Abstract
The iron- and 2-oxoglutarate-dependent oxygenases constitute a phylogenetically conserved class of enzymes that catalyze hydroxylation reactions in humans by acting on various types of substrates, including metabolic intermediates, amino acid residues in different proteins and various types of nucleic acids. The discovery of jumonji (Jmj), the founding member of a class of Jmj-type chromatin modifying enzymes and transcriptional regulators, has culminated in the discovery of several branches of histone lysine demethylases, with essential functions in regulating the epigenetic landscape of the chromatin environment. This work has now been considerably expanded into other aspects of epigenetic biology and includes the discovery of enzymatic steps required for methyl-cytosine demethylation as well as modification of RNA and ribosomal proteins. This overview aims to summarize the current knowledge on the human Jmj-type enzymes and their involvement in human pathological processes, including development, cancer, inflammation and metabolic diseases.
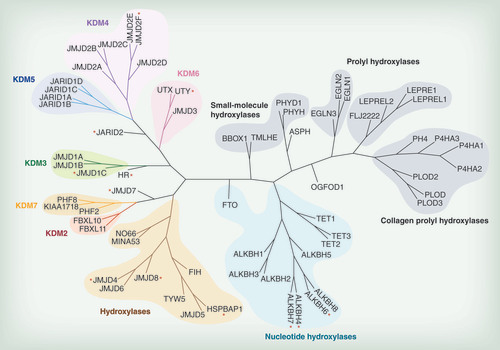
Different subfamilies discussed in the text are highlighted in various colors. Red asterisks indicate members for which no enzymatic activity has been determined yet.
(A) Jmj prototype member JmjD2A (PDB ID: 2OQ7) in complex with Ni2+ (which replaces the endogenous Fe2+) and the 2-oxoglutarate competitive inhibitor N-oxalyl glycine (NOG). The double-stranded β-helical core elements are labeled I–VIII and colored cyan, the additional β-strands in blue and the helices in red. Ni2+ is shown as a green sphere and NOG as yellow sticks. (B) Overlay of the catalytic core (displayed are the active site metal, the Glu-His triad of active site residues and NOG) of human JmjD2A (green) compared to human ALKBH2 (PDB ID: 3BTX; light blue), indicating similar folding patterns of the catalytic domain. (C) Catalytic core of human methyladenosine demethylase FTO (PDB ID: 3LFM [Citation14]) demonstrating the double-stranded β-helical fold and including the active site metal (blue sphere).
![Figure 2. The overall fold of the catalytic JmjC domain in iron- and 2-oxoglutarate-dependent histone demethylases and nucleotide hydroxylases. (A) Jmj prototype member JmjD2A (PDB ID: 2OQ7) in complex with Ni2+ (which replaces the endogenous Fe2+) and the 2-oxoglutarate competitive inhibitor N-oxalyl glycine (NOG). The double-stranded β-helical core elements are labeled I–VIII and colored cyan, the additional β-strands in blue and the helices in red. Ni2+ is shown as a green sphere and NOG as yellow sticks. (B) Overlay of the catalytic core (displayed are the active site metal, the Glu-His triad of active site residues and NOG) of human JmjD2A (green) compared to human ALKBH2 (PDB ID: 3BTX; light blue), indicating similar folding patterns of the catalytic domain. (C) Catalytic core of human methyladenosine demethylase FTO (PDB ID: 3LFM [Citation14]) demonstrating the double-stranded β-helical fold and including the active site metal (blue sphere).](/cms/asset/451d13bd-7376-448e-b2e5-6e26169b2c0e/iepi_a_12324875_f0002.jpg)
Background
Several landmark discoveries have defined the Jumonji (Jmj) oxygenase protein family and provided important connections to chromatin and human biology. This family of iron (Fe2+)- and 2-oxoglutarate (2-OG)-dependent enzymes has its phylogenetic roots in prokaryotes, thus highlighting the functional versatility and critical importance of this protein class for life in an oxygen atmosphere [Citation1]. Regarding their roles in chromatin biology, the first breakthrough discovery was a gene trap approach to identify novel factors involved in mouse embryonic development – this study defined the jmj gene (named JARID2 in humans) as a novel class of transcriptional regulators. The gene was given the Japanese name jumonji, literally translated as ’cruciform’, based on the cross-like shape found during neural groove development of jmj mutant mice [Citation2,Citation3]. Analysis of the domain arrangements and primary structure of this gene identified an ARID/Bright domain, found in many DNA-binding proteins, immediately suggesting chromatin association of the gene product, as well as novel, so-called Jmj N and C domains (JmjN and JmjC, respectively) [Citation3,Citation4]. Further bioinformatic analysis of the JmjC domain revealed this to be an evolutionarily conserved protein fold belonging to the cupin family [Citation5].
Mechanistic features of Jmj enzymes
Cupins are ancient protein domains, found in archaea, bacteria and eukarya, and are often metalloenzymes with metal ion-containing active sites based around a histidine cluster – hence the jumonji C (JmjC) enzymes belong to a large family of metalloproteins that, despite low sequence similarities, share a common Fe2+- and 2-OG-dependent catalytic core. The human 2-OG oxygenase family consists of over 60 members, to which the different Jmj-type lysine demethylases contribute significantly (). All members of this family share few conserved sequence motifs necessary for iron and cofactor binding (with exceptions discussed below), and are mechanistically defined by their ability to hydroxylate their specific substrates using an activated oxo-ferryl intermediate [Citation6]. In brief, the catalytic sequence proceeds through distinct steps of 2-OG cofactor and molecular oxygen binding and activation, resulting in a highly reactive oxo-ferryl intermediate that reacts with the specific substrate atom, usually resulting in a hydroxylated substrate and concomitant CO2 and succinate formation. In terms of lysine demethylase reactions, the hydroxylated substrate is an unstable hemiaminal intermediate derived from the lysine methyl hydroxylation of the N∊- side-chain [Citation6]. This intermediate then fragments into formaldehyde and a lysine side-chain decreased by a methyl group. Importantly, this mechanism allows to demethylate all possible N∊-methyl states (mono-, di- and tri-) found in methylated histone lysine residues and thus differs significantly from the mechanism of monoamine oxidases such as LSD1 and LSD2 (KDM 1A and B) [Citation7] which are important mediators of chromatin function, and which can only demethylate mono- or di-methyl lysine states. Other important functions found in the human 2-OG family comprise hydroxylation of metabolic intermediates (e.g. phytanoic acid degradation [PHYH] [Citation8], or involvement in carnitine synthesis [BBOX, TMLH] [Citation9]), amino acids found, for example, in collagen (e.g., PLOD enzymes ()) [Citation10], or transcription factors (e.g., HIFα by 2-OG oxygenases such as FIH and PHD enzymes).
Structural features of 2-OG enzymes
The JmjC domain is a double-stranded β-helical (DSBH) fold also called the jelly-roll fold or double Greek motif. The DSBH fold is composed of eight β-strands that form a β-sandwich structure comprised of two four-stranded antiparallel β-sheets () [Citation11]. Most 2-OG oxygenases also contain additional secondary structure elements that surround the DSBH and define the different subfamilies. This includes additional β-strands that extend the DSBH, at least one helix on the C-terminal side of the DSBH, and inserts between the fourth and fifth β-strand. This DSBH core fold provides a rigid scaffold for the cosubstrates 2-OG and Fe2+, which are located in the more open end of the barrel (). The iron is coordinated by two histidinyl residues together with a glutamyl or aspartyl residue in a conserved HxE/DxH motif found among 2-OG oxygenases. The 2-OG binding is less well conserved, and the cofactor coordinates the iron in a bidentate manner via its 2-oxo group and one of its 1-carboxylate oxygens, whereas the 5-carboxylate is usually bound to the side-chain of a basic residue (Arg/Lys) and to a hydroxyl group from a Ser/Thr or Tyr residue. The substrate binding varies considerably among the different subfamilies of 2-OG oxygenases/demethylases and the specific interactions identified thus far are beyond the scope of this review. However, crystallographic studies have revealed that it often involves residues from the first and second β-strand, together with strands and loops that extend the DSBH. Other protein domain modules, such as a plant homeodomain (PHD), Tudor, AT-rich interaction domain (ARID) or CxxC zinc finger motifs that vary between the different subfamilies, are usually also required for interaction with DNA/RNA or chromatin protein substrates [Citation11–13]. An overview of the different protein domains as well as annotation of substrate specificities found in Jmj enzymes with defined or hypothetical roles in chromatin biology is given in .
Jmj-enzymes as transcriptional regulators
Paradoxically, the founding member of the Jmj family, JARID2, lacks essential residues necessary for catalytic activity but nevertheless has been shown in various studies to be a key factor in mammalian development [Citation3]. JARID2 shows sequence homology with the JARID1 (KDM 5) family which is defined by the presence of JmjC/JmjN and ARID domains. The molecular mechanism by which JARID2 modulates mammalian development is not well understood, however it is now established that it associates with Polycomb group (PcG) proteins in several cell types, including embryonic stem (ES) cells, and is also critical for ES cell differentiation [Citation62–65]. PcG proteins were identified more than 30 years ago as regulators of homeobox (Hox) genes and development in Drosophila melanogaster [Citation66,Citation67] and also have a critical role in mammalian adult tissue homeostasis and cancer [Citation68].
Importantly, these studies highlight the fact that most, if not all Jmj-type chromatin enzymes function as parts of transcriptional or chromatin protein complexes and in addition to their catalytic roles exert their function as scaffold proteins. The second major breakthrough in Jmj enzyme biochemistry and understanding as transcriptional regulators was therefore the discovery that members of the Jmj family indeed possess catalytic activity towards methyl-lysine histone substrates. This effort was spawned by the discovery of the first lysine demethylase, LSD1 [Citation7,Citation69] (which belongs to the amine oxidase family) followed by the characterization of distinct members of the Jmj family displaying methyl-lysine demethylase activity. These chronological discoveries led to the current and systematic nomenclature system of lysine demethylases (KDM) [Citation70].
Identification of methyl-lysine demethylases was important, because post-translational modifications of N-terminal, unstructured tails of histone proteins had been previously recognized as key-components in the regulation and signaling of functional states of the epigenomic landscape. Accordingly, reversal of these modifications has important consequences in processes such as gene regulation or genome stability; for example, trimethylated lysine 9 of histone 3 (H3K9me3) indicates heterochromatic or repetitive regions, whereas H3K4me3 marks regulatory elements associated with active promoters or transcription start sites, and H3K27me3 often specifies developmentally repressed genes [Citation71]. With the discovery of histone demethylation, lysine methylation of histone residues is now considered reversible and to respond dynamically to changes in metabolism or chromatin environment [Citation72–74]. At present several classes of histone modifications and their respective enzymatic modification systems have been identified [Citation72,Citation75] and amongst their epigenetic substrate marks, lysine and arginine modifications are probably the best studied: acetylation and methylation of lysine residues, as well as methylation of arginine [Citation72,Citation75–76]. Whereas acetylation of histone tails is correlated with gene activation, the influence of histone methylation on regulating gene transcription depends on the exact residue methylated and the number of added methyl groups, both for arginine and lysine residues [Citation72,Citation75–76].
This review focuses on the description of Jmj-type chromatin hydroxylases and their involvement in human chromatin biology and associated pathologies. We will focus our description on the different KDM subfamilies, but also include the nucleotide hydroxylases as well as several further subfamily members with suspected nuclear or chromatin functions (). For completeness, we will also briefly describe the monoamine oxidases of the KDM1 family despite being mechanistically distinct. It is worth noting that since the discovery of Jmj enzymes as mediators of histone demethylation [Citation15] with its important consequences in chromatin biology and transcriptional regulation, not in every case have sufficiently stringent methods (e.g., product identification through mass spectrometry) been employed to properly characterize these enzymes. Therefore in several instances it remains to be established what their true endogenous substrates and inferred biological functions are.
KDM1 subfamily
Starting with the groundbreaking discovery of LSD1 (KDM1A) as methyl-lysine demethylase [Citation7], a series of studies revealed the enzymatic and biological roles of this amine oxidase that was able to demethylate histone methyl-lysine residues H3K4me2/1 and H3K9me2/1 in an FADH-dependent reaction (summarized in [Citation73,Citation77]). Following this work, the paralog LSD2 (KDM1B) with activity towards H3Kme2/1 was discovered [Citation78,Citation79]. Analyses of mice with targeted deletion of LSD1 or LSD2 revealed their essential roles in development [Citation78,Citation80], and other studies highlight the essential role of LSD1 in embryonic [Citation81,Citation82] and cancer stem cell biology [Citation83,Citation84]. Several investigations found that both KDM1 forms are aberrantly expressed in a variety of human tumor types (see [Citation73]) and, taken together, provide the rationale for current inhibitor development. LSD1 is found in a variety of chromatin complexes, which include components such as HDAC/CoREST, BRAF35, BHC80 and noncoding RNA [Citation85], and possibly the NuRD remodeling complex [Citation86]; in these cases LSD1 functions as a repressor through its H3K4me2 demethylase activity. In contrast, LSD1 can associate with nuclear hormone receptors such as the androgen or estrogen receptor [Citation69,Citation87], and in these situations LSD1 shows H3K9me2 demethylase activity, thereby promoting target gene transcription. Taken together, the KDM1 enzymes are involved in various context-specific functions (e.g., in transcription initiation, enhancer modification or chromatin remodeling [Citation73]). Interestingly, LSD1 is able to demethylate other nonhistone, chromatin-associated factors such as p53 or DNA methyltransferase 1 (DNMT1) [Citation80,Citation88], indicating that lysine demethylase substrates are not restricted to histone molecules.
KDM2 subfamily
Soon after the breakthrough discovery of histone lysine demethylation mediated by LSD1, mechanistically different members of the Jmj family were shown to catalyze removal of methyl-lysine histone marks [Citation15]. The first enzymes characterized within this family were human FBXL10 and FBXL11 (KDM2B and KDM2A, respectively), catalyzing the demethylation of H3K36me2/1 histone chromatin marks [Citation15], an activity that orthologous forms display as well. The full-length proteins show a domain organization comprising JmjC, CxxC, PHD, Fbox and leucine-rich repeats (). Fbox-containing domains are involved in formation of cullin-Fbox protein-E3 ubiquitin ligase complexes, which suggests a link between KDM2, histone ubiquitination and proteasomal degradation. The CxxC zinc finger domains are modules that recognize unmethylated cytosine residues in a CpG dinucleotide context. Usually unmethylated, contiguous stretches of CpG elements are often found overlapping with a large fraction (up to 70%) of promoter and transcriptional start sites (so-called CpG islands [CGIs]), enforcing the notion that they contribute to gene regulation by providing a suitable environment for chromatin complexes [Citation89]. Elegant work has provided evidence that FBXL10 and FBXL11 are recruited to the majority of mammalian unmethylated CGIs [Citation90–93] through their CxxC-Zn finger domains. KDM2 enzymes thereby contribute to a specific chromatin environment surrounding CpG sites, independent of the transcriptional states of the particular gene. For example, FBXL11 is bound to 90% of annotated CpG sites in murine ES cells, creates a unique H3K36me2 depleted signature that distinguishes CpG/promoter structures from bulk chromatin, and possibly provides a permissive environment allowing subsequently chromatin elements such as regulatory and transcription factors to bind [Citation90]. Interestingly, FBXL10 shows a largely overlapping occupancy of binding sites when compared to FBXL11 [Citation91–93] and, in addition, defines a CGI subset through binding to Polycomb repressive complex 1 (PRC1), which mediates histone 2A (H2A K119) ubiquitylation through the RING1B component of this noncanonical PRC1 complex. This is in line with data about the activity of CGI1033, the Drosophila KDM2 ortholog that regulates PRC1-mediated H2A monoubiquitylation, independent of its demethylase function [Citation94] and, collectively, provides novel insights how PRC1 is directed to its target genes. The localization of KDM2 enzymes to unmethylated DNA regions is consistent with mass spectrometry studies using nucleosome preparations, demonstrating enrichment at H3K9me3/unmethylated DNA nucleosomes [Citation95]. Other chromatin functions of KDM2 forms may be related to heterochromatin formation and ribosomal gene transcription [Citation16,Citation96].
These studies give evidence for the essential role of KDM2 genes during development and in general for an emerging picture of chromatin architecture and function. The general targeting of KDM2 enzyme forms to CpG-containing promoter regions can explain many of the various phenotypes observed in several biological systems. For example, by being part of the Oct4/Sox2 regulatory axis, KDM2 enzymes can regulate reprogramming, pluripotency and stem cell properties [Citation97–102], associate with corepressors such as BCL-6 corepressor (BCOR) [Citation98,Citation103], or control senescence and proliferation through repression of the Ink4b locus [Citation99,Citation104–105] and hence KDM2 involvement in oncogenic processes has been analyzed [Citation106–108]. From these studies, it is apparent that KDM2 enzymes exert their critical importance through enzymatic (i.e., H3K36me demethylase activity) and nonenzymatic scaffolding functions (i.e., as being part of chromatin protein complexes). In addition, it has been shown that FBXL11 regulates the activity of the transcription factor NF-κB through direct demethylation of methylated Lys218 and Lys221 in NF-κB, a modification that is installed through the methyltransferase NSD1 [Citation17].
