Abstract
Determining the mechanisms of enzymatic regulation is central to the study of cellular metabolism. Regulation of enzyme activity via polymerization-mediated strategies has been shown to be widespread, and plays a vital role in mediating cellular homeostasis. In this review, we begin with an overview of the filamentation of CTP synthase, which forms filamentous structures termed cytoophidia. We then highlight other important examples of the phenomenon. Moreover, we discuss recent data relating to the regulation of enzyme activity by compartmentalization into cytoophidia. Finally, we hypothesize potential roles for enzyme filament formation in the regulation of metabolism, development and disease.
Introduction
The traditional view of cell biology in which the cytoplasm represents a largely disordered collection of freely diffusible proteins and metabolites has been extensively challenged in recent years. Advances in microscopy and imaging technologies has led to a greater appreciation of the presence of high order cytoskeletal organization as well as numerous novel intracellular compartments and bodies. Furthermore, it is increasingly apparent that many of these subcellular compartments are dynamic, undergoing considerable reorganization in response to various extrinsic and intrinsic stimuli.
A large-scale screen of a yeast GFP library showed that a surprising number of metabolic proteins undergo spatial reorganization during nutrient stress induced quiescence, with over 20% of the strains examined displaying novel punctate structures (Narayanaswamy et al., Citation2009). Similarly, the unexpected compartmentalization of a large number of metabolic proteins was observed in a localization screen in the asymmetric bacterium, Caulobacter crescentus (Werner et al., Citation2009). These studies indicate that control of metabolic flux by compartmentalization may be a more widespread phenomenon than previously realized. Although several of these cellular compartments have since been independently corroborated, fluorescence-based localization studies must interpreted with some caution, as the presence of fluorescent fusion proteins has been known to result in aberrant aggregation, sometimes leading to the false identification of novel cytoplasmic bodies. For example, In the case of Clp proteases, which were thought to localize to biologically relevant bodies until examined further by orthogonal methods (Kain et al., Citation2008; Landgraf et al., Citation2012; Simmons et al., Citation2008). Despite these limitations, it has become apparent in recent years that a large number of these self-organizing cytoplasmic structures mediate novel biochemical functions, many of which are involved in fundamental metabolic processes.
It has been hypothesized that the assembly of intracellular bodies contributes to the regulation of metabolism by controlling flux through a particular pathway. This phenomenon has been most comprehensively demonstrated for several of the enzymes involved in the de novo biosynthesis of purine nucleotides. Key enzymes in this pathway have been shown to reversibly co-localize to discrete cytoplasmic bodies known as purinosomes, which are responsive to changing purine concentrations (An et al., Citation2008). It has been demonstrated that the close association of these enzymes facilitates efficient production of purines, presumably through a substrate channeling mechanism (Zhao et al., Citation2013). Conflicting evidence has been presented casting doubt on the validity of these observations and questioning whether the observed bodies are in fact non-functional protein aggregates (Zhao et al., Citation2014). A time-lapse study shows that the formation of purinosome is closely linked to the cell cycle (Chan et al., Citation2015). Super-resolution microscopy reveals that purinosomes locate in close proximity of mitochondria (French et al., Citation2016). Dysregulation of mitochondria caused an increase in the number of purinosomes and the association of these two types of organelles is mediated by mTOR (French et al., Citation2016).
In contrast to the many subcellular “bodies” observed that have punctate or amorphous structures, several intracellular compartments with filamentous morphologies have more recently been identified. These filaments have in many cases been identified as being distinct from the canonical cytoskeleton, being frequently comprised of or containing metabolic enzymes. The contribution of the majority of these cellular filaments to the regulation of metabolism has been poorly understood. However, the mechanisms by which these enzyme filaments assemble and regulate enzymatic activity have begun to be better characterized. These insights have led to a greater understanding of the role of enzyme filaments in cell and developmental biology, metabolic homeostasis and disease biology.
