K pathway and lower estrogen-receptor (ER) levels and activity in ER+ breast cancerBreast Cancer Res2010123R4020569503
- LiuLGregerJShiHNovel mechanism of lapatinib resistance in HER2-positive breast tumor cells: activation of AXLCancer Res200969176871687819671800
- XiaWBacusSHegdePA model of acquired autoresistance to a potent ErbB2 tyrosine kinase inhibitor and a therapeutic strategy to prevent its onset in breast cancerProc Natl Acad Sci U S A2006103207795780016682622
- MontemurroFRossiVCossu RoccaMHormone-receptor expression and activity of trastuzumab with chemotherapy in HER2-positive advanced breast cancer patientsCancer20121181172621598238
- AndreFDieciMVDubskyPMolecular pathways: involvement of immune pathways in the therapeutic response and outcome in breast cancerClin Cancer Res2013191283323258741
- RexerBNArteagaCLIntrinsic and acquired resistance to HER2-targeted therapies in HER2 gene-amplified breast cancer: mechanisms and clinical implicationsCrit Rev Oncog201217111622471661
- O’ReganROzgurogluMustafaAndreFabricePhase III, randomized, double-blind, placebo-controlled multicenter trial of daily everolimus plus weekly trastuzumab and vinorelbine in trastuzumab-resistant, advanced breast cancer (BOLERO-3)J Clin Oncol312013 (suppl; abstr 505)
- HurvitzSAFabriceABurrisHABOLERO-1: A random-ized, phase III, double-blind, placebo-controlled multicenter trial of everolimus in combination with trastuzumab and paclitaxel as first-line therapy in women with HER2-positive (HER2+), locally advanced or metastatic breast cancer (BC)J Clin Oncol302012 (suppl; abstr TPS648.)
- WhitesellLLindquistSLHSP90 and the chaperoning of cancerNat Rev Cancer200551076177216175177
- XuWMimnaughERosserMFSensitivity of mature Erbb2 to geldanamycin is conferred by its kinase domain and is mediated by the chaperone protein Hsp90J Biol Chem20012763702270811071886
- WorkmanPBurrowsFNeckersLRosenNDrugging the cancer chaperone HSP90: combinatorial therapeutic exploitation of oncogene addiction and tumor stressAnn N Y Acad Sci2007111320221617513464
- JhaveriKTaldoneTModiSChiosisGAdvances in the clinical development of heat shock protein 90 (Hsp90) inhibitors in cancersBiochim Biophys Acta20121823374275522062686
- ModiSStopeckATGordonMSCombination of trastuzumab and tanespimycin (17-AAG, KOS-953) is safe and active in trastuzumab-refractory HER-2 overexpressing breast cancer: a phase I dose-escalation studyJ Clin Oncol200725345410541718048823
- ModiSStopeckALindenHHSP90 inhibition is effective in breast cancer: a phase II trial oftanespimycin (17-AAG) plus trastuzumab in patients with HER2-positive metastatic breast cancer progressing on trastuzumabClin Cancer Res201117155132513921558407
- ZagouriFSergentanisTNChrysikosDPapadimitriouCADimopoulosMAPsaltopoulouTHsp90 inhibitors in breast cancer: A systematic reviewBreast201322556957823870456
- ChandarlapatySScaltritiMAngeliniPInhibitors of HSP90 block p95-HER2 signaling in Trastuzumab-resistant tumors and suppress their growthOncogene201029332533419855434
- LiuPChengHRobertsTMZhaoJJTargeting the phosphoinositide 3-kinase pathway in cancerNat Rev Drug Discov20098862764419644473
- BoyaultSDrouetYNavarroCMutational characterization of individual breast tumors: TP53 and PI3K pathway genes are frequently and distinctively mutated in different subtypesBreast Cancer Res Treat20121321293921512767
- BachmanKEArganiPSamuelsYThe PIK3CA gene is mutated with high frequency in human breast cancersCancer Biol Ther20043877277515254419
- CarptenJDFaberALHornCA transforming mutation in the pleckstrin homology domain of AKT1 in cancerNature2007448715243944417611497
- GarciaJMSilvaJMDominguezGAllelic loss of the PTEN region (10q23) in breast carcinomas of poor pathophenotypeBreast Cancer Res Treat199957323724310617300
- GarcíaJMSilvaJPeñaCPromoter methylation of the PTEN gene is a common molecular change in breast cancerGenes Chromosomes Cancer200441211712415287024
- BernsKHorlingsHMHennessyBTA functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancerCancer Cell200712439540217936563
- WangLZhangQZhangJPI3K pathway activation results in low efficacy of both trastuzumab and lapatinibBMC Cancer20111124821676217
- GallardoALermaEEscuinDIncreased signalling of EGFR and IGF1R, and deregulation of PTEN/PI3K/Akt pathway are related with trastuzumab resistance in HER2 breast carcinomasBr J Cancer201210681367137322454081
- NagataYLanKHZhouXPTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patientsCancer Cell20046211712715324695
- MassarwehSOsborneCKCreightonCJTamoxifen resistance in breast tumors is driven by growth factor receptor signaling with repression of classic estrogen receptor genomic functionCancer Res200868382683318245484