KDM3 subfamily
In humans the evolutionarily conserved KDM3 subfamily comprises four proteins, JmjD1A-C and the related hairless (HR) gene product. These proteins differ considerably in amino acid chain-length but have two regions in common that are both required for demethylase activity: a C2HC4-zinc finger-like domain followed by a C-terminal ∼220-residue-long JmjC-domain that shares approximately 65% overall similarities [Citation18–19]. Both full-length and truncated versions of JmjD1A and B specifically demethylate H3K9me2/me1 substrates while no demethylase activity has been described for human JmjD1C or HR yet. The demethylase activity of JmjD1A is inhibited by iron chelators and divalent metals (Co2+ and Ni2+) [Citation109] that replace the ferrous iron at the catalytic site, as well as by NO that forms a nitrosyl-iron complex in the catalytic pocket [Citation110]. JmjD1A-C enzymes are expressed in many cell lines or tissues and are primarily localized to the nucleus. Several phosphorylation sites have been reported for all members in the KDM3 subfamily that could be important for substrate recognition and signaling, but the impact of these modifications is unknown at present [Citation19,Citation111].
JmjD1A was identified by its interaction with the androgen receptor (AR) and acts as a coactivator of AR-mediated transcription [Citation18]. The protein is also known as KDM3A, JHDM2 or testis-specific gene A (TSGA) since it is highly and dynamically expressed during spermatogenesis. Male knockout mice are infertile with small testes and a severe reduction in sperm count, which suggests that JmjD1A positively regulates the expression of genes involved in sperm chromatin condensation and maturation [Citation112,Citation113]. JmjD1A-knockout mice also exhibit an adult obesity phenotype, which implies an important role in transcriptional control of metabolic genes in muscle and adipose tissue [Citation114,Citation115]. The protein is upregulated in human cells exposed to hypoxia and its expression is regulated by HIF-1α that binds to a specific hypoxia responsive element in the JmjD1A promoter [Citation116]. The hypoxia signaling pathway plays an important role in tumor progression, and JmjD1A has recently been associated with many types of cancers. Significantly elevated levels have been reported in several human cancer cells [Citation117] including bladder, lung, hepatocellular [Citation118], prostate [Citation119], colorectal [Citation120] and renal cell carcinoma [Citation121], and treatment of cancer cell lines with siRNA targeting JmjD1A results in significant suppression of proliferation [Citation118], tumor angiogenesis and macrophage infiltration into tumor tissues [Citation117]. Although the exact role of JmjD1A in tumor progression remains to be elucidated it is considered a potential therapeutic target in the treatment of cancer.
JmjD1B also has the synonyms KDM3B/JHDM2B/5qNCA. In humans, the gene is located in the 5q31 chromosomal region, which is frequently deleted in myeloid leukemias [Citation122] and in breast cancer [Citation123]. JmjD1B is a suggested tumor suppressor [Citation124] that regulates the expression of the leukemic oncogene lmo2 [Citation125] and it is also found to specifically interact with the transcriptional repressor suppressor of cancer cell invasion (SCAI) [Citation19]. JmjD1C/JHDM2C was originally referred to as TRIP8 [Citation126]. Two transcripts have been reported for this gene where the major transcript (variant 2) consists of a TRI8H1 domain with the zinc finger motif, a TRI8H2 domain with a thyroid hormone receptor β-binding region, and a C-terminal JmjC domain [Citation127,Citation128]. JmjD1C is similar to JmjD1A, an AR-interacting coactivator (see above) [Citation129], that is suggested to perform a transcriptional regulatory function in testes development by regulating the expression of the steroidogenesis marker p450c17, via SF-1-mediated interactions [Citation130]. JmjD1C is expressed in a variety of human tissues and its reduced expression in human breast cancer tumors compared to normal breast tissues suggests a putative role in tumor suppression [Citation129]. No H3K9me2/1 demethylase activity has thus far been detected for the human protein [Citation19], although all the conserved residues known to be important for enzymatic activity are conserved. A recent report that used both mutational analysis of single residues and domain swapping of both the JmjC- and the Zn-finger domain with JmjD1A/B revealed that both the sequence identity within the JmjC domain and in the sequence N-terminal of the JmjC domain are important for enzyme activity [Citation19]. In other KDMs it has been shown that single amino acid substitutions are sufficient to completely abrogate enzymatic activity [Citation15] and it is possible that thus far unidentified residues involved in substrate binding differ between the different KDM3 homologs. Further studies are needed to elucidate whether JmjD1C utilizes other substrates or cofactors, or if it has a predominant scaffolding function.
The HR protein plays a critical role in the maintenance of hair growth but the underlying mechanisms are still unclear. The protein is expressed in the skin but also the CNS [Citation131,Citation132] where it is suggested to function as a corepressor for multiple nuclear receptors [Citation133–135] including the thyroid hormone receptor, the retinoic acid receptor-related orphan receptors and the vitamin D receptor [Citation136]. HR also interacts with histone deacetylases, including HDACs 1, 3 and 5, and this interaction supposedly is responsible for the corepressor activity observed for HR [Citation137]. Mutations in the HR protein (C622G, N970S, D1012N, V1136D) are associated with congenital alopecia or atrichia with papular lesions [Citation131,Citation138]. These pathogenic mutants occur primarily within the JmjC and the Zn-finger domain and appear to be essential for its co-repressor activity [Citation136].
KDM4 subfamily
The KDM4 subfamily of histone demethylases comprises four members, JmjD2A –D. The proteins are defined by an N-terminal JmjN and JmjC domain that in JmjD2A-C is followed by two tandem PHD and tudor domains (). These enzymes catalyze the demethylation of H3K9me3/me2 with a preference for the tri-methyl state, a histone mark associated with gene repression and found in heterochromatin. In addition, JmjD2A-C catalyze the demethylation of H3K36me3 albeit at a lower rate [Citation20]. A less well-studied repressive histone mark, H1.4K27me, is also reported to be demethylated by all KDM4 members [Citation22]. JmjD2A-C isoforms are also found to catalyze demethylation of trimethyl-lysine peptides of chromatin repressors WIZ, CDYL1, CSB and G9a proteins [Citation21], indicating that KDM4 members can antagonize chromatin repressive marks and regulate gene transcription. A further two members of the human KDM4 subfamily (JmjD2E and JmjD2F) are known to exist but are currently considered to be pseudogenes [Citation139] due to lack of intronic sequences in their genes. However, in vitro studies demonstrate that at least the catalytic domain of human JmjD2E is highly active and shows similar substrate specificities as JmjD2D [Citation20].
The involvement of JmjD2A, B and C in cancer is now unequivocally established, highlighting the importance of these enzymes as potential targets for drug discovery (reviewed recently [Citation140,Citation141]). Many studies over the past 6 years have focused on the role of JmjD2 demethylases in progression of hormone responsive and hormone non-responsive cancers in both prostate and breast, underpinning the coregulation with nuclear hormone receptors. For example, Cloos et al. [Citation142] showed that JmjD2A, B and C are increased in prostate cancer tissues followed by studies demonstrating KDM4 enzymes to be coactivators of the androgen receptor [Citation143,Citation144], in addition to roles in androgen independent proliferation of prostate cancer cells [Citation145]. More recently [Citation146,Citation147] it was demonstrated by siRNA approaches that JmjD2B regulates AR transcriptional activity and controls stability of AR by prevention of proteosomal degradation of the receptor. JmjD2C (GASC1) has been shown to be amplified in triple-negative (estrogen, progesterone and HER2 receptor-negative) breast cancers [Citation148]. Gene silencing studies demonstrate clear antiproliferative effects of JmjD2A knockdown in (estrogen receptor [ER]-negative breast cancer cells [Citation149], and downregulation of JmjD2A in ER-positive breast cancer cells leads to reduced cell proliferation associated with decreased levels of cyclin D1 [Citation150]. Yang et al. [Citation151] showed that JmjD2B is highly expressed in ER-positive primary breast cancer and that JmjD2B is a target gene for ER itself, underlining the evidence for the importance of JmjD2B in the etiology of ER-positive breast cancers [Citation152,Citation153]. JmjD2B and JmjD2C are targets for the hypoxia inducible transcription factor HIF1α in cancer cell lines [Citation154], and hence are of possible importance in regulating transcriptional responses in a hypoxic tumor environment. A role for the lipopolysaccharide (LPS)- and TNF-inducible enzyme JmjD2D in regulating H3K9me3 levels, and hence activity of upstream enhancer elements in a variety of cell types, was recently demonstrated [Citation155].
Given the wealth of publications on the KDM4 family and cancer, relatively little has been published on their role in epigenetic control of other key processes such as embryonic development or normal tissue remodeling. Recent studies revealed a function for JmjD2A in specification of neural crest cells in the early mouse embryo [Citation156]. Conditional gene ablation studies and transgenic studies in mice provide further insights into the function of KDM4 enzymes in development, for example, the generation of mice with a knockout or overexpression of JmjD2A in the heart shows a role for JmjD2A in cardiac hypertrophy in response to cardiac stresses [Citation157]. More recent studies provide insights into how KDM4 enzymes regulate cell fate determination. In vitro and in vivo studies [Citation158] have demonstrated a role for JmjD2B in commitment of mesenchymal stem cells (MSC) to the osteogenic lineage by removing the repressive H3K9me3 marks from osteogenic lineage-selective genes such as DLX5, thus favoring osteoblastogenesis over adipogenesis. JmjD2B has also been shown to be a target gene for the transcription factor C/EBPβ, a transcription factor that is involved in preadipocyte proliferation and differentiation. In this study, a knockdown of JmjD2B inhibited mitotic cell expansion and terminal differentiation of the preadipocyte cell line, 3T3-L1 [Citation159].
More recently, several studies demonstrated a role for the H3K9me3 demethylase JmjD2D (KDM4D) in oncology, inflammation and drug metabolism. In mice, JmjD2D assists the nuclear receptor constitutive androstane receptor (CAR), a key regulator of drug metabolizing enzymes in induction of a neonatal metabolic gene program persisting through adult life [Citation160]. JmjD2D also interacts with the tumor suppressor p53 and controls proliferation [Citation140,Citation161], and activity of cell-type specific enhancer regions in immune cells through H3K9me3 demethylation [Citation155].
KDM5 subfamily
As with many of the JmjC lysine demethylases, the KDM5 family is composed of multidomain members. These are defined by the presence of both JmjC and JmjN domains but also a DNA-binding, ARID domain [Citation162] and a C5HC2 zinc finger motif as well as methyl-lysine- or methyl-arginine-binding PHD domains involved in histone-substrate recognition () [Citation163]. There are four genes described within the human family, namely JARID1A (KDM5A/RBP2), JARID1B (KDM5B/PLU1), JARID1C (KDM5C/SMCX) and JARID1D (KDM5D/SMCY). All KDM5 members demethylate the H3K4me3 histone mark [Citation23–27], a signature indicative of transcriptional activation, and hence KDM5 are considered transcriptional corepressors. One important function appears to be as members of chromatin complexes, for example, JARID1A is found associated to the PRC2 Polycomb complex, important in establishing repressive chromatin marks during development, and JARID1C can be identified in complexes with other repressive chromatin modulators such as HDAC1/2/REST, or the H3K9 methyltransferase G9a [Citation77,Citation164]. KDM5 enzymes are found around the transcriptional start sites of a large set of target genes, where they are thought to fine-tune the transcriptional output, as indicated by modest transcriptional responses upon depletion of, for example, JARID1B – effects, which nevertheless are critical for cellular responses in development and disease [Citation73].
JARID1A was discovered as an interaction partner of the retinoblastoma protein (Rb), a key regulator of cell cycle control and differentiation [Citation165], hence its alias as Rb-binding protein (RBP2). Interactions between the two proteins can promote cellular differentiation [Citation166], in addition to a role for JARID1A and demethylation of H3K4 in the control of cellular senescence [Citation167]. JARID1A regulates HOX gene activity during development [Citation23], underlining the importance of KDM5 enzymes in stem cell biology. The role of JARID1A also extends to other areas of cellular control, with an example being modification of the circadian clock period [Citation168]. In cancer, the NUP98/JARID1A gene fusion has been described as a cryptic translocation involved in pediatric acute megakaryoblastic leukemia [Citation169], and additionally the JARID1A locus was identified in a recent GWAS analysis as a susceptibility gene in ankylosing spondylitis [Citation170]. Importantly, JARID1A has been implicated in facilitating an altered chromatin state that promotes drug-tolerant subpopulations of cancer cells [Citation171].
JARID1B displays an important yet complex role in stem cell biology by blocking differentiation in embryonic and hematopoietic stem cells [Citation172,Citation173], however in a different study it was found to be dispensable for embryonic stem cell maintenance and critical for neural differentiation [Citation174]. JARID1B is also implicated in the control of cellular senescence [Citation167]. It is a deregulated factor in several cancer types, for example, it can suppress tumor progression in breast cancer cells [Citation175], and is specifically expressed in melanoma [Citation176], as well as breast cancer [Citation177]. The involvement of JARID1B in cancer stem cell maintenance highlights its role in tumorigenesis [Citation178,Citation179].
JARID1C and JARID1D are located on the X- and Y-chromosomes respectively and may well have diverged from a single ancestral sex chromosome [Citation180]. Intriguingly, this segregation to the gonosomes offers some interesting prospects regarding sex-specific gene regulation, however, it should be noted that JARID1C is not subject to X-inactivation [Citation181]. The ubiquitously transcribed JARID1C is known for its role in X-linked intellectual disability (XLID). It is highly expressed in brain tissue and the gene is found to be frequently mutated in this condition of male cognitive impairment [Citation180]. There is also a potential role for JARID1C variants in autism spectrum disorder [Citation182]. Taken together, these data present evidence for an important role for JARID1C in normal brain function, by regulating expression of important neuronal genes [Citation164]. In tumor biology, JARID1C interacts with the von-Hippel–Lindau protein and has been shown to regulate gene expression and to suppress tumor growth [Citation183].
JARID1D transcripts are detected in a wide range of male tissues and a JARID1D-derived peptide has been identified as the male-specific histocompatibility antigen (H-Y) [Citation184–186]. The JARID1D genomic locus is associated with a region of the Y-chromosome designated azoospermia factor region; a region involved in spermatogenesis. It is thought that JARID1D interacts with the meiosis-regulatory protein MSH5 during this process [Citation187]. Furthermore, deletion of this gene has been detected in prostate cancer [Citation188]. It has been shown that PCGF6/MBLR can directly interact with JARID1D to enhance its demethylase activity via a C-terminal domain [Citation26].
KDM 6 subfamily
Since the identification of JmjD3 and UTX as H3K27 demethylases in 2007 [Citation28–32], numerous publications have emerged describing the functions of H3K27 demethylase enzymes in various biological contexts such as development, inflammation and oncology. The human KDM6 subfamily consists of three distinct members, namely JmjD3, UTX and UTY with documented histone lysine demethylase activities for UTX and JmjD3 (KDM6A and B). Whereas UTX and UTY are located on X and Y chromosomes, respectively, JmjD3 is found on chromosome 17. All KDM6 members have a JmjC domain followed by a GATA-like DNA-binding domain, whereas UTX and UTY also have N-terminal located protein interaction (tetratricopeptide repeat [TPR]) domains () [Citation189]. Whereas the catalytic role of JmjD3 and UTX is undisputed, critical scaffolding and protein interaction functions of all KDM6 members have been recognized. For example, Sola et al. reported that during mouse neural stem cell differentiation, the N-terminal region of JmjD3 stabilizes the key transcription factor p53 in a demethylase-independent manner and contributes to nuclear localization of p53 [Citation190], or during T-cell development JmjD3 associates with Tbox proteins independent of its catalytic role [Citation191,Citation192]. However, a key mechanism of JmjD3 and UTX appears to be their ability to remove H3K27me3 chromatin marks which anchor Polycomb repressive complexes and are often found in ’bivalent’ chromatin domains thought to be in a transcriptionally ’poised’ state [Citation193].