In this review, we provide an overview of our current knowledge of filament-forming enzymes. We go on to highlight remaining questions in the field, and advance novel hypotheses regarding the overall impact of metabolic filament formation in cell biology based on evidence in the literature.
CTP synthase and the cytoophidium
To date, the best characterized example of enzymatic activity mediated by filament formation is that of CTP synthase (CTPS). CTPS is responsible for catalyzing the ATP dependent conversion of UTP to CTP, and as such acts as a critical rate limiting step for both the de novo and salvage pyrimidine synthesis pathways (Lauritsen et al., Citation2011; Long et al., Citation1970). The concentration of CTP synthase affects the equilibrium between its monomeric, dimeric, and tetrameric forms (Robertson, Citation1995). Each monomer contains two functional domains: the kinase ammonia ligase (ALase) domain and the glutamine amidotransferase (GAT) domain (Massiere & Badet-Denisot, Citation1998; Zalkin, Citation1993; Zalkin & Smith, Citation1998). The GAT domain catalyzes GTP-activated glutamine hydrolysis, while the ALase domain mediates Mg2+-ATP-dependent phosphorylation of the UTP uracil O4 atom and displacement of the uracil O4 phosphate by ammonia (Levitzki & Koshland, Citation1976; Weng & Zalkin, Citation1987). CTPS is therefore vital for synthesizing CTP nucleotides required for cellular growth and proliferation.
In 2010, three groups independently reported that CTPS compartmentalizes into filamentous cytoplasmic structures in bacteria, yeast and Drosophila cells (Ingerson-Mahar et al., Citation2010; Liu, Citation2010; Noree et al., Citation2010) (). These intracellular structures have been termed cytoophidia, meaning cellular serpents (Liu, Citation2010). The terms “CTPS filaments” (Ingerson-Mahar et al., Citation2010; Noree et al., Citation2010) and “cytoplasmic rods and rings” (Carcamo et al., Citation2011) have also been used to describe equivalent structures.
Figure 1. Cytoophidia exist in various organisms. CTPS has been found forming filamentous structures in fruit flies (Liu, Citation2010), bacteria (Ingerson-Mahar et al., Citation2010), budding yeast (Noree et al., Citation2010), fission yeast (Zhang et al., Citation2014) and human cells (Carcamo et al., Citation2011; Chen et al., Citation2011). (A) In the fruit fly (Drosophila melanogaster) follicle cells, overexpressing CTPS-GFP (green) leads to long cytoophidia. (B) Cytoophidia, labeled by CTPS-GFP (green), is detectable in budding yeast (Saccharomyces cerevisiae). (C) Cytoophidia, labeled by CTPS-GFP (green), is detectable in fission yeast (Schizosaccharomyces pombe). (D) CTPS1-GFP (green) and IMPDH (red) form filamentous cytoophidia in human (Homo sapiens) HEK293T cells. Image in D is kindly provided by Chia Chun Chang and Li-Ying Sung from National Taiwan University. Nuclei are labeled by DNA dyes (magenta in A–C; blue in D). Scale bars, 10 μm. (see colour version of this figure at www.informahealthcare.com/bmg)
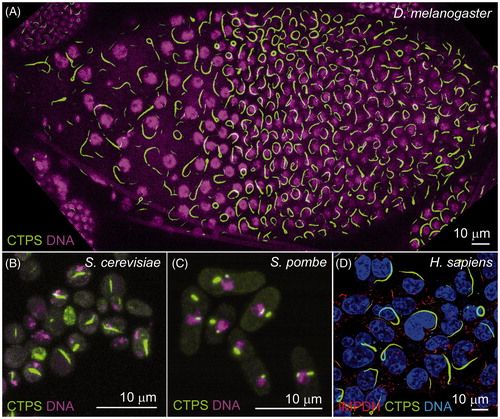
The presence of this feature across such diverse species () suggests that the formation of cytoophidia represents a common regulatory strategy for mediating the production of CTP nucleotides. Multiple lines of evidence have previously indicated that CTPS incorporated into cytoophidia is catalytically inactive, thereby implicating the structure in the downregulation of enzymatic activity. For example, it has been extensively reported that treatment with glutamine analogs such as 6-diazo-5-oxo-L-norleucine (DON), an irreversible competitive inhibitor of CTPS, induces the formation of cytoophidia in Drosophila tissues and human cell lines (Carcamo et al., Citation2011; Chen et al., Citation2011; Gou et al., Citation2014). Similarly, treatment of yeast cells with CTP (an allosterically binding end-product inhibitor of CTPS), caused an increase in filament formation, whilst point mutations at the allosteric CTP binding site resulted in disruption of filaments (Noree et al., Citation2010). However, in contrast to these data, treatment of the bacteria C. crescentus with DON caused CTPS filaments to disassemble (Ingerson-Mahar et al., Citation2010).