- CavazzoniABonelliMAFumarolaCOvercoming acquired resistance to letrozole by targeting the PI3K/AKT/mTOR pathway in breast cancer cell clonesCancer Lett20123231778722484466
- MillerWRLarionovARenshawLGene expression profiles differentiating between breast cancers clinically responsive or resistant to letrozoleJ Clin Oncol20092791382138719224856
- MairaSMPecchiSHuangAIdentification and characterization of NVP-BKM120, an orally available pan-class I PI3-kinase inhibitorMol Cancer Ther201211231732822188813
- Novartis PharmaceuticalsPhase III Study of BKM120/Placebo With Fulvestrant in Postmenopausal Patients With Hormone Receptor Positive HER2-negative Locally Advanced or Metastatic Breast Cancer Refractory to Aromatase Inhibitor (BELLE-2) Available from http://clinicaltrials.gov/show/NCT01610284. Identifier NCT01610284Accessed November 4, 2013
- Novartis PharmaceuticalsA Phase III Study of BKM120 With Fulvestrant in Patients With HR+,HER2-, AI Treated, Locally Advanced or Metastatic Breast Cancer Who Progressed on or After mTORi (BELLE-3) Available from http://clinicaltrials.gov/show/NCT01633060. Identifier NCT01633060Accessed November 4, 2013
- SauraCBendellJJerusalemGPhase I/II study of BKM120 in combination with trastuzumab in patients with HER2-overexpressing meta-static breast cancer resistant to trastuzumab-containing therapySan Antonio Breast Cancer Conference 2011 [abstract PD09-03]
- O’BrienCWallinJJSampathDPredictive biomarkers of sensitivity to the phosphatidylinositol 3′ kinase inhibitor GDC-0941 in breast cancer preclinical modelsClin Cancer Res201016143670368320453058
- ChakrabartyASánchezVKubaMGRinehartCArteagaCLFeedback upregulation of HER3 (ErbB3) expression and activity attenuates antitumor effect of PI3K inhibitorsProc Natl Acad Sci U S A201210982718272321368164
- BrachmannSMHofmannISchnellCSpecific apoptosis induction by the dual PI3K/mTor inhibitor NVP-BEZ235 in HER2 amplified and PIK3CA mutant breast cancer cellsProc Natl Acad Sci U S A200910652222992230420007781
- JankuFWhelerJJWestinSNPI3K/AKT/mTOR inhibitors in patients with breast and gynecological malignancies harboring PIK3CA mutationsJ Clin Oncol201230877778222271473
- JuricDBaselgaJTumor genetic testing for patient selection in phase I clinical trials: the case of PI3K inhibitorsJ Clin Oncol201230876576622271482
- SerraVScaltritiMPrudkinLPI3K inhibition results in enhanced HER signaling and acquired ERK dependency in HER2-overexpressing breast cancerOncogene201130222547255721278786
- TurnerNGroseRFibroblast growth factor signalling: from development to cancerNat Rev Cancer201010211612920094046
- TenhagenMvan DiestPJIvanovaIAvan der WallEvan der GroepPFibroblast growth factor receptors in breast cancer: expression, downstream effects, and possible drug targetsEndocr Relat Cancer2012194R115R12922508544
- TurnerNPearsonASharpeRFGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancerCancer Res20107052085209420179196
- TurnerNLambrosMBHorlingsHMIntegrative molecular profiling of triple negative breast cancers identifies amplicon drivers and potential therapeutic targetsOncogene201029142013202320101236
- MassabeauCSigal-ZafraniBBelinLThe fibroblast growth factor receptor 1 (FGFR1), a marker of response to chemoradiotherapy in breast cancer?Breast Cancer Res Treat2012134125926622438050
- Elbauomy ElsheikhSGreenARLambrosMBFGFR1 amplification in breast carcinomas: a chromogenic in situ hybridisation analysisBreast Cancer Res200792R2317397528
- SunSJiangYZhangGIncreased expression of fibroblastic growth factor receptor 2 is correlated with poor prognosis in patients with breast cancerJ Surg Oncol2012105877377922006548
- TomlinsonDKnowlesMASpeirsVMechanisms of FGFR3 actions in endocrine resistant breast cancerInt J Cancer2012130122857286621792889
- MeijerDSieuwertsAMLookMPFibroblast growth factor receptor 4 predicts failure on tamoxifen therapy in patients with recurrent breast cancerEndocr Relat Cancer200815110111118310279
- AndréFBachelotTCamponeMTargeting FGFR with dovitinib (TKI258): preclinical and clinical data in breast cancerClin Cancer Res201319133693370223658459
- BaselgaJBradburyIEidtmannHNeoALTTO Study TeamLapatinib with trastuzumab for HER2-positive early breast cancer (NeoALTTO): a randomised, open-label, multicentre, phase 3 trialLancet2012379981663364022257673
- GianniLBianchiniGValagussaPAdaptive immune system and immune checkpoints are associated with response to pertuzumab (P) and trastuzumab (H) in the NeoSphere studyCancer Research201272S6S7
- BlackwellKLBursteinHJStornioloAMOverall survival benefit with lapatinib in combination with trastuzumab for patients with human epidermal growth factor receptor 2-positive metastatic breast cancer: final results from the EGF104900 StudyJ Clin Oncol201230212585259222689807