JmjD3 is expressed in human, rhesus monkey and mouse oocytes during metaphase [Citation194], and plays a role in oocyte preimplantation (e.g., in bovine fertilization), since knockdown of JmjD3 in oocytes inhibited H3K27me3 demethylation and impacted blastocyst [Citation195], and later endoderm and mesoderm development. This is accomplished through methylation of bivalent chromatin domains and their poised transcriptional states [Citation71], by removal of H3K27me3 marks, thus enabling expression of key developmental genes such as WNT3, DKK1 or FOXH1 involved in Wnt and Nodal pathways [Citation196,Citation197]. Multiple studies highlight the key role of H3K27 demethylases in cellular differentiation, for example, in later stages of embryo development, JmjD3 is largely involved in regulation of genes responsible for body and limb morphogenesis, including HOX and T-box transcription factors [Citation198,Citation199]. However, here the enzymatic function of JmjD3 appears to be at least in part dispensable. The importance of H3K27 demethylases is further underpinned by their roles in induced pluripotent stem cell (iPS) biology. Silencing of JmjD3 in mouse embryonic fibroblasts (MEFs) during iPS reprogramming enhanced iPS formation, whereas ectopic expression of JmjD3 inhibited reprogramming, an effect that is mediated through both demethylase-dependent and -independent pathways [Citation200]. Whereas the demethylase-dependent function mediates removal of H3K27me3 repressive marks on genes involved in reprogramming such as Ink4/Arf, the demethylase-independent pathway involves PHF20 for ubiquitination, thereby leading to degradation [Citation200]. Other work has shown that JmjD3 is involved in bone cell differentiation, for example, knockdown of JmjD3 caused significantly reduced osteogenic differentiation of mesenchymal stem cells to osteoblasts [Citation158]. Osteoclast development is partly regulated by H3K27 demethylation, by controlling H3K27me3 levels at the promoter of the key osteoclastogenic transcription factor NFATc1 [Citation201]. A number of studies indicated that JmjD3 also plays an important role in neural differentiation and commitment [Citation202,Citation203]. Here, JmjD3 is one of the early neural differentiation regulators modulated by BMP4 in neural stem cells and is crucial to activate TGFβ-responsive genes [Citation204,Citation205]. In retinoic acid (RA)-induced neural differentiation, an increased expression level of JmjD3 was shown to remove H3K27me3 at the HOX gene regulatory elements [Citation199]. Mash1, a key gene in RA neuronal differentiation, is also regulated by a similar mechanism [Citation206], whereas silencing of an RNA binding protein induces expression levels of JmjD3 in RA neurogenesis [Citation207]. Importantly, a JmjD3-knockout in mouse resulted in perinatal lethality, affecting the pre-Botzinger complex (PBC) complex, which is the pacemaker of the CNS respiratory rhythm generator [Citation202]. The critical role of UTX in developmental effects is underscored by the discovery of mutations in the human gene associated with Kabuki syndrome, an intellectual disability syndrome also associated with mutations in the H3K4 MLL methyltransferase [Citation208,Citation209].
A role in inflammation was found for KDM6 enzymes, since JmjD3 expression in macrophages is regulated through the NF-κB signalling pathway in LPS-stimulated macrophages [Citation28]. It was found that JmjD3 associates with 70% of LPS-inducible genes [Citation210,Citation211], though it is unclear whether JmjD3 is directly involved in transcription of these genes [Citation212]. A direct effect on regulating TNF promoter K27 methylation and transcription was demonstrated in a study using a H3K27 demethylase inhibitor [Citation189]. Stimulation of mouse macrophages by IL-4 induces JmjD3 via the STAT6 pathway and leads to activation of several anti-inflammatory genes [Citation213]. In addition, in mice, the polarization of anti-inflammatory macrophages during helminth infection is modulated by JmjD3 [Citation214]. In T cells, JmjD3 is involved in T-helper cell lineage commitment [Citation191], through interaction with T-box transcription factors. Expression of HPK1/MAP4K1, a kinase that negatively regulates T-cell immune response, is influenced by JmjD3-mediated demethylation of H3K27 [Citation215]. Overexpression of JmjD3 causes upregulation of HPK1, which in turn suppresses T-cell responses. Hence, blocking of JmjD3 activity could be a potential therapeutic approach for the treatment of systemic lupus erythematosus [Citation215]. During wound healing, JmjD3 and UTX are upregulated at the wound site and control epidermal differentiation [Citation216,Citation217]. JmjD3 is also overexpressed in vasculitis [Citation218] where it regulates expression of Itaga2b and Mpl in megakaryocytes [Citation219].
Unsurprisingly, the critical function of H3K27 demethylases (KDM6 enzymes) in controlling key developmental factors is reflected also through their involvement in oncology. Importantly, the particular role (either as tumor suppressor or oncogene) depends on the particular cellular context. Numerous studies have demonstrated overexpression of JmjD3 in cancer cell lines or tumor biopsies, also associated with increased proliferation or control of anti-apoptotic genes such as Bcl-2 [Citation211,Citation220–223], or for example aberrant repressive PcG methylation patterns in medulloblastoma [Citation224,Citation225].
However, in several instances KDM6 enzymes can also be regarded as tumor suppressor genes by counteracting the oncogenic function of PcG proteins, for example by controlling the INK4A/ARF locus that comprises tumor suppressor genes p16INK4A and p14ARF involved in cellular senescence. For example, in cells undergoing oncogenic stress, JmjD3 is induced by RAS/RAF signaling pathways and activates P16/INK4a and p14/ARF, causing p53-dependent cell cycle arrest [Citation226–228].
JmjD3 is involved in the epithelial–mesenchymal transition (EMT), and is overexpressed in invasive breast carcinoma. Knockdown of JmjD3 prevented breast cancer infiltration, and overexpression of JmjD3 affected the expression of a range of adhesion molecules, including downregulation of E-cadherin and upregulation of N-cadherin and fibronectin. Detailed analysis revealed that during TGFβ stimulated EMT, JmjD3 removes H3K27me3 at the promoter of an EMT-inducer gene, SNAI1, thus allowing transcription of the gene [Citation229]. In SW480-ADH colon cancer cells, calcitrol (vitamin D3, 1,25-(OH)2D3 induces JmjD3 expression thus controlling critical target genes. Knockdown of JmjD3 induced SNAI1-mediated EMT, upregulation of mesenchymal markers and downregulation of epithelial proteins [Citation230,Citation231]. Expression of JmjD3 correlates with expression of Vitamin D receptor but is inversely correlated with expression of SNAI1 in 96 human colon tumors [Citation230,Citation231]. Lastly, mutations found in the UTX or JmjD3 genes suggest a possible role in the oncogenic process [Citation232,Citation233].
Little is currently known about the function of UTY, a gene showing a large number of splice variants [Citation234], but encodes a minor HLA-B8 HY-antigen, implicated in graft/host interactions [Citation235]. A recent genetic study identified downregulation of UTY in the predisposing haplogroup I of the Y chromosome in coronary artery disease [Citation236].
KDM7 subfamily
The human KDM7 subfamily consists of three members: KIAA1718 (KDM7A), PHF8 (KDM7B) and PHF2 (KDM7C), showing a characteristic domain organization with an N-terminal PHD domain, and a catalytic Jmj domain ( & ). The C-termini are comprised of coiled–coiled domains and structurally undetermined amino acid stretches. These C-terminal regions have been shown to be important for association with RNA polymerase or transcription factors such as the retinoic acid receptor (RAR) and possibly cell cycle regulators such as E2F1, and contain nuclear localization signals or phosphorylation sites [Citation237]. Several reports unequivocally demonstrate a catalytic role of KDM7 members in demethylation of repressive histone marks such as H3K9me1/2, H3K27me1/2 or H4K20me1 (summarized in [Citation237]) with distinct differences in substrate specificities among the various enzymes. The critical importance of these activities in mediating transcriptional coactivation is highlighted through association with H3K4me3 marks (through the conserved PHD modules [Citation34]), and oxidative removal of repressive histone marks. This creates a permissive chromatin environment by facilitating transcription through subsequent acetylation of lysine residues after methyl mark removal which is accomplished through KDM7 enzymes [Citation237]. The cooperative role of histone marks in KDM7 mediated coactivation is further emphasized through phosphorylation of Ser 3 of histone 3 (H3S3), an important histone mark during mitotic progression, which also prevents KDM7 binding to mitotic chromosomes [Citation238,Citation239], in addition to the cell cycle-dependent phosphorylation of, for example, PHF8, a critical event in cell-cycle progression [Citation240]. The importance of KDM7 members in gene transcription is demonstrated through chromatin immunoprecipitation followed by next-generation sequencing, showing promoter occupancy at a large fraction of transcriptionally active genes [Citation33,Citation239–241]. Besides their roles in RNA polymerase II (RNA Pol II)-mediated transcription, at least PHF8 and PHF2 are also involved in RNA Pol I-mediated RNA gene transcription [Citation33,Citation35,Citation242]. The pathophysiological roles of KDM7 members include involvement in cancer, such as RARα coactivation in pro-myelocytic leukemia [Citation243], regulation of proinflammatory responses [Citation244] and development (e.g., of neural tissues) [Citation245]. PHF2, a H3K9me2 demethylase [Citation33], has been shown to play a role in breast cancer [Citation246]. Several mutations described in PHF8 are causative of Hamel–Siderius syndrome [Citation247–249], an X-linked mental retardation often accompanied by developmental malformation such as cleft lip/cleft palate [Citation250] or microdeletions in autism disorders [Citation251], in line with observations on the critical roles of KDM7 enzymes during neuronal development [Citation36]. Interestingly, another 2-OG enzyme, trimethyllysine hydroxylase found within the small molecule branch (TMLHE, ), was identified along with PHF8 in an X-chromosome exome sequencing study of autism and intellectual disability cases [Citation252].
Other putative Jmj-type demethylases & amino acid hydroxylases
This group (see ) of human 2-OG oxygenases comprises a functionally diverse group of enzymes several of which have well-described roles, such as factor inhibiting HIF (FIH), or which have been rarely studied to date (e.g., JmjD8). These enzymes have, beside the Jmj core domains, few predicted or structurally verified additional domains, the lack of chromatin-binding domains especially suggests potential functions other than histone modification. Indeed, with few studies conducted thus far, the role in histone modification appears, in most instances, secondary at best. Nevertheless, for several of these 2-OG enzymes, hydroxylation of amino acids (e.g., asparagine or histidine residues) in nonhistone proteins has been clearly shown by using mass spectrometry techniques.
Among the best-studied members of the Jmj family is FIH, an asparaginyl hydroxylase that modifies an asparagine residue (Asp803) in the C-terminal transactivation domain of HIFα proteins. These transcription factors regulate key programs involved in metabolic adaptation to low O2 tension, such as energy metabolism, redox and pH homeostasis, and O2 supply, as well as many other functions [Citation253]. FIH-mediated HIFα hydroxylation is important to regulate interaction with the transcriptional coactivator p300, a function that is impaired under low oxygen levels [Citation254]. HIFα activity is regulated by FIH in concert with prolyl hydroxylases (see ) such as PHD1 and PHD2, which regulate proteasomal degradation of HIFα under normoxic conditions. In normoxia, PHD and FIH enzymes act synergistically to degrade and inactivate HIF-1α. Under hypoxic conditions, the PHD enzymes no longer hydroxylate HIFα, leading to stabilization and accumulation of the HIF-1α subunit. FIH remains active at this stage and continues to repress HIFα activity until conditions of severe hypoxia occur, where FIH ceases to hydroxylate the asparagine residue in the C-terminal domain and releases HIFα repression. FIH-null mice do not display a phenotype related to increased HIF or hypoxia responses, but show significant metabolic changes such as reduced body weight, elevated metabolic rate and resistance to effects of high-fat diet [Citation255,Citation256]. FIH is important in several cancer types, for example, FIH expression is regulated by miRNA-31 in head and neck squamous cell carcinoma [Citation257]. Low nuclear expression of FIH is a prognostic factor for poor overall survival in clear cell renal cell carcinoma [Citation258]. Inhibition of FIH-1 in cell cultures of cell renal cell carcinoma decreases cells expansion and increases apoptosis by regulating HIFα [Citation259]. Overexpression of FIH and PHD HIF hydroxylases in non-small-cell lung cancer is indicative of a poor prognosis [Citation260]. FIH is also expressed in invasive breast carcinomas [Citation261], and is found to be inhibited by gankyrin, an oncoprotein expressed in hepatocellular carcinomas, causing an increased HIF and VEGF expression resulting in hemangioma [Citation262].
More recently, it has been demonstrated that FIH can also modify asparagine, aspartate and histidine residues in ankyrin repeat proteins such as NF-κB or Notch [Citation48–50] indicating hydroxylation of amino acid residues as a widespread function that awaits further investigations.
MINA53 is closely linked to expression of the transcription factor c-Myc [Citation263], a gene associated with cell growth, proliferation, loss of differentiation and apoptosis [Citation264]. MINA53 has been reported to demethylate H3K9me3 [Citation265], but this activity is controversial since a lack of demethylase activity has been reported by several groups. More recently it was shown that MINA53 catalyses histidyl hydroxylation of ribosomal protein L27a, suggesting a function in ribosomal assembly and function [Citation39]. A series of papers have confirmed MINA53 effects on cell differentiation, and overexpression of MINA53 in various cancers appears to be linked to oncogenesis [Citation266–273]. The role of MINA53 has been especially well characterized in lung cancer, making it a potential prognostic biomarker for the disease [Citation270,Citation273]. MINA53 expression was also found to be induced by silica particles [Citation273], and the protein plays a key regulatory role in immunity, for example, in allergen-induced inflammatory response [Citation274] and, importantly, in differentiation of proinflammatory TH17 cells [Citation275]. Taken together these studies conclude that MINA53 is an important regulator in inflammation and oncology, however the mechanistic link between its enzymatic activity and those cellular phenotypes awaits clarification.
The closely related oxygenase NO66 (also known as MAPJD) was first identified and characterized in 2004 by Eilbracht et al. in purified nucleoli from Xenopus laevis oocytes [Citation276]. NO66 shows significant sequence homology to MINA53 (34% sequence identity) and is reported to demethylate H3K4 and H3K36 [Citation40], and similar to MINA53, to catalyse ribosomal histidinyl hydroxylation, in this case on ribosomal protein L8 [Citation39]. During mouse embryonic stem cell differentiation NO66 is recruited by PHF19, a component of the Polycomb repressive complex PRC2, to stem cell genes resulting in loss of H3K36me3, transcriptional silencing and PRC2-dependent methylation of H3K27 [Citation277]. NO66 regulates osteoblast differentiation and bone formation by inhibiting Osterix (Osx) [Citation40], a key transcription factor essential at later stages of bone development, accordingly Osx-null mice have a normal cartilage development, but lack bone formation. Conversely, knockdown of NO66 in differentiating osteoblasts derived from mesenchymal stem cells causes accelerated differentiation and mineralization of osteoblasts [Citation40]. The interaction between NO66 and Osx was recently mapped to a conserved hinge domain, that links the N-terminal JmjC domain and C-terminal wHTH domain of NO66 () [Citation278]. The protein is overexpressed in non-small-cell lung cancer, and postulated to be a potential drug target [Citation279].
JmjD4 is a poorly characterized oxygenase whose enzymatic activity has not been characterized, and shows approximately 30% sequence similarity to JmjD6. JmjD5 (also classified as KDM8) plays a critical role in the regulation of cell cycle by inhibition of HDAC recruitment and by activating the cyclin A1 locus, postulated through demethylation of histone H3K36me2 [Citation42], although this activity needs further investigation. Instead, a hydroxylase activity of JmjD5 appears to control the half-life of transcriptional regulators such as NFATc1 through promotion of E3-ligase association and proteasomal degradation of the transcription factor [Citation43]. JmjD5 deficiency results in a short-period circadian phenotype both in mammalian cell cultures and Arabidopsis plants [Citation280]. JmjD5 is widely overexpressed in several types of tumors (e.g., in leukemia and breast cancer [Citation281]) and a knockout in a MCF7 breast cancer cell line resulted in reduced proliferation [Citation42], and its function appears to be interrelated with the tumor suppressor p53. Ablation of murine JmjD5 causes severe growth retardation and ends in embryonic lethality [Citation281,Citation282].
JmjD6, initially characterized as phosphatidyl serine receptor [Citation283], a function that together with its postulated methyl arginine demethylase activity [Citation44] has not yet been confirmed, catalyzes the hydroxylation on the carbon side-chain of lysyl residues located in arginine–serine rich regions of, for example, splicing factors [Citation45,Citation284] or might modify single-stranded RNA [Citation285], indicating involvement in RNA modification and function. In mice, the gene is indispensable for normal development of many tissues such as brain, eyes, lung, kidney, liver and intestine at different stages of embryogenesis [Citation286–288]. Furthermore, JmjD6 plays an important role during heart development since ablation of its function is associated with complex cardiopulmonary malformations that resemble the human congenital heart syndrome tetralogy of Fallot [Citation289]. Overexpression of JmjD6 protein has now been strongly linked to poor prognosis in breast cancer [Citation290] and in lung adenocarcinoma [Citation92]. JmjD7 is a poorly characterized enzyme located on chromosome 15; a fusion with phospholipase represents a novel isoform of this lipid metabolizing enzyme [Citation291]. JmjD8 is linked to cell proliferation and cancer but its enzymatic and cellular functions remain unknown. JmjD8 siRNA knockdown in SCC23/MET cells, a cellular model for squamous cell carcinoma, inhibited invasion of the cells by 80%, suggesting an inhibitory effect on cell proliferation [Citation292]. The rat ortholog of human HSPBAP1 (named PASS1) was identified in 2000 by Liu et al., associated with the heat shock protein hsp27 [Citation293]. PASS1 is expressed in various tissues, but shows high expression in the testis and kidney, whereas human HSPBAP1 is abundant in the thymus and pancreas, as indicated by reverse transcription-PCR analysis [Citation294]. HSPBAP1 inhibits the function of hsp27, which has been shown to be neuroprotective in animal models of motor neuron disease and peripheral nerve injury [Citation295,Citation296]. HSPBAP1 is expressed in neuronal and glial cells in the temporal lobe of patients with IE, with no brain expression in control tissues [Citation296]. A follow-up study tested samples from the temporal neocortex taken from intractable epilepsy patients showing significant expression of HSPBAP gene over the controls [Citation297], however the enzymatic role of HSPBAP1 remains unclear. TYW5 acts as a tRNA hydroxylase and at present a biological function has not been reported [Citation52].