Cytoophidia have been shown to be widespread in various tissues in vivo, across which a surprising amount of morphological variation and specific spatial distribution is observed. Most strikingly, the germline cells of the Drosophila ovary reliably contain very large cytoophidia, often exceeding 20 μm in length (Aughey et al., Citation2016; Azzam & Liu, Citation2013; Liu, Citation2010; Strochlic et al., Citation2014; Wang et al., Citation2015) (). Other tissues in which these structures are frequently observed include: the larval lymph gland (the Drosophila hematopoietic stem cell niche), in which filaments are largely observed as closed circles (Liu, Citation2010); primary spermatocytes in the testis, and the optic lobe of the larval CNS (Chen et al., Citation2011; Tastan & Liu, Citation2015). In rat hippocampal neurons, CTPS filaments have a polarized distribution, being restricted to axons (Noree et al., Citation2010). Moreover, CTPS has been found to form cytoophidia both in the cytoplasm and nucleus in fission yeast () and mammalian cells (Gou et al., Citation2014; Zhang et al., Citation2014).
Cytoophidia typically exhibit the snake-shaped, filamentous, and elongated forms, which lend them their name. In some cases, filaments have been observed with toroidal structures, which seem to be more prevalent in certain tissues or cell lines than others (Aughey et al., Citation2014; Liu, Citation2010; Noree et al., Citation2010). Macroscopic cytoophidia are comprised of small CTPS polymers, which associate into larger filaments. The relationship between small and large filaments is that smaller cytoophidia can fuse together to form larger ones, while large filaments can break apart (fission) to form small ones (Gou et al., Citation2014; Liu, Citation2010). The mechanism by which enzyme polymers associate into larger filaments is yet to be fully elucidated. It is still unclear whether the varying morphologies and localizations of cytoophidia are important for mediating specific functions in vivo.
Metabolic regulation by enzyme polymerization
Recently, the relationship between polymerization and CTPS enzyme activity has been more extensively elucidated. Using a novel light-scattering and absorbance assay to simultaneously monitor CTPS activity and filament assembly, it was shown that CTPS polymerization is strongly attenuated by increasing CTP concentration in vitro, providing further evidence that filament formation inhibits enzyme activity (Barry et al., Citation2014). This result is supported by the previously reported observation that mutation of a critical residue for CTP feedback inhibition (E160K) caused a disruption of filament assembly in yeast (Noree et al., Citation2010, Citation2014). Similarly, Aughey et al. (Citation2014) have shown that order of magnitude increases in the levels of intracellular CTPS are accompanied by corresponding increases in length and numbers of cytoophidia, but only moderate increases in CTP concentration in Drosophila (Aughey et al., Citation2014). The consensus view from these three studies is that polymerization of CTPS is a novel mechanism which acts to downregulate enzymatic activity, although the exact mechanism by which this occurs is less well understood.