Nucleotide hydroxylases
The distantly related group of nucleotide hydroxylases comprises, in humans, the ALKBH members (ALKBH 1–8) and the TET 1–3 and FTO enzymes (see ). Despite low sequence identities they share the conserved sequence and structural elements found within the 2-OG oxygenase family (). ALKBH enzymes were initially identified as mammalian orthologs of the E.coli AlkB DNA repair enzyme [Citation53], important in the adaptive repair response to alkylated DNA residues such as 1-methyladenosine and 3-methylcytosine in single-stranded DNA [Citation53,Citation54]. However, more recent investigations have demonstrated that their physiological role in humans extends well beyond DNA damage repair (see below).
ALKBH 1–3 have been studied extensively and their role in DNA and RNA damage repair has been established [Citation54,Citation298–303]. Although they display distinct but overlapping subcellular distribution and substrate specificities, they are critically involved in various types of cancer and indicative of cancer drug responses [Citation304–311]. Human ALKBH4 supposedly interacts with chromatin-binding proteins such as the histone acetyltransferase p300 [Citation312], however, no enzymatic function with nucleotide substrates has been demonstrated yet. Recently, it was found that demethylation of a monomethylated lysine residue in actin by ALKBH4 regulates actin–myosin interactions, of importance in cytokinesis and cell migration [Citation55].
The landmark discoveries of two Jmj members as RNA demethylases have opened new avenues in understanding the relationships between RNA metabolism, gene regulation and human physiology and disease [Citation313,Citation314]. Initially found in FTO [Citation315] and later extended to ALKBH5 [Citation56], demethylation of N6-methyladenosine (m6A) residues found in several types of RNA species was identified, with a reaction sequence analogous to N∊ demethylation of lysine residues [Citation313]. The discovery of reversible methylation of m6A, a prevalent methyl modification found in mRNA and noncoding RNA, the effects of chemical inhibition of RNA methylation on transcription, processing, translation and the analysis of animal or clinical phenotypes therefore suggest a fundamental role of m6A demethylation in human physiology [Citation313,Citation314]. The critical role of these enzymes was already suggested by the identification of FTO variants as risk alleles for BMI and obesity [Citation316]. Subsequently, the in vitro activity as demethylase for 3-methylthymine and 3-methyluracil found in single-stranded DNA and RNA was established, indicating the distant relationship with the ALKBH-type nucleotide hydroxylases [Citation60,Citation61]. Although the subject of in vivo activity is still a matter of debate, data indicate that m6A is a physiological substrate of FTO, as evidenced from knockdown studies [Citation315]. The role of FTO in maintenance and regulation of energy homeostasis and food intake has been demonstrated in animal models [Citation317] and through GWAS relationships to insulin resistance and obesity [Citation318,Citation319], Alzheimer’s disease [Citation320], cardiovascular disease [Citation321] and renal failure [Citation322], as well as breast and colorectal cancer [Citation323,Citation324] have been established. A loss-of-function homozygous mutation in the FTO gene shows a defect in normal brain and cardiac development [Citation325], in line with previous observations in animal studies.
Other currently uncharacterized members of the ALKBH subfamily comprise members ALKBH6 and 7. Human ALKBH8 has been recently identified as a methyltransferase acting upon 5-carboxymethyl-uridine and catalyzes an important step in tRNA synthesis, is involved in survival after DNA damage [Citation57], and may contribute to cancer progression [Citation326].
The subgroup of TET enzymes () was named after the ten-eleven translocation (t10;11)(q22;q23) found in rare cases of acute myeloid and lymphocytic leukemias [Citation327,Citation328]. The translocation fuses the mixed-lineage leukemia (MLL) gene on chromosome 10 with the TET1 gene on chromosome 11. Subsequently the relationship to the 2-OG family and the activity as hydroxylases of 5-methylcytosine with the consecutive formation of 5-hydroxymethylcytosine, 5-formylcytosine and 5-carboxycytosine was established in a series of elegant studies [Citation58,Citation59]. Identification of TET enzymes as 2-OG members was a challenging task, due to the large insertions found with the catalytic domains () [Citation58], that made correct assignment challenging. Oxidized methylcytosine marks are found at promoters, enhancers and gene bodies, depending on the tissue examined. The chemical modification sequence of oxidation of methylcytosine forms the basis for an enzymatic mechanism to remove the DNA methylcytosine modification, shown to be critical for epigenetic regulation of gene transcription. Several mechanisms of methylcytosine demethylation might apply, including passive demethylation during cell division, or active enzymatic mechanisms acting on the oxidized intermediates. The TET-modified cytosine residues can be actively removed through a base-excision mechanism using DNA glycosylases, or might involve activation-induced cytidine deaminase (AID) and APOBEC constituting a deamination reaction of 5-hydroxymethylcytosine to 5-hydroxyuracil, followed by glycosylase and cytosine replacement. Other mechanisms might involve enzymatic decarboxylation by a yet unidentified decarboxylase, or dehydroxy-methylation by DNA methyltransferases [Citation329]. The importance of the TET enzymes in stem cell biology and development (summarized in [Citation329]) is at present not completely understood, however, especially in myeloproliferative types of cancers, their critical roles have been now established [Citation327–328,Citation330].
Future perspective
Over the last decade we have witnessed significant progress in understanding the function that many members of the Jmj family have in human physiology and disease. This advancement went hand-in-hand with technological improvements in, for example, allowing interrogation of epigenomic changes at single base-pair resolution, providing unprecedented details and mechanisms how chromatin marks sculpt and regulate cellular phenotypes and features such as proliferation, differentiation, metabolism and genomic stability, among others. Undoubtedly, the discovery of hydroxylation as a mechanism to revert N-methylation found in nucleic acids or proteins is a milestone that has triggered substantial progress in identifying the role of this post-translational modification in chromatin biology, and defining epigenomic elements in genome biology in general.
We have here attempted to summarize the current knowledge of human Jmj enzymes with particular emphasis on histone lysine modification and their roles in human disease. It is apparent from this body of research literature that the involvement of N∊-methyl lysine demethylases in disease is partially related to putative functions of the epigenomic elements they modify, however, recent data suggest that several histone demethylases bind to a large number of genes, but display only modest effects on target gene expression when depleted, suggesting a ’fine-tuning’ effect of these enzymes [Citation73]. Moreover, they are often found bound to chromatin without their substrate mark present, suggesting a ’guardian’ function to ’protect’ chromatin against aberrant modifications [Citation73]. This indicates that the KDM-Jmj hydroxylases are not necessarily involved as on–off switches (as found, for example, in the kinase-phosphatase paradigm of signal transduction), but support other roles, highlighted by the fact that a scaffolding function is of critical importance in various settings, (e.g., as observed for JARID2 or the KDM6 enzymes). However, a substantial fraction of the Jmj enzymes () remains enigmatic in terms of substrate specificity or enzymatic activity, calling for further systematic screening activities, including non-histone substrates. In fact, the discovery of Jmj-mediated oxidation of nucleic acids or nonhistone proteins suggests that the substrate spectrum is much larger than anticipated. Furthermore, there might have been substantial misleads in identifying substrate specificities by employing inappropriate assays, so it appears mandatory to stringently evaluate those biochemical parameters to deduce cellular functions. Laudable efforts to map the cellular ’lysine-methylome’ [Citation332] have recently demonstrated that nuclear lysine methylation constitutes approximately 40% of cellular N∊-lysine methylation, including transcription factors and chromatin modifiers besides histone proteins, thus providing a wealth of testable novel substrate hypotheses for Jmj enzymes.
An important aspect deserving further research efforts is to understand the regulation of Jmj enzymes themselves. Intriguingly, some of the KDM enzymes are inducible by extracellular stimuli (e.g., LPS or inflammatory cytokines and growth factors) exemplified by JmjD3 or JmjD2D, and post-translational modifications or domain arrangements of the enzymes have an impact on enzyme activity as seen with PHF2, indicating that substrate analyses need evaluation in the context of cellular expression and specific growth conditions. It is also becoming increasingly clear, that regulation of Jmj activity through levels (normoxic vs hypoxic) of the cosubstrate O2, or indirectly through HIFα-mediated responses, reactive oxygen species or metabolic intermediates is of importance [Citation154,Citation253,Citation333–334]. Taken together, these research lines will undoubtedly reveal novel facets in regulation of the epigenetic landscape and its impact on human disease. However, to evaluate and harness the potential of Jmj-enzymes as potential drug targets, chemical biology efforts to provide tools for better understanding of biology are mandatory [Citation335]; fortunately recent efforts from several laboratories reveal that this class of enzymes indeed is chemically tractable, expanding the chemical toolbox of epigenetic inhibitors and possibly allowing to modulate disease outcome by targeting Jmj-type oxygenases.
Table 1. Domain organization and substrates of human Jmj-type oxygenases.
Table 2. Involvement of Jmj-type demethylases and other oxygenases in human physiology and disease.
Jumonji proteins were identified as transcriptional regulators in mouse knockout models, and constitute a large, evolutionarily conserved family of iron and 2-oxoglutarate-dependent oxygenase enzymes.
The human 2-oxoglutarate family consists of over 60 members, sharing within the catalytic domains few conserved sequence motifs and a similar structural folding pattern. The enzymatically active members of this class of metalloenzymes mediate hydroxylation of several types of amino acid residues found in chromatin (e.g., methyl-lysine in histones, thereby mediating demthylation reactions), transcription factors, ribosomal and structural proteins, as well as small molecules such as lipids or metabolic intermediates. Other important functions extend to nucleotide hydroxylations, of importance in DNA repair, and in reversal of 5-methylcytosine or N6-methyladenosine modifications, which are critical for epigenetic gene regulation and regulation of RNA functions, respectively.
Acknowledgements
The authors are grateful to Michael McDonough and Chris Schofield (Department of Chemistry, Oxford, UK) and Susanne Muller-Knapp (SGC Oxford, UK) for fruitful discussions and collaborations.
Financial & competing interests disclosure
Research in the authors’ laboratories is funded by the Oxford NIHR BRU, the Rosetrees Foundation, Bayer Healthcare, MRC, BBSRC, ARUK and GlaxoSmithKline. The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from Abbvie, Boehringer Ingelheim, the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Eli Lilly and Company, Genome Canada, GlaxoSmithKline, the Ontario Ministry of Economic Development and Innovation, Janssen, the Novartis Research Foundation, Pfizer, Takeda and the Wellcome Trust. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Additional information
Funding
References
- Loenarz C , SchofieldCJ . Physiological and biochemical aspects of hydroxylations and demethylations catalyzed by human 2-oxoglutarate oxygenases . Trends Biochem. Sci.36 ( 1 ), 7 – 18 ( 2010 ).
- Takeuchi T , YamazakiY , Katoh-FukuiYet al. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation . Genes Dev.9 ( 10 ), 1211 – 1222 ( 1995 ).
- Takeuchi T , WatanabeY , Takano-ShimizuT , KondoS . Roles of jumonji and jumonji family genes in chromatin regulation and development . Dev. Dyn.235 ( 9 ), 2449 – 2459 ( 2006 ).
- Balciunas D , RonneH . Evidence of domain swapping within the jumonji family of transcription factors . Trends Biochem. Sci.25 ( 6 ), 274 – 276 ( 2000 ).
- Clissold PM , PontingCP . Jmj C: cupin metalloenzyme-like domains in jumonji, hairless and phospholipase A2beta . Trends Biochem Sci.26 ( 1 ), 7 – 9 ( 2001 ).
- Ng SS , KavanaghKL , McdonoughMAet al. Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity . Nature448 ( 7149 ), 87 – 91 ( 2007 ).
- Shi Y , LanF , MatsonCet al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1 . Cell119 ( 7 ), 941 – 953 ( 2004 ).
- Mcdonough MA , KavanaghKL , ButlerD , SearlsT , OppermannU , SchofieldCJ . Structure of human phytanoyl-CoA 2-hydroxylase identifies molecular mechanisms of Refsum disease . J. Biol. Chem.280 ( 49 ), 41101 – 41110 ( 2005 ).
- Leung IK , KrojerTJ , KochanGTet al. Structural and mechanistic studies on gamma-butyrobetaine hydroxylase . Chem. Biol17 ( 12 ), 1316 – 1324 ( 2010 ).
- Hautala T , ByersMG , EddyRL , ShowsTB , KivirikkoKI , MyllylaR . Cloning of human lysyl hydroxylase: complete cDNA-derived amino acid sequence and assignment of the gene (PLOD) to chromosome 1p36.3—-p36.2 . Genomics13 ( 1 ), 62 – 69 ( 1992 ).
- Clifton IJ , McdonoughMA , EhrismannD , KershawNJ , GranatinoN , SchofieldCJ . Structural studies on 2-oxoglutarate oxygenases and related double-stranded beta-helix fold proteins . J. Inorganic Biochem.100 ( 4 ), 644 – 669 ( 2006 ).
- Klose RJ , KallinEM , ZhangY . JmjC-domain-containing proteins and histone demethylation . Nat. Rev. Genet.7 ( 9 ), 715 – 727 ( 2006 ).
- Mcdonough MA , LoenarzC , ChowdhuryR , CliftonIJ , SchofieldCJ . Structural studies on human 2-oxoglutarate dependent oxygenases . Curr. Opin Struct. Biol.20 ( 6 ), 659 – 672 ( 2010 ).
- Han Z , NiuT , ChangJet al. Crystal structure of the FTO protein reveals basis for its substrate specificity . Nature464 ( 7292 ), 1205 – 1209 ( 2010 ).
- Tsukada Y , FangJ , Erdjument-BromageHet al. Histone demethylation by a family of JmjC domain-containing proteins . Nature439 ( 7078 ), 811 – 816 ( 2006 ).
- Frescas D , GuardavaccaroD , BassermannF , Koyama-NasuR , PaganoM . JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes . Nature450 ( 7167 ), 309 – 313 ( 2007 ).
- Lu T , JacksonMw , WangBet al. Regulation of NF-kappaB by NSD1/FBXL11-dependent reversible lysine methylation of p65 . Proc. Natl Acad. Sci. USA107 ( 1 ), 46 – 51 ( 2010 ).
- Yamane K , ToumazouC , TsukadaYet al. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor . Cell125 ( 3 ), 483 – 495 ( 2006 ).
- Brauchle M , YaoZ , AroraRet al. Protein Complex Interactor Analysis and Differential Activity of KDM3 Subfamily Members Towards H3K9 Methylation . PLoS ONE8 ( 4 ), e60549 ( 2013 ).
- Hillringhaus L , YueWW , RoseNRet al. Structural and evolutionary basis for the dual substrate selectivity of human KDM4 histone demethylase family . J. Biol. Chem.286 ( 48 ), 41616 – 41625 ( 2011 ).
- Ponnaluri VK , VavilalaDT , PuttyS , GutheilWG , MukherjiM . Identification of non-histone substrates for JMJD2A-C histone demethylases . Biochem. Biophys. Res. Commun.390 ( 2 ), 280 – 284 ( 2009 ).
- Trojer P , ZhangJ , YonezawaMet al. Dynamic histone H1 isotype 4 methylation and demethylation by histone lysine methyltransferase G9a/KMT1C and the Jumonji domain-containing JMJD2/KDM4 proteins . J Biol Chem284 ( 13 ), 8395 – 8405 ( 2009 ).
- Christensen J , AggerK , CloosPaet al. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3 . Cell128 ( 6 ), 1063 – 1076 ( 2007 ).
- Iwase S , LanF , BaylissPet al. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases . Cell128 ( 6 ), 1077 – 1088 ( 2007 ).
- Klose RJ , YanQ , TothovaZet al. The retinoblastoma binding protein RBP2 is an H3K4 demethylase . Cell128 ( 5 ), 889 – 900 ( 2007 ).
- Lee MG , NormanJ , ShilatifardA , ShiekhattarR . Physical and functional association of a trimethyl H3K4 demethylase and Ring6a/MBLR, a polycomb-like protein . Cell128 ( 5 ), 877 – 887 ( 2007 ).
- Seward DJ , CubberleyG , KimSet al. Demethylation of trimethylated histone H3 Lys4 in vivo by JARID1 JmjC proteins . Nat. Struct. Mol. Biol.14 ( 3 ), 240 – 242 ( 2007 ).
- De Santa F , TotaroMG , ProsperiniE , NotarbartoloS , TestaG , NatoliG . The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing . Cell130 ( 6 ), 1083 – 1094 ( 2007 ).
- Hong S , ChoYW , YuLR , YuH , VeenstraTD , GeK . Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases . Proc. Natl Acad. Sci. USA104 ( 47 ), 18439 – 18444 ( 2007 ).
- Lan F , BaylissPE , RinnJLet al. A histone H3 lysine 27 demethylase regulates animal posterior development . Nature449 ( 7163 ), 689 – 694 ( 2007 ).
- Lee MG , VillaR , TrojerPet al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination . Science318 ( 5849 ), 447 – 450 ( 2007 ).
- Xiang Y , ZhuZ , HanG , LinH , XuL , ChenCD . JMJD3 is a histone H3K27 demethylase . Cell Res.17 ( 10 ), 850 – 857 ( 2007 ).
- Wen H , LiJ , SongTet al. Recognition of histone H3K4 trimethylation by the plant homeodomain of PHF2 modulates histone demethylation . J. Biol. Chem.285 ( 13 ), 9322 – 9326 ( 2010 ).
- Horton JR , UpadhyayAK , QiHH , ZhangX , ShiY , ChengX . Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases . Nat. Struct. Mol. Biol.17 ( 1 ), 38 – 43 ( 2010 ).
- Zhu Z , WangY , LiXet al. PHF8 is a histone H3K9me2 demethylase regulating rRNA synthesis . Cell Res.20 ( 7 ), 794 – 801 ( 2010 ).
- Qi HH , SarkissianM , HuGGet al. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development . Nature466 ( 7305 ), 503 – 507 ( 2010 ).
- Tsukada Y , IshitaniT , NakayamaKI . KDM7 is a dual demethylase for histone H3 Lys 9 and Lys 27 and functions in brain development . Genes Dev.24 ( 5 ), 432 – 437 ( 2010 ).
- Yue Ww , HozjanV , GeWet al. Crystal structure of the PHF8 Jumonji domain, an Nepsilon-methyl lysine demethylase . FEBS Lett.584 ( 4 ), 825 – 830 ( 2010 ).
- Ge W , WolfA , FengTet al. Oxygenase-catalyzed ribosome hydroxylation occurs in prokaryotes and humans . Nat. Chem. Biol.8 ( 12 ), 960 – 962 ( 2012 ).
- Sinha KM , YasudaH , CoombesMM , DentSY , DeCrombrugghe B . Regulation of the osteoblast-specific transcription factor Osterix by NO66, a Jumonji family histone demethylase . EMBO J.29 ( 1 ), 68 – 79 ( 2010 ).
- Lu Y , BeezholdK , ChangQet al. Lung cancer-associated JmjC domain protein mdig suppresses formation of tri-methyl lysine 9 of histone H3 . Cell Cycle8 ( 13 ), 2101 – 2109 ( 2009 ).
- Hsia DA , TepperCG , PochampalliMRet al. KDM8, a H3K36me2 histone demethylase that acts in the cyclin A1 coding region to regulate cancer cell proliferation . Proc. Natl Acad. Sci. USA107 ( 21 ), 9671 – 9676 ( 2010 ).
- Youn M-Y , YokoyamaA , Fujiyama-NakamuraSet al. JMJD5, a Jumonji C (JmjC) domain-containing protein, negatively regulates osteoclastogenesis by facilitating NFATc1 protein degradation . J. Biol. Chem.287 ( 16 ), 12994 – 13004 ( 2012 ).
- Chang B , ChenY , ZhaoY , BruickRK . JMJD6 is a histone arginine demethylase . Science318 ( 5849 ), 444 – 447 ( 2007 ).
- Webby Cj , WolfA , GromakNet al. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing . Science325 ( 5936 ), 90 – 93 ( 2009 ).
- Mantri M , LoikND , HamedRB , ClaridgeTDW , MccullaghJSO , SchofieldCJ . The 2-oxoglutarate-dependent oxygenase JMJD6 catalyses oxidation of lysine residues to give 5S-hydroxylysine residues . ChemBioChem12 ( 4 ), 531 – 534 ( 2011 ).
- Unoki M , MasudaA , DohmaeNet al. Lysyl 5-hydroxylation, a novel hstone modification, by Jumonji domain containing 6 (JMJD6) . J. Biol. Chem.288 ( 9 ), 6053 – 6062 ( 2013 ).
- Yang M , ChowdhuryR , GeWet al. Factor-inhibiting hypoxia-inducible factor (FIH) catalyses the post-translational hydroxylation of histidinyl residues within ankyrin repeat domains . FEBS J.278 ( 7 ), 1086 – 1097 ( 2011 ).
- Yang M , GeW , ChowdhuryRet al. Asparagine and aspartate hydroxylation of the cytoskeletal ankyrin family is catalyzed by factor-inhibiting hypoxia-inducible factor . J. Biol. Chem.286 ( 9 ), 7648 – 7660 ( 2011 ).
- Cockman ME , LancasterDE , StolzeIPet al. Posttranslational hydroxylation of ankyrin repeats in IkappaB proteins by the hypoxia-inducible factor (HIF) asparaginyl hydroxylase, factor inhibiting HIF (FIH) . Proc. Natl Acad. Sci. USA103 ( 40 ), 14767 – 14772 ( 2006 ).
- Lando D , PeetDJ , GormanJJ , WhelanDA , WhitelawML , BruickRK . FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor . Genes Dev.16 ( 12 ), 1466 – 1471 ( 2002 ).
- Noma A , IshitaniR , KatoM , NagaoA , NurekiO , SuzukiT . Expanding role of the jumonji C domain as an RNA hydroxylase . J. Biol. Chem.285 ( 45 ), 34503 – 34507 ( 2010 ).
- Sedgwick B . Repairing DNA-methylation damage . Nat. Rev. Mol. Cell Biol.5 ( 2 ), 148 – 157 ( 2004 ).
- Duncan T , TrewickSC , KoivistoP , BatesPA , LindahlT , SedgwickB . Reversal of DNA alkylation damage by two human dioxygenases . Proc. Natl Acad. Sci. USA99 ( 26 ), 16660 – 16665 ( 2002 ).
- Li Mm , NilsenA , ShiYet al. ALKBH4-dependent demethylation of actin regulates actomyosin dynamics . Nat. Commun.4 , 1832 ( 2013 ).
- Zheng G , DahlJA , NiuYet al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility . Mol. Cell49 ( 1 ), 18 – 29 ( 2012 ).
- Fu D , BrophyJA , ChanCTet al. Human AlkB homolog ABH8 Is a tRNA methyltransferase required for wobble uridine modification and DNA damage survival . Mol. Cell Biol.30 ( 10 ), 2449 – 2459 ( 2010 ).
- Tahiliani M , KohKP , ShenYet al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1 . Science324 ( 5929 ), 930 – 935 ( 2009 ).
- Ito S , ShenL , DaiQet al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine . Science333 ( 6047 ), 1300 – 1303 ( 2011 ).
- Gerken T , GirardCA , TungYCet al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase . Science318 ( 5855 ), 1469 – 1472 ( 2007 ).
- Jia G , YangCg , YangSet al. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO . FEBS Lett.582 ( 23–24 ), 3313 – 3319 ( 2008 ).
- Landeira D , SauerS , PootRet al. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators . Nat. Cell. Biol.12 ( 6 ), 618 – 624 ( 2010 ).
- Pasini D , CloosPA , WalfridssonJet al. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells . Nature464 ( 7286 ), 306 – 310 ( 2010 ).
- Peng JC , ValouevA , SwigutTet al. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells . Cell139 ( 7 ), 1290 – 1302 ( 2009 ).
- Shen X , KimW , FujiwaraYet al. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells . Cell139 ( 7 ), 1303 – 1314 ( 2009 ).
- Lewis EB . A gene complex controlling segmentation in Drosophila . Nature276 ( 5688 ), 565 – 570 ( 1978 ).
- Struhl G . A gene product required for correct initiation of segmental determination in Drosophila . Nature293 ( 5827 ), 36 – 41 ( 1981 ).
- Bracken AP , HelinK . Polycomb group proteins: navigators of lineage pathways led astray in cancer . Nat. Rev. Cancer9 ( 11 ), 773 – 784 ( 2009 ).
- Metzger E , WissmannM , YinNet al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription . Nature437 ( 7057 ), 436 – 439 ( 2005 ).
- Allis CD , BergerSL , CoteJet al. New nomenclature for chromatin-modifying enzymes . Cell131 ( 4 ), 633 – 636 ( 2007 ).
- Bernstein BE , BirneyE , DunhamI , GreenED , GunterC , SnyderM . An integrated encyclopedia of DNA elements in the human genome . Nature489 ( 7414 ), 57 – 74 ( 2012 ).
- Ng SS , YueWW , OppermannU , KloseRJ . Dynamic protein methylation in chromatin biology . Cell. Mol. Life Sci.66 ( 3 ), 407 – 422 ( 2009 ).
- Kooistra SM , HelinK . Molecular mechanisms and potential functions of histone demethylases . Nat. Rev. Mol. Cell Biol.13 ( 5 ), 297 – 311 ( 2012 ).
- Pedersen MT , HelinK . Histone demethylases in development and disease . Trends Cell Biol.20 ( 11 ), 662 – 671 ( 2010 ).
- Dawson MA , KouzaridesT . Cancer epigenetics: from mechanism to therapy . Cell150 ( 1 ), 12 – 27 ( 2012 ).
- Greer El , ShiY . Histone methylation: a dynamic mark in health, disease and inheritance . Nat. Rev. Genet.13 ( 5 ), 343 – 357 ( 2012 ).
- Mosammaparast N , ShiY . Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases . Annu. Rev. Biochem.79 , 155 – 179 ( 2010 ).
- Ciccone DN , SuH , HeviSet al. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints . Nature461 ( 7262 ), 415 – 418 ( 2009 ).
- Karytinos A , FornerisF , ProfumoAet al. A novel mammalian flavin-dependent histone demethylase . J. Biol. Chem.284 ( 26 ), 17775 – 17782 ( 2009 ).
- Wang J , HeviS , KurashJKet al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation . Nat. Genet.41 ( 1 ), 125 – 129 ( 2009 ).
- Kerenyi Ma , ShaoZ , HsuYJet al. Histone demethylase Lsd1 represses hematopoietic stem and progenitor cell signatures during blood cell maturation . Elife2 , e00633 ( 2013 ).
- Sprussel A , SchulteJH , WeberSet al. Lysine-specific demethylase 1 restricts hematopoietic progenitor proliferation and is essential for terminal differentiation . Leukemia26 ( 9 ), 2039 – 2051 ( 2012 ).
- Schenk T , ChenWc , GollnerSet al. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia . Nat Med18 ( 4 ), 605 – 611 ( 2012 ).
- Wang J , LuF , RenQet al. Novel histone demethylase LSD1 inhibitors selectively target cancer cells with pluripotent stem cell properties . Cancer Res71 ( 23 ), 7238 – 7249 ( 2011 ).
- Tsai Mc , ManorO , WanYet al. Long noncoding RNA as modular scaffold of histone modification complexes . Science329 ( 5992 ), 689 – 693 ( 2010 ).
- Wang Y , ZhangH , ChenYet al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer . Cell138 ( 4 ), 660 – 672 ( 2009 ).
- Garcia-Bassets I , KwonYs , TeleseFet al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors . Cell128 ( 3 ), 505 – 518 ( 2007 ).
- Huang J , SenguptaR , EspejoABet al. p53 is regulated by the lysine demethylase LSD1 . Nature449 ( 7158 ), 105 – 108 ( 2007 ).
- Deaton Am , BirdA . CpG islands and the regulation of transcription . Genes Dev25 ( 10 ), 1010 – 1022 ( 2011 ).
- Blackledge NP , ZhouJC , TolstorukovMY , FarcasAM , ParkPJ , KloseRJ . CpG islands recruit a histone H3 lysine 36 demethylase . Molecular cell38 ( 2 ), 179 – 190 ( 2010 ).
- Farcas Am , BlackledgeNp , SudberyIet al. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands . Elife1 , e00205 ( 2012 ).
- He J , ShenL , WanM , TaranovaO , WuH , ZhangY . Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes . Nat. Cell Biol.15 ( 4 ), 373 – 384 ( 2013 ).
- Wu X , JohansenJV , HelinK . Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation . Mol. Cell49 ( 6 ), 1134 – 1146 ( 2013 ).
- Lagarou A , Mohd-SaripA , MoshkinYMet al. dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing . Genes Dev.22 ( 20 ), 2799 – 2810 ( 2008 ).
- Bartke T , VermeulenM , XhemalceB , RobsonSC , MannM , KouzaridesT . Nucleosome-interacting proteins regulated by DNA and histone methylation . Cell143 ( 3 ), 470 – 484 ( 2010 ).
- Tanaka Y , OkamotoK , TeyeKet al. JmjC enzyme KDM2A is a regulator of rRNA transcription in response to starvation . EMBO J.29 ( 9 ), 1510 – 1522 ( 2010 ).
- Barrero MJ , IzpisuaBelmonte JC . Polycomb complex recruitment in pluripotent stem cells . Nat. Cell Biol.15 ( 4 ), 348 – 350 ( 2013 ).
- Du J , MaY , MaP , WangS , FanZ . Demethylation of epiregulin gene by histone demethylase FBXL11 and BCL6 corepressor inhibits osteo/dentinogenic differentiation . Stem Cells31 ( 1 ), 126 – 136 ( 2013 ).
- Gao R , DongR , DuJ , MaP , WangS , FanZ . Depletion of histone demethylase KDM2A inhibited cell proliferation of stem cells from apical papilla by de-repression of p15(INK4B) and p27 (Kip1.) . Mol. Cellular Biochemistry379 ( 1–2 ), 115 – 122 ( 2013 ).
- Iuchi S , GreenH . Lysine-specific demethylase 2A (KDM2A) normalizes human embryonic stem cell derived keratinocytes . Proc. Natl Acad. Sci. USA109 ( 24 ), 9442 – 9447 ( 2012 ).
- Liang G , HeJ , ZhangY . Kdm2b promotes induced pluripotent stem cell generation by facilitating gene activation early in reprogramming . Nat. Cell. Biol.14 ( 5 ), 457 – 466 ( 2012 ).
- Wang T , ChenK , ZengXet al. The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner . Cell Stem Cell9 ( 6 ), 575 – 587 ( 2011 ).
- Gearhart MD , CorcoranCM , WamstadJA , BardwellVJ . Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets . Mol. Cell Biol.26 ( 18 ), 6880 – 6889 ( 2006 ).
- He J , KallinEM , TsukadaY , ZhangY . The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b) . Nat. Struct. Mol. Biol.15 ( 11 ), 1169 – 1175 ( 2008 ).
- Tzatsos A , PfauR , KampranisSC , TsichlisPN . Ndy1/KDM2B immortalizes mouse embryonic fibroblasts by repressing the Ink4a/Arf locus . Proc. Natl Acad. Sci. USA106 ( 8 ), 2641 – 2646 ( 2009 ).
- He J , NguyenAt , ZhangY . KDM2b/JHDM1b, an H3K36me2-specific demethylase, is required for initiation and maintenance of acute myeloid leukemia . Blood117 ( 14 ), 3869 – 3880 ( 2011 ).
- Kottakis F , PolytarchouC , FoltopoulouP , SanidasI , KampranisSC , TsichlisPN . FGF-2 regulates cell proliferation, migration, and angiogenesis through an NDY1/KDM2B-miR-101-EZH2 pathway . Mol. Cell43 ( 2 ), 285 – 298 ( 2011 ).
- Tzatsos A , PaskalevaP , FerrariFet al. KDM2B promotes pancreatic cancer via Polycomb-dependent and -independent transcriptional programs . J. Clin. Invest.123 ( 2 ), 727 – 739 ( 2013 ).
- Chen H , GiriNC , ZhangRet al. Nickel ions inhibit histone demethylase JMJD1A and DNA repair enzyme ABH2 by replacing the ferrous iron in the catalytic centers . J. Biol. Chem.285 ( 10 ), 7374 – 7383 ( 2010 ).
- Hickok JR , VasudevanD , AntholineWE , ThomasDD . Nitric oxide modifies global histone methylation by Inhibiting Jumonji C domain-containing demethylases . J. Biol. Chem.288 ( 22 ), 16004 – 16015 ( 2013 ).
- Yang F , StenoienDL , StrittmatterEFet al. Phosphoproteome profiling of human skin fibroblast cells in response to low- and high-dose irradiation . J. Proteome Res.5 ( 5 ), 1252 – 1260 ( 2006 ).
- Liu Z , ZhouS , LiaoL , ChenX , MeistrichM , XuJ . Jmjd1a demethylase-regulated histone modification is essential for cAMP-response element modulator-regulated gene expression and spermatogenesis . J. Biol. Chem.285 ( 4 ), 2758 – 2770 ( 2010 ).
- Okada Y , ScottG , RayMK , MishinaY , ZhangY . Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis . Nature450 ( 7166 ), 119 – 123 ( 2007 ).
- Inagaki T , TachibanaM , MagooriKet al. Obesity and metabolic syndrome in histone demethylase JHDM2a-deficient mice . Genes Cells14 ( 8 ), 991 – 1001 ( 2009 ).
- Tateishi K , OkadaY , KallinEm , ZhangY . Role of Jhdm2a in regulating metabolic gene expression and obesity resistance . Nature458 ( 7239 ), 757 – 761 ( 2009 ).
- Wellmann S , BettkoberM , ZelmerAet al. Hypoxia upregulates the histone demethylase JMJD1A via HIF-1 . Biochem. Biophys. Res. Commun.372 ( 4 ), 892 – 897 ( 2008 ).
- Osawa T , TsuchidaR , MuramatsuMet al. Inhibition of histone demethylase JMJD1A improves anti-angiogenic therapy and reduces tumor-associated macrophages . Cancer Res.73 ( 10 ), 3019 – 3028 ( 2013 ).
- Yamada D , KobayashiS , YamamotoHet al. Role of the hypoxia-related gene, JMJD1A, in hepatocellular carcinoma: clinical impact on recurrence after hepatic resection . Ann. Surg. Oncol.19 ( Suppl. 3 ), S355 – 364 ( 2012 ).
- Qi J , NakayamaK , CardiffRdet al. Siah2-dependent concerted activity of HIF and FoxA2 regulates formation of neuroendocrine phenotype and neuroendocrine prostate tumors . Cancer Cell18 ( 1 ), 23 – 38 ( 2010 ).
- Uemura M , YamamotoH , TakemasaIet al. Jumonji domain containing 1A is a novel prognostic marker for colorectal cancer: in vivo identification from hypoxic tumor cells . Clin. Cancer Res.16 ( 18 ), 4636 – 4646 ( 2010 ).
- Guo X , ShiM , SunLet al. The expression of histone demethylase JMJD1A in renal cell carcinoma . Neoplasma58 ( 2 ), 153 – 157 ( 2011 ).
- Mackinnon RN , KannourakisG , WallM , CampbellLJ . A cryptic deletion in 5q31.2 provides further evidence for a minimally deleted region in myelodysplastic syndromes . Cancer Genet.204 ( 4 ), 187 – 194 ( 2011 ).
- Tang MH , VaradanV , KamalakaranS , ZhangMG , DimitrovaN , HicksJ . Major chromosomal breakpoint intervals in breast cancer co-localize with differentially methylated regions . Front. Oncol.2 , 197 ( 2012 ).
- Hu Z , GomesI , HorriganSKet al. A novel nuclear protein, 5qNCA (LOC51780) is a candidate for the myeloid leukemia tumor suppressor gene on chromosome 5 band q31 . Oncogene20 ( 47 ), 6946 – 6954 ( 2001 ).
- Kim JY , KimKB , EomGHet al. KDM3B is the H3K9 demethylase involved in transcriptional activation of lmo2 in leukemia . Mol. Cell Biol.32 ( 14 ), 2917 – 2933 ( 2012 ).
- Lee JW , ChoiHS , GyurisJ , BrentR , MooreDD . Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor . Mol. Endocrinol.9 ( 2 ), 243 – 254 ( 1995 ).
- Katoh M . Identification and characterization of TRIP8 gene in silico . Int. J. Mol. Med.12 ( 5 ), 817 – 821 ( 2003 ).
- Katoh M . Comparative integromics on JMJD1C gene encoding histone demethylase: conserved POU5F1 binding site elucidating mechanism of JMJD1C expression in undifferentiated ES cells and diffuse-type gastric cancer . Int. J. Oncol.31 ( 1 ), 219 – 223 ( 2007 ).
- Wolf SS , PatchevVK , ObendorfM . A novel variant of the putative demethylase gene, s-JMJD1C, is a coactivator of the AR . Arch. Biochem. Biophys.460 ( 1 ), 56 – 66 ( 2007 ).
- Kim SM , KimJY , ChoeNWet al. Regulation of mouse steroidogenesis by WHISTLE and JMJD1C through histone methylation balance . Nucleic Acids Res.38 ( 19 ), 6389 – 6403 ( 2010 ).
- Cachon-Gonzalez MB , FennerS , CoffinJM , MoranC , BestS , StoyeJP . Structure and expression of the hairless gene of mice . Proc. Natl Acad. Sci. USA91 ( 16 ), 7717 – 7721 ( 1994 ).
- Thompson CC . Thyroid hormone-responsive genes in developing cerebellum include a novel synaptotagmin and a hairless homolog . J. Neurosci.16 ( 24 ), 7832 – 7840 ( 1996 ).
- Hsieh JC , SiskJM , JurutkaPWet al. Physical and functional interaction between the vitamin D receptor and hairless corepressor, two proteins required for hair cycling . J. Biol. Chem.278 ( 40 ), 38665 – 38674 ( 2003 ).
- Moraitis AN , GiguereV , ThompsonCC . Novel mechanism of nuclear receptor corepressor interaction dictated by activation function 2 helix determinants . Mol. Cell Biol.22 ( 19 ), 6831 – 6841 ( 2002 ).
- Potter GB , BeaudoinGM3rd , DerenzoCL , ZarachJM , ChenSH , ThompsonCC . The hairless gene mutated in congenital hair loss disorders encodes a novel nuclear receptor corepressor . Genes Dev.15 ( 20 ), 2687 – 2701 ( 2001 ).
- Mi Y , ZhangY , ShenYf . Mechanism of JmjC-containing protein Hairless in the regulation of vitamin D receptor function . Biochim. Biophys. Acta1812 ( 12 ), 1675 – 1680 ( 2011 ).
- Potter GB , ZarachJM , SiskJM , ThompsonCC . The thyroid hormone-regulated corepressor hairless associates with histone deacetylases in neonatal rat brain . Mol. Endocrinol.16 ( 11 ), 2547 – 2560 ( 2002 ).
- Ahmad W , Faiyaz UL , Haque M , BrancoliniVet al. Alopecia universalis associated with a mutation in the human hairless gene . Science279 ( 5351 ), 720 – 724 ( 1998 ).
- Katoh Y , KatohM . Comparative integromics on JMJD2A, JMJD2B and JMJD2C: preferential expression of JMJD2C in undifferentiated ES cells . Int. J. Mol. Med.20 ( 2 ), 269 – 273 ( 2007 ).
- Berry WL , JanknechtR . KDM4/JMJD2 histone demethylases: epigenetic regulators in cancer cells . Cancer Res.73 ( 10 ), 2936 – 2942 ( 2013 ).
- Crea F , SunL , MaiAet al. The emerging role of histone lysine demethylases in prostate cancer . Mol. Cancer11 , 52 ( 2012 ).
- Cloos PA , ChristensenJ , AggerKet al. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3 . Nature442 ( 7100 ), 307 – 311 ( 2006 ).
- Shin S , JanknechtR . Activation of androgen receptor by histone demethylases JMJD2A and JMJD2D . Biochem. Biophys. Res. Commun.359 ( 3 ), 742 – 746 ( 2007 ).
- Wissmann M , YinN , MullerJmet al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression . Nat. Cell. Biol.9 ( 3 ), 347 – 353 ( 2007 ).
- Suikki He , KujalaPm , TammelaTl , VanWeerden Wm , VessellaRl , VisakorpiT . Genetic alterations and changes in expression of histone demethylases in prostate cancer . Prostate70 ( 8 ), 889 – 898 ( 2010 ).
- Coffey K , RogersonL , Ryan-MundenCet al. The lysine demethylase, KDM4B, is a key molecule in androgen receptor signalling and turnover . Nucleic Acids Res.41 ( 8 ), 4433 – 4446 ( 2013 ).
- Gaughan L , StockleyJ , CoffeyKet al. KDM4B is a master regulator of the estrogen receptor signalling cascade . Nucleic Acids Res.41 ( 14 ), 6892 – 6904 ( 2013 ).
- Liu G , Bollig-FischerA , KreikeBet al. Genomic amplification and oncogenic properties of the GASC1 histone demethylase gene in breast cancer . Oncogene28 ( 50 ), 4491 – 4500 ( 2009 ).
- Li BX , ZhangMC , LuoCLet al. Effects of RNA interference-mediated gene silencing of JMJD2A on human breast cancer cell line MDA-MB-231 in vitro . J. Exp. Clin. Cancer Res.30 , 90 ( 2011 ).
- Berry WL , ShinS , LightfootSA , JanknechtR . Oncogenic features of the JMJD2A histone demethylase in breast cancer . Int. J. Oncol.41 ( 5 ), 1701 – 1706 ( 2012 ).
- Yang J , JubbAM , PikeLet al. The histone demethylase JMJD2B is regulated by estrogen receptor alpha and hypoxia, and is a key mediator of estrogen induced growth . Cancer Res.70 ( 16 ), 6456 – 6466 ( 2010 ).
- Kawazu M , SasoK , TongKiet al. Histone demethylase JMJD2B functions as a co-factor of estrogen receptor in breast cancer proliferation and mammary gland development . PLoS ONE6 ( 3 ), e17830 ( 2011 ).
- Shi L , SunL , LiQet al. Histone demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes hormonally responsive breast carcinogenesis . Proc. Natl Acad. Sci. USA108 ( 18 ), 7541 – 7546 ( 2011 ).
- Pollard PJ , LoenarzC , MoleDRet al. Regulation of Jumonji-domain-containing histone demethylases by hypoxia-inducible factor (HIF)-1alpha . Biochem. J.416 ( 3 ), 387 – 394 ( 2008 ).
- Zhu Y , Van EssenD , SaccaniS . Cell-type-specific control of enhancer activity by H3K9 trimethylation . Mol. Cell.46 ( 4 ), 408 – 423 ( 2012 ).
- Strobl-Mazzulla PH , Sauka-SpenglerT , Bronner-FraserM . Histone demethylase JmjD2A regulates neural crest specification . Dev. Cell19 ( 3 ), 460 – 468
- Zhang QJ , ChenHZ , WangL , LiuDP , HillJA , LiuZP . The histone trimethyllysine demethylase JMJD2A promotes cardiac hypertrophy in response to hypertrophic stimuli in mice . J. Clin. Invest.121 ( 6 ), 2447 – 2456 ( 2011 ).
- Ye L , FanZ , YuBet al. Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human MSCs . Cell Stem Cell11 ( 1 ), 50 – 61 ( 2012 ).
- Guo L , LiX , HuangJXet al. Histone demethylase Kdm4b functions as a co-factor of C/EBPbeta to promote mitotic clonal expansion during differentiation of 3T3-L1 preadipocytes . Cell Death Differ.19 ( 12 ), 1917 – 1927 ( 2012 ).
- Chen Wd , FuX , DongBet al. Neonatal activation of the nuclear receptor CAR results in epigenetic memory and permanent change of drug metabolism in mouse liver . Hepatology56 ( 4 ), 1499 – 1509 ( 2012 ).
- Kim TD , OhS , ShinS , JanknechtR . Regulation of tumor suppressor p53 and HCT116 cell physiology by histone demethylase JMJD2D/KDM4D . PLoS ONE7 ( 4 ), e34618 ( 2012 ).
- Wilsker D , ProbstL , WainHM , MaltaisL , TuckerPW , MoranE . Nomenclature of the ARID family of DNA-binding proteins . Genomics86 ( 2 ), 242 – 251 ( 2005 ).
- Wysocka J , SwigutT , XiaoHet al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling . Nature442 ( 7098 ), 86 – 90 ( 2006 ).
- Tahiliani M , MeiP , FangRet al. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation . Nature447 ( 7144 ), 601 – 605 ( 2007 ).
- Defeo-Jones D , HuangPS , JonesREet al. Cloning of cDNAs for cellular proteins that bind to the retinoblastoma gene product . Nature352 ( 6332 ), 251 – 254 ( 1991 ).
- Benevolenskaya EV , MurrayHL , BrantonP , YoungRA , KaelinW G Jr. Binding of pRB to the PHD protein RBP2 promotes cellular differentiation . Mol. Cell18 ( 6 ), 623 – 635 ( 2005 ).
- Chicas A , KapoorA , WangXet al. H3K4 demethylation by Jarid1a and Jarid1b contributes to retinoblastoma-mediated gene silencing during cellular senescence . Proc. Natl Acad. Sci. USA109 ( 23 ), 8971 – 8976 ( 2012 ).
- Ditacchio L , LeHD , VollmersCet al. Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock . Science333 ( 6051 ), 1881 – 1885 ( 2011 ).
- De Rooij JD , HollinkIH , Arentsen-PetersSTet al. NUP98/JARID1A is a novel recurrent abnormality in pediatric acute megakaryoblastic leukemia with a distinct HOX gene expression pattern . Leukemia27 ( 12 ), 2280 – 2288 ( 2013 ).
- Pointon JJ , HarveyD , KaraderiT , AppletonLH , FarrarC , WordsworthBP . The histone demethylase JARID1A is associated with susceptibility to ankylosing spondylitis . Genes Immun.12 ( 5 ), 395 – 398 ( 2011 ).
- Sharma SV , LeeDY , LiBet al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations . Cell141 ( 1 ), 69 – 80 ( 2010 ).
- Cellot S , HopeKJ , ChagraouiJet al. RNAi screen identifies Jarid1b as a major regulator of mouse HSC activity . Blood122 ( 9 ), 1545 – 1555 ( 2013 ).
- Dey BK , StalkerL , SchnerchA , BhatiaM , Taylor-PapidimitriouJ , WynderC . The histone demethylase KDM5b/JARID1b plays a role in cell fate decisions by blocking terminal differentiation . Mol. Cell Biol28 ( 17 ), 5312 – 5327 ( 2008 ).
- Schmitz Su , AlbertM , MalatestaMet al. Jarid1b targets genes regulating development and is involved in neural differentiation . EMBO J.30 ( 22 ), 4586 – 4600 ( 2011 ).
- Li Q , ShiL , GuiBet al. Binding of the JmjC demethylase JARID1B to LSD1/NuRD suppresses angiogenesis and metastasis in breast cancer cells by repressing chemokine CCL14 . Cancer Res.71 ( 21 ), 6899 – 6908 ( 2011 ).
- Kuzbicki L , LangeD , Straczynska-NiemiecA , ChwirotBW . JARID1B expression in human melanoma and benign melanocytic skin lesions . Melanoma Res.23 ( 1 ), 8 – 12 ( 2013 ).
- Lu Pj , SundquistK , BaeckstromDet al. A novel gene (PLU-1) containing highly conserved putative DNA/chromatin binding motifs is specifically up-regulated in breast cancer . J. Biol. Chem.274 ( 22 ), 15633 – 15645 ( 1999 ).
- Roesch A , Fukunaga-KalabisM , SchmidtECet al. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth . Cell141 ( 4 ), 583 – 594 ( 2010 ).
- Wouters J , StasM , GremeauxLet al. The human melanoma side population displays molecular and functional characteristics of enriched chemoresistance and tumorigenesis . PLoS ONE8 ( 10 ), e76550 ( 2013 ).
- Jensen LR , AmendeM , GurokUet al. Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation . Am. J. Hum. Genet.76 ( 2 ), 227 – 236 ( 2005 ).
- Agulnik Ai , MitchellMj , MatteiMget al. A novel X gene with a widely transcribed Y-linked homologue escapes X-inactivation in mouse and human . Hum. Mol. Genet.3 ( 6 ), 879 – 884 ( 1994 ).
- Adegbola A , GaoH , SommerS , BrowningM . A novel mutation in JARID1C/SMCX in a patient with autism spectrum disorder (ASD) . Am. J. Med. Genet. Part A146A ( 4 ), 505 – 511 ( 2008 ).
- Niu X , ZhangT , LiaoLet al. The von Hippel-Lindau tumor suppressor protein regulates gene expression and tumor growth through histone demethylase JARID1C . Oncogene31 ( 6 ), 776 – 786 ( 2012 ).
- Agulnik Ai , MitchellMj , LernerJl , WoodsDr , BishopCe . A mouse Y chromosome gene encoded by a region essential for spermatogenesis and expression of male-specific minor histocompatibility antigens . Hum. Mol. Genet.3 ( 6 ), 873 – 878 ( 1994 ).
- Scott DM , EhrmannIE , EllisPSet al. Identification of a mouse male-specific transplantation antigen, H-Y . Nature376 ( 6542 ), 695 – 698 ( 1995 ).
- Wang W , MeadowsLR , DenHaan JMet al. Human H-Y: a male-specific histocompatibility antigen derived from the SMCY protein . Science269 ( 5230 ), 1588 – 1590 ( 1995 ).
- Akimoto C , KitagawaH , MatsumotoT , KatoS . Spermatogenesis-specific association of SMCY and MSH5 . Genes Cells13 ( 6 ), 623 – 633 ( 2008 ).
- Perinchery G , SasakiM , AnganA , KumarV , CarrollP , DahiyaR . Deletion of Y-chromosome specific genes in human prostate cancer . J. Urol.163 ( 4 ), 1339 – 1342 ( 2000 ).
- Kruidenier L , ChungCW , ChengZet al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response . Nature488 ( 7411 ), 404 – 408 ( 2012 ).
- Sola S , XavierJM , SantosDMet al. p53 interaction with JMJD3 results in its nuclear distribution during mouse neural stem cell differentiation . PLoS ONE6 ( 3 ), e18421 ( 2011 ).
- Miller SA , MohnSE , WeinmannAS . Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression . Mol. Cell40 ( 4 ), 594 – 605 ( 2010 ).
- Miller SA , WeinmannAS . Molecular mechanisms by which T-bet regulates T-helper cell commitment . Immunol. Rev.238 ( 1 ), 233 – 246 ( 2010 ).
- Bernstein BE , MikkelsenTS , XieXet al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells . Cell125 ( 2 ), 315 – 326 ( 2006 ).
- Awe JP , ByrneJA . Identifying candidate oocyte reprogramming factors using cross-species global transcriptional analysis . Cell Reprogram.15 ( 2 ), 126 – 133 ( 2013 ).
- Canovas S , CibelliJB , RossPJ . Jumonji domain-containing protein 3 regulates histone 3 lysine 27 methylation during bovine preimplantation development . Proc. Natl Acad. Sci. USA109 ( 7 ), 2400 – 2405 ( 2012 ).
- Jiang W , WangJ , ZhangY . Histone H3K27me3 demethylases KDM6A and KDM6B modulate definitive endoderm differentiation from human ESCs by regulating WNT signaling pathway . Cell Res.23 ( 1 ), 122 – 130 ( 2013 ).
- Kim SW , YoonSJ , ChuongEet al. Chromatin and transcriptional signatures for Nodal signaling during endoderm formation in hESCs . Dev. Biol.357 ( 2 ), 492 – 504 ( 2011 ).
- Agger K , CloosPA , ChristensenJet al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development . Nature449 ( 7163 ), 731 – 734 ( 2007 ).
- Shahhoseini M , TaghizadehZ , HatamiM , BaharvandH . Retinoic acid dependent histone 3 demethylation of the clustered HOX genes during neural differentiation of human embryonic stem cells . Biochem. Cell Biol.91 ( 2 ), 116 – 122 ( 2013 ).
- Zhao W , LiQ , AyersSet al. Jmjd3 inhibits reprogramming by upregulating expression of INK4a/Arf and targeting PHF20 for ubiquitination . Cell152 ( 5 ), 1037 – 1050 ( 2013 ).
- Yasui T , HiroseJ , TsutsumiS , NakamuraK , AburataniH , TanakaS . Epigenetic regulation of osteoclast differentiation: possible involvement of Jmjd3 in the histone demethylation of Nfatc1 . J. Bone Miner. Res.26 ( 11 ), 2665 – 2671 ( 2011 ).
- Burgold T , VoituronN , CaganovaMet al. The H3K27 demethylase JMJD3 is required for maintenance of the embryonic respiratory neuronal network, neonatal breathing, and survival . Cell Rep.2 ( 5 ), 1244 – 1258 ( 2012 ).
- Jepsen K , SolumD , ZhouTet al. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron . Nature450 ( 7168 ), 415 – 419 ( 2007 ).
- Estaras C , AkizuN , GarciaA , BeltranS , De La CruzX , Martinez-BalbasMA . Genome-wide analysis reveals that Smad3 and JMJD3 HDM co-activate the neural developmental program . Development139 ( 15 ), 2681 – 2691 ( 2012 ).
- Fei T , XiaK , LiZet al. Genome-wide mapping of SMAD target genes reveals the role of BMP signaling in embryonic stem cell fate determination . Genome Res.20 ( 1 ), 36 – 44 ( 2010 ).
- Dai JP , LuJY , ZhangY , ShenYF . Jmjd3 activates Mash1 gene in RA-induced neuronal differentiation of P19 cells . J. Cell Biochem.110 ( 6 ), 1457 – 1463 ( 2010 ).
- Giovarelli M , BucciG , PaseroM , GherziR , BriataP . KSRP silencing favors neural differentiation of P19 teratocarcinoma cells . Biochim. Biophys. Acta1829 ( 5 ), 469 – 479 ( 2013 ).
- Lederer D , GrisartB , DigilioMCet al. Deletion of KDM6A, a histone demethylase interacting with MLL2, in three patients with Kabuki syndrome . Am. J. Human Genet.90 ( 1 ), 119 – 124 ( 2011 ).
- Priolo M , MicaleL , AugelloBet al. Absence of deletion and duplication of MLL2 and KDM6A genes in a large cohort of patients with Kabuki syndrome . Mol. Genet. Metab.107 ( 3 ), 627 – 629 ( 2012 ).
- Das ND , JungKH , ChoiMR , YoonHS , KimSH , ChaiYG . Gene networking and inflammatory pathway analysis in a JMJD3 knockdown human monocytic cell line . Cell Biochem. Funct.30 ( 3 ), 224 – 232 ( 2012 ).
- Wei Y , ChenR , DimicoliSet al. Global H3K4me3 genome mapping reveals alterations of innate immunity signaling and overexpression of JMJD3 in human myelodysplastic syndrome CD34+ cells . Leukemia27 ( 11 ), 2177 – 2186 ( 2013 ).
- De Santa F , NarangV , YapZHet al. Jmjd3 contributes to the control of gene expression in LPS-activated macrophages . EMBO J.28 ( 21 ), 3341 – 3352 ( 2009 ).
- Ishii M , WenH , CorsaCAet al. Epigenetic regulation of the alternatively activated macrophage phenotype . Blood114 ( 15 ), 3244 – 3254 ( 2009 ).
- Satoh T , TakeuchiO , VandenbonAet al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection . Nat. Immunol.11 ( 10 ), 936 – 944 ( 2010 ).
- Zhang Q , LongH , LiaoJet al. Inhibited expression of hematopoietic progenitor kinase 1 associated with loss of jumonji domain containing 3 promoter binding contributes to autoimmunity in systemic lupus erythematosus . J. Autoimmun.37 ( 3 ), 180 – 189 ( 2011 ).
- Sen Gl , WebsterDE , BarraganDI , ChangHY , KhavariPA . Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3 . Genes Dev.22 ( 14 ), 1865 – 1870 ( 2008 ).
- Shaw T , MartinP . Epigenetic reprogramming during wound healing. loss of polycomb-mediated silencing may enable upregulation of repair genes . EMBO Rep.10 ( 8 ), 881 – 886 ( 2009 ).
- Ciavatta DJ , YangJ , PrestonGAet al. Epigenetic basis for aberrant upregulation of autoantigen genes in humans with ANCA vasculitis . J. Clin. Invest.120 ( 9 ), 3209 – 3219 ( 2010 ).
- Dumon S , WaltonDS , VolpeGet al. Itga2b regulation at the onset of definitive hematopoiesis and commitment to differentiation . PLoS ONE7 ( 8 ), e43300 ( 2012 ).
- Anderton JA , BoseS , VockerodtMet al. The H3K27me3 demethylase, KDM6B, is induced by Epstein-Barr virus and over-expressed in Hodgkin’s Lymphoma . Oncogene30 ( 17 ), 2037 – 2043 ( 2011 ).
- Mclaughlin-Drubin ME , CrumCP , MungerK . Human papillomavirus E7 oncoprotein induces KDM6A and KDM6B histone demethylase expression and causes epigenetic reprogramming . Proc. Natl Acad. Sci. USA108 ( 5 ), 2130 – 2135 ( 2011 ).
- Shen Y , GuoX , WangYet al. Expression and significance of histone H3K27 demethylases in renal cell carcinoma . BMC Cancer12 , 470 ( 2012 ).
- Svotelis A , BiancoS , MadoreJet al. H3K27 demethylation by JMJD3 at a poised enhancer of anti-apoptotic gene BCL2 determines ERalpha ligand dependency . EMBO J.30 ( 19 ), 3947 – 3961 ( 2011 ).
- Bunt J , HasseltNA , ZwijnenburgDA , KosterJ , VersteegR , KoolM . OTX2 sustains a bivalent-like state of OTX2-bound promoters in medulloblastoma by maintaining their H3K27me3 levels . Acta Neuropathol.125 ( 3 ), 385 – 394 ( 2012 ).
- Dubuc Am , RemkeM , KorshunovAet al. Aberrant patterns of H3K4 and H3K27 histone lysine methylation occur across subgroups in medulloblastoma . Acta Neuropathol.125 ( 3 ), 373 – 384 ( 2012 ).
- Agger K , CloosPA , RudkjaerLet al. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence . Genes Dev.23 ( 10 ), 1171 – 1176 ( 2009 ).
- Agherbi H , Gaussmann-WengerA , VerthuyC , ChassonL , SerranoM , DjabaliM . Polycomb mediated epigenetic silencing and replication timing at the INK4a/ARF locus during senescence . PLoS ONE4 ( 5 ), e5622 ( 2009 ).
- Barradas M , AndertonE , AcostaJCet al. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS . Genes Dev.23 ( 10 ), 1177 – 1182 ( 2009 ).
- Ramadoss S , ChenX , WangCy . Histone demethylase KDM6B promotes epithelial-mesenchymal transition . J. Biol. Chem.287 ( 53 ), 44508 – 44517 ( 2012 ).
- Pereira F , BarbachanoA , SilvaJet al. KDM6B/JMJD3 histone demethylase is induced by vitamin D and modulates its effects in colon cancer cells . Hum. Mol. Genet.20 ( 23 ), 4655 – 4665 ( 2011 ).
- Pereira F , BarbachanoA , SinghPK , CampbellMJ , MunozA , LarribaMJ . Vitamin D has wide regulatory effects on histone demethylase genes . Cell Cycle11 ( 6 ), 1081 – 1089 ( 2012 ).
- Chen C , BartenhagenC , GombertMet al. Next-generation-sequencing-based risk stratification and identification of new genes involved in structural and sequence variations in near haploid lymphoblastic leukemia . Genes Chromosomes Cancer52 ( 6 ), 564 – 579 ( 2013 ).
- Van Haaften G , DalglieshGL , DaviesHet al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer . Nat. Genet.41 ( 5 ), 521 – 523 ( 2009 ).
- Laaser I , TheisFJ , DeAngelis MH , KolbHJ , AdamskiJ . Huge splicing frequency in human Y chromosomal UTY gene . OMICS15 ( 3 ), 141 – 154 ( 2011 ).
- Warren EH , GavinMA , SimpsonEet al. The human UTY gene encodes a novel HLA-B8-restricted H-Y antigen . J. Immunol.164 ( 5 ), 2807 – 2814 ( 2000 ).
- Bloomer LD , NelsonCP , EalesJet al. Male-specific region of the Y chromosome and cardiovascular risk: phylogenetic analysis and gene expression studies . Arterioscler. Thromb. Vasc. Biol.33 ( 7 ), 1722 – 1727 ( 2013 ).
- Fortschegger K , ShiekhattarR . Plant homeodomain fingers form a helping hand for transcription . Epigenetics6 ( 1 ), 4 – 8 ( 2010 ).
- Dai J , SultanS , TaylorSS , HigginsJM . The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment . Genes Dev.19 ( 4 ), 472 – 488 ( 2005 ).
- Fortschegger K , DeGraaf P , OutchkourovNs , Van SchaikFM , TimmersHT , ShiekhattarR . PHF8 targets histone methylation and RNA polymerase II to activate transcription . Mol. Cell Biol.30 ( 13 ), 3286 – 3298 ( 2010 ).
- Liu W , TanasaB , TyurinaOVet al. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression . Nature466 ( 7305 ), 508 – 512 ( 2010 ).
- Kleine-Kohlbrecher D , ChristensenJ , VandammeJet al. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation . Mol. Cell38 ( 2 ), 165 – 178 ( 2010 ).
- Feng W , YonezawaM , YeJ , JenuweinT , GrummtI . PHF8 activates transcription of rRNA genes through H3K4me3 binding and H3K9me1/2 demethylation . Nat. Struct. Mol. Biol.17 ( 4 ), 445 – 450 ( 2010 ).
- Arteaga MF , MikeschJH , QiuJet al. The histone demethylase PHF8 governs retinoic acid response in acute promyelocytic leukemia . Cancer Cell23 ( 3 ), 376 – 389 ( 2013 ).
- Stender Jd , PascualG , LiuWet al. Control of proinflammatory gene programs by regulated trimethylation and demethylation of histone H4K20 . Mol. Cell48 ( 1 ), 28 – 38 ( 2012 ).
- Huang C , XiangY , WangYet al. Dual-specificity histone demethylase KIAA1718 (KDM7A) regulates neural differentiation through FGF4 . Cell Res.20 ( 2 ), 154 – 165 ( 2010 ).
- Sinha S , SinghRK , AlamN , RoyA , RoychoudhuryS , PandaCK . Alterations in candidate genes PHF2, FANCC, PTCH1 and XPA at chromosomal 9q22.3 region. pathological significance in early- and late-onset breast carcinoma . Mol. Cancer7 , 84 ( 2008 ).
- Abidi F , MianoM , MurrayJ , SchwartzC . A novel mutation in the PHF8 gene is associated with X-linked mental retardation with cleft lip/cleft palate . Clin. Genet.72 ( 1 ), 19 – 22 ( 2007 ).
- Koivisto AM , Ala-MelloS , LemmelaS , KomuHA , RautioJ , JarvelaI . Screening of mutations in the PHF8 gene and identification of a novel mutation in a Finnish family with XLMR and cleft lip/cleft palate . Clin. Genet.72 ( 2 ), 145 – 149 ( 2007 ).
- Laumonnier F , HolbertS , RonceNet al. Mutations in PHF8 are associated with X linked mental retardation and cleft lip/cleft palate . J. Med. Genet.42 ( 10 ), 780 – 786 ( 2005 ).
- Siderius LE , HamelBC , Van BokhovenHet al. X-linked mental retardation associated with cleft lip/palate maps to Xp11.3-q21.3 . Am. J. Med. Genet.85 ( 3 ), 216 – 220 ( 1999 ).
- Qiao Y , LiuX , HarvardCet al. Autism-associated familial microdeletion of Xp11.22 . Clin. Genet.74 ( 2 ), 134 – 144 ( 2008 ).
- Nava C , LamariF , HeronDet al. Analysis of the chromosome X exome in patients with autism spectrum disorders identified novel candidate genes, including TMLHE . Transl Psychiatry2 , e179 ( 2012 ).
- Aragones J , FraislP , BaesM , CarmelietP . Oxygen sensors at the crossroad of metabolism . Cell Metab.9 ( 1 ), 11 – 22 ( 2009 ).
- Schofield CJ , RatcliffePJ . Oxygen sensing by HIF hydroxylases . Nat. Rev. Mol. Cell Biol.5 ( 5 ), 343 – 354 ( 2004 ).
- Melvin A , MudieS , RochaS . The chromatin remodeler ISWI regulates the cellular response to hypoxia: role of FIH . Mol. Biol. 22 ( 21 ), 4171 – 4181 ( 2011 ).
- Zhang N , FuZ , LinkeSet al. The asparaginyl hydroxylase factor inhibiting HIF-1α is an essential regulator of metabolism . Cell Metabolism11 ( 5 ), 364 – 378 ( 2010 ).
- Liu C-J , TsaiM-M , HungP-Set al. miR-31 ablates expression of the HIF regulatory factor FIH to activate the HIF pathway in head and neck carcinoma . Cancer Research70 ( 4 ), 1635 – 1644 ( 2010 ).
- Kroeze SGC , VermaatJS , Van BrusselAet al. Expression of nuclear FIH independently predicts overall survival of clear cell renal cell carcinoma patients . Eur. J. Cancer46 ( 18 ), 3375 – 3382 ( 2010 ).
- Khan MN , BhattacharyyaT , AndrikopoulosPet al. Factor inhibiting HIF (FIH-1) promotes renal cancer cell survival by protecting cells from HIF-1[alpha]-mediated apoptosis . Br. J. Cancer104 ( 7 ), 1151 – 1159 ( 2011 ).
- Andersen S , DonnemT , StenvoldHet al. Overexpression of the HIF hydroxylases PHD1, PHD2, PHD3 and FIH are individually and collectively unfavorable prognosticators for NSCLC survival . PLoS ONE6 ( 8 ), e23847 ( 2011 ).
- Hyseni A , GroepP , WallE , DiestP . Subcellular FIH-1 expression patterns in invasive breast cancer in relation to HIF-1α expression . Cell Oncol.34 ( 6 ), 565 – 570 ( 2011 ).
- Liu Y , HigashitsujiH , HigashitsujiHet al. Overexpression of gankyrin in mouse hepatocytes induces hemangioma by suppressing factor inhibiting hypoxia-inducible factor-1 (FIH-1) and activating hypoxia-inducible factor-1 . Biochem. Biophys. Res. Comm.432 ( 1 ), 22 – 27 ( 2013 ).
- Tsuneoka M , KodaY , SoejimaM , TeyeK , KimuraH . A novel myc target gene, mina53, that is involved in cell proliferation . J. Biol. Chem.277 ( 38 ), 35450 – 35459 ( 2002 ).
- Hoffman B , LiebermannDA . Apoptotic signaling by c-MYC . Oncogene27 ( 50 ), 6462 – 6472 ( 2008 ).
- Lu Y , ChangQ , ZhangYet al. Lung cancer-associated JmjC domain protein mdig suppresses formation of tri-methyl lysine 9 of histone H3 . Cell Cycle8 ( 13 ), 2101 – 2109 ( 2009 ).
- Fukahori S , YanoH , TsuneokaMet al. Immunohistochemical expressions of Cap43 and Mina53 proteins in neuroblastoma . J. Pediatric Surg.42 ( 11 ), 1831 – 1840 ( 2007 ).
- Ishizaki H , YanoH , TsuneokaMet al. Overexpression of the myc target gene Mina53 in advanced renal cell carcinoma . Pathol. Int.57 ( 10 ), 672 – 680 ( 2007 ).
- Komiya K , Sueoka-AraganeN , SatoAet al. Expression of Mina53, a novel c-Myc target gene, is a favorable prognostic marker in early stage lung cancer . Lung Cancer69 ( 2 ), 232 – 238 ( 2010 ).
- Tan XP , ZhangQ , DongWG , LeiXW , YangZR . Upregulated expression of Mina53 in cholangiocarcinoma and its clinical significance . Oncol. Lett.3 ( 5 ), 1037 – 1041 ( 2012 ).
- Teye K , TsuneokaM , ArimaNet al. Increased expression of a Myc target gene Mina53 in human colon cancer . Am. J. Pathol.164 ( 1 ), 205 – 216 ( 2004 ).
- Tsuneoka M , FujitaH , ArimaNet al. Mina53 as a Potential Prognostic Factor for Esophageal Squamous Cell Carcinoma . Clin. Cancer Res.10 ( 21 ), 7347 – 7356 ( 2004 ).
- Tsuneoka M , NishimuneY , OhtaKet al. Expression of Mina53, a product of a Myc target gene in mouse testis . Int. J. Androl.29 ( 2 ), 323 – 330 ( 2006 ).
- Zhang Y , LuY , YuanBZet al. The Human mineral dust-induced gene, mdig, is a cell growth regulating gene associated with lung cancer . Oncogene24 ( 31 ), 4873 – 4882 ( 2005 ).
- Mori T , OkamotoK , TanakaYet al. Ablation of mina53 in mice reduces allergic response in the airways . Cell Struct. Funct.38 ( 2 ), 155 – 167 ( 2013 ).
- Yosef N , ShalekAk , GaublommeJTet al. Dynamic regulatory network controlling TH17 cell differentiation . Nature496 ( 7446 ), 461 – 468 ( 2013 ).
- Eilbracht J , ReichenzellerM , HergtMet al. NO66, a highly conserved dual location protein in the nucleolus and in a special type of synchronously replicating chromatin . Mol Biol Cell15 ( 4 ), 1816 – 1832 ( 2004 ).
- Brien GL , GamberoG , O’ConnellDJet al. Polycomb PHF19 binds H3K36me3 and recruits PRC2 and demethylase NO66 to embryonic stem cell genes during differentiation . Nat. Struct. Mol. Biol.19 ( 12 ), 1273 – 1281 ( 2012 ).
- Tao Y , WuM , ZhouXet al. Structural Insights into histone demethylase NO66 in interaction with osteoblast-specific transcription factor Osterix and gene repression . J. Biol. Chem.288 ( 23 ), 16430 – 16437 ( 2013 ).
- Suzuki C , TakahashiK , HayamaSet al. Identification of Myc-associated protein with JmjC domain as a novel therapeutic target oncogene for lung cancer . Mol. Cancer Ther.6 ( 2 ), 542 – 551 ( 2007 ).
- Jones MA , CovingtonMF , DitacchioL , VollmersC , PandaS , HarmerSl . Jumonji domain protein JMJD5 functions in both the plant and human circadian systems . Proc. Natl Acad. Sci. USA107 ( 50 ), 21623 – 21628 ( 2010 ).
- Oh S , JanknechtR . Histone demethylase JMJD5 is essential for embryonic development . Biochem. Biophys. Res. Comm.420 ( 1 ), 61 – 65 ( 2012 ).
- Ishimura A , MinehataK-I , TerashimaM , KondohG , HaraT , SuzukiT . Jmjd5, an H3K36me2 histone demethylase, modulates embryonic cell proliferation through the regulation of Cdkn1a expression . Development139 ( 4 ), 749 – 759 ( 2012 ).
- Cikala M , AlexandrovaO , DavidCNet al. The phosphatidylserine receptor from Hydra is a nuclear protein with potential Fe(II) dependent oxygenase activity . BMC Cell Biol.5 , 26 ( 2004 ).
- Mantri M , KrojerT , BaggEAet al. Crystal structure of the 2-oxoglutarate- and Fe(II)-dependent lysyl hydroxylase JMJD6 . J. Mol. Biol.401 ( 2 ), 211 – 222 ( 2010 ).
- Hong X , ZangJ , WhiteJet al. Interaction of JMJD6 with single-stranded RNA . Proc. Natl Acad. Sci. USA107 ( 33 ), 14568 – 14572 ( 2010 ).
- Bose J , GruberAD , HelmingLet al. The phosphatidylserine receptor has essential functions during embryogenesis but not in apoptotic cell removal . J. Biol.3 ( 4 ), 15 ( 2004 ).
- Kunisaki Y , MasukoS , NodaMet al. Defective fetal liver erythropoiesis and T lymphopoiesis in mice lacking the phosphatidylserine receptor . Blood103 ( 9 ), 3362 – 3364 ( 2004 ).
- Schneider JE , BoseJ , BamforthSDet al. Identification of cardiac malformations in mice lacking Ptdsr using a novel high-throughput magnetic resonance imaging technique . BMC Dev. Biol.4 , 16 ( 2004 ).
- Hahn P , WegenerI , BurrellsAet al. Analysis of Jmjd6 cellular localization and testing for its involvement in histone demethylation . PLoS ONE5 ( 10 ), e13769 ( 2010 ).
- Lee Y , MillerL , ChanXet al. JMJD6 is a driver of cellular proliferation and motility and a marker of poor prognosis in breast cancer . Breast Cancer Res.14 ( 3 ), R85 ( 2012 ).
- Ghosh M , LoperR , GelbMh , LeslieCc . Identification of the expressed form of human cytosolic phospholipase A2beta (cPLA2beta): cPLA2beta3 is a novel variant localized to mitochondria and early endosomes . J Biol Chem281 ( 24 ), 16615 – 16624 ( 2006 ).
- Ding X , PanH , LiJet al. Epigenetic activation of AP1 promotes squamous cell carcinoma metastasis . Sci. Signal.6 ( 273 ), ra28 21–13, S20–S15 ( 2013 ).
- Liu C , GilmontRr , BenndorfR , WelshMJ . Identification and characterization of a novel protein from Sertoli cells, PASS1, that associates with mammalian small stress protein hsp27 . J. Biol. Chem.275 ( 25 ), 18724 – 18731 ( 2000 ).
- Jiang M , MaY , ChengHet al. Molecular cloning and characterization of a novel human gene (HSPBAP1) from human fetal brain . Cytogenet. Cell Genet.95 ( 1–2 ), 48 – 51 ( 2001 ).
- Srivastava AK , RenuschSR , NaimanNEet al. Mutant HSPB1 overexpression in neurons is sufficient to cause age-related motor neuronopathy in mice . Neurobiol. Dis.47 ( 2 ), 163 – 173 ( 2012 ).
- Xi ZQ , SunJJ , WangXFet al. HSPBAP1 is found extensively in the anterior temporal neocortex of patients with intractable epilepsy . Synapse61 ( 9 ), 741 – 747 ( 2007 ).
- Xi ZQ , XiaoF , YuanJet al. Gene expression analysis on anterior temporal neocortex of patients with intractable epilepsy . Synapse63 ( 11 ), 1017 – 1028 ( 2009 ).
- Aas PA , OtterleiM , FalnesPOet al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA . Nature421 ( 6925 ), 859 – 863 ( 2003 ).
- Dango S , MosammaparastN , SowaMEet al. DNA unwinding by ASCC3 helicase is coupled to ALKBH3-dependent DNA alkylation repair and cancer cell proliferation . Mol. Cell44 ( 3 ), 373 – 384 ( 2011 ).
- Fu D , SamsonLD . Direct repair of 3,N(4)-ethenocytosine by the human ALKBH2 dioxygenase is blocked by the AAG/MPG glycosylase . DNA Repair (Amst.)11 ( 1 ), 46 – 52 ( 2012 ).
- Lee DH , JinSG , CaiS , ChenY , PfeiferGP , O’connorTR . Repair of methylation damage in DNA and RNA by mammalian AlkB homologues . J. Biol. Chem.280 ( 47 ), 39448 – 39459 ( 2005 ).
- Muller TA , MeekK , HausingerRP . Human AlkB homologue 1 (ABH1) exhibits DNA lyase activity at abasic sites . DNA Repair (Amst.)9 ( 1 ), 58 – 65 ( 2010 ).
- Westbye MP , FeyziE , AasPAet al. Human AlkB homolog 1 is a mitochondrial protein that demethylates 3-methylcytosine in DNA and RNA . J. Biol. Chem.283 ( 36 ), 25046 – 25056 ( 2008 ).
- Bethke L , MurrayA , WebbEet al. Comprehensive analysis of DNA repair gene variants and risk of meningioma . J. Natl Cancer Inst.100 ( 4 ), 270 – 276 ( 2008 ).
- Fujii T , ShimadaK , AnaiS , FujimotoK , KonishiN . ALKBH2, a novel AlkB homologue, contributes to human bladder cancer progression by regulating MUC1 expression . Cancer Sci.104 ( 3 ), 321 – 327 ( 2013 ).
- Hu L , WuC , ZhaoXet al. Genome-wide association study of prognosis in advanced non-small cell lung cancer patients receiving platinum-based chemotherapy . Clin. Cancer Res.18 ( 19 ), 5507 – 5514 ( 2012 ).
- Johannessen TC , PrestegardenL , GrudicA , HegiME , TysnesBB , BjerkvigR . The DNA repair protein ALKBH2 mediates temozolomide resistance in human glioblastoma cells . Neuro Oncol.15 ( 3 ), 269 – 278 ( 2013 ).
- Koike K , UedaY , HaseHet al. anti-tumor effect of AlkB homolog 3 knockdown in hormone- independent prostate cancer cells . Curr. Cancer Drug Targets12 ( 7 ), 847 – 856 ( 2012 ).
- Shimada K , FujiiT , TsujikawaK , AnaiS , FujimotoK , KonishiN . ALKBH3 contributes to survival and angiogenesis of human urothelial carcinoma cells through NADPH oxidase and tweak/Fn14/VEGF signals . Clin. Cancer Res.18 ( 19 ), 5247 – 5255 ( 2012 ).
- Tasaki M , ShimadaK , KimuraH , TsujikawaK , KonishiN . ALKBH3, a human AlkB homologue, contributes to cell survival in human non-small-cell lung cancer . Br. J. Cancer104 ( 4 ), 700 – 706 ( 2011 ).
- Yamato I , ShoM , ShimadaKet al. PCA-1/ALKBH3 contributes to pancreatic cancer by supporting apoptotic resistance and angiogenesis . Cancer Res.72 ( 18 ), 4829 – 4839 ( 2012 ).
- Bjornstad LG , MezaTJ , OtterleiM , OlafsrudSM , Meza-ZepedaLA , FalnesPO . Human ALKBH4 interacts with proteins associated with transcription . PLoS ONE7 ( 11 ), e49045 ( 2012 ).
- Fu Y , HeC . Nucleic acid modifications with epigenetic significance . Curr. Opin. Chem. Biol.16 ( 5–6 ), 516 – 524 ( 2012 ).
- Jia G , FuY , HeC . Reversible RNA adenosine methylation in biological regulation . Trends Genet.29 ( 2 ), 108 – 115 ( 2013 ).
- Jia G , FuY , ZhaoXet al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO . Nat. Chem. Biol.7 ( 12 ), 885 – 887 ( 2011 ).
- Frayling TM , TimpsonNJ , WeedonMNet al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity . Science316 ( 5826 ), 889 – 894 ( 2007 ).
- Gao X , ShinYH , LiM , WangF , TongQ , ZhangP . The fat mass and obesity associated gene FTO functions in the brain to regulate postnatal growth in mice . PLoS ONE5 ( 11 ), e14005 ( 2010 ).
- Do R , BaileySD , DesbiensKet al. Genetic variants of FTO influence adiposity, insulin sensitivity, leptin levels, and resting metabolic rate in the Quebec Family Study . Diabetes57 ( 4 ), 1147 – 1150 ( 2008 ).
- Jacobsson JA , KlovinsJ , KapaIet al. Novel genetic variant in FTO influences insulin levels and insulin resistance in severely obese children and adolescents . Int. J. Obes. (Lond.)32 ( 11 ), 1730 – 1735 ( 2008 ).
- Keller L , XuW , WangHx , WinbladB , FratiglioniL , GraffC . The obesity related gene, FTO, interacts with APOE, and is associated with Alzheimer’s disease risk: a prospective cohort study . J. Alzheimers Dis.23 ( 3 ), 461 – 469 ( 2011 ).
- Lappalainen T , KolehmainenM , SchwabUSet al. Association of the FTO gene variant (rs9939609) with cardiovascular disease in men with abnormal glucose metabolism–the Finnish Diabetes Prevention Study . Nutr Metab Cardiovasc Dis21 ( 9 ), 691 – 698 ( 2011 ).
- Hubacek JA , ViklickyO , DlouhaDet al. The FTO gene polymorphism is associated with end-stage renal disease: two large independent case-control studies in a general population . Nephrol. Dial. Transplant27 ( 3 ), 1030 – 1035 ( 2012 ).
- Kaklamani V , YiN , SadimMet al. The role of the fat mass and obesity associated gene (FTO) in breast cancer risk . BMC Med. Genet.12 , 52 ( 2011 ).
- Tarabra E , ActisGC , FaddaMet al. The obesity gene and colorectal cancer risk: a population study in Northern Italy . Eur. J. Intern. Med.23 ( 1 ), 65 – 69 ( 2012 ).
- Boissel S , ReishO , ProulxKet al. Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations . Am. J. Hum. Genet.85 ( 1 ), 106 – 111 ( 2009 ).
- Shimada K , NakamuraM , AnaiSet al. A novel human AlkB homologue, ALKBH8, contributes to human bladder cancer progression . Cancer Res.69 ( 7 ), 3157 – 3164 ( 2009 ).
- Lorsbach RB , MooreJ , MathewS , RaimondiSC , MukatiraST , DowningJR . TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23) . Leukemia17 ( 3 ), 637 – 641 ( 2003 ).
- Ono R , TakiT , TaketaniT , TaniwakiM , KobayashiH , HayashiY . LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23) . Cancer Res.62 ( 14 ), 4075 – 4080 ( 2002 ).
- Pastor Wa , AravindL , RaoA . TETonic shift: biological roles of TET proteins in DNA demethylation and transcription . Nat. Rev. Mol. Cell Biol.14 ( 6 ), 341 – 356 ( 2013 ).
- Abdel-Wahab O , MullallyA , HedvatCet al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies . Blood114 ( 1 ), 144 – 147 ( 2009 ).
- Comuzzie Ag , ColeSa , LastonSlet al. Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population . PLoS ONE7 ( 12 ), e51954 ( 2012 ).
- Cao XJ , ArnaudoAM , GarciaBA . Large-scale global identification of protein lysine methylation in vivo . Epigenetics8 ( 5 ), 477 – 485 ( 2013 ).
- Chowdhury R , YeohKK , TianYMet al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases . EMBO Rep.12 ( 5 ), 463 – 469 ( 2011 ).
- Watson JA , WatsonCJ , MccannA , BaughJ . Epigenetics, the epicenter of the hypoxic response . Epigenetics5 ( 4 ), 293 – 296 ( 2010 ).
- Edwards AM , IsserlinR , BaderGD , FryeSV , WillsonTM , YuFH . Too many roads not taken . Nature470 ( 7333 ), 163 – 165 ( 2011 ).