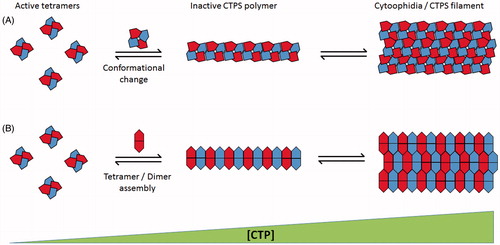
Using cryo-electron microscopy, the structure of CTPS polymers has been determined, from which a novel mechanism of enzymatic regulation has been proposed (Barry et al., Citation2014). According to the model proposed by Barry et al., symmetrical cross-shaped CTPS homotetramers assemble with a novel interdigitated conformation, mediated by interactions between adjacent linker regions (connecting the GATase and ALase domains). It is thought physical constraints to the conformation of tetramers, imposed by polymerization are responsible for the inhibition of the CTP synthesis reaction (). This model of filament assembly is unusual as most previously described examples of filament forming proteins rely on horizontal stacking of (loosely ring-shaped) homo-oligomers in which interacting residues are multiplied through the oligomer’s radial symmetry (O'Connell et al., Citation2012). In conflict with this model, data has been presented indicating that the oligomer subunits of CTPS polymers may be catalytically inactive dimers instead of tetramers (). Mutations at regions of S. cerevisiae or Drosophila CTPS thought to destabilize the tetramer conformation were shown to result in longer filaments, indicating that CTPS tetramers are not incorporated into cytoophidia (Aughey et al., Citation2014; Noree et al., Citation2014). This discrepancy may be explained by the possibility that the mechanism of CTPS polymer assembly has diverged between eukaryotes and prokaryotes.
Figure 2. Schematic representation of proposed mechanisms of CTPS polymer assembly. (A) Mechanism demonstrated by Barry et al. (Citation2014). Active CTPS tetramers (left) undergo conformational change dependant on CTP concentration leading to polymer formation of interdigitated tetramer subunits. Multiple polymers associate into cytoplasmic filaments (right). (B) Mechanism proposed by Aughey et al. (Citation2014) and Noree et al. (Citation2014). Polymerization is dependent on dimerization/tetramerization state of CTPS. Catalytically active tetramers (left) dissociate into constituent dimers for inclusion into inactive cytoplasmic filaments (right). Both mechanisms rely on increasing CTP concentration to promote filament assembly (increasing left to right). (see colour version of this figure at www.informahealthcare.com/bmg)
The regulation of CTPS by filament formation provides an additional mechanism of metabolic control to an enzyme that has already been shown to be subject to regulation by a plethora of different strategies. These include covalent modification, (i.e. phosphorylation by multiple kinases), allosteric interactions, feedback inhibition and transcriptional control (Meng et al., Citation2004). Given that CTPS activity is tightly coordinated by multiple independent regulatory strategies, it is unclear why CTPS requires this additional level of regulation. Although this question remains unanswered, it is possible to speculate on possible reasons for the presence of this well conserved feature of CTPS.
It is possible that the formation of a centralized “storage depot” of CTPS allows for faster re-activation of the enzyme following periods of low nutrient availability to quickly increase the intracellular CTP pool. A filamentous structure has an advantage over a non-linear protein aggregate in this respect due to its larger surface area, therefore providing easier access for regulatory small molecules (such as CTP) or kinases to stimulate filament dissociation and enzyme reactivation. Furthermore, the model demonstrated by Barry et al. (Citation2014) implies that the polymer is apolar (unlike the majority of biological polymers such as microtubules), therefore assembly and disassembly are presumably not end-limiting, allowing for faster state transitions. The incorporation of tetramers into the polymer would be advantageous to such a strategy as they would be catalytically “ready” upon filament depolymerization. It has also been suggested that having multiple levels of regulation for a single enzyme allows for regulation of CTP production over a wider range of kinetic parameters, which may be important for an enzyme such as CTPS that is rate limiting for an important anabolic pathway (Barry et al., Citation2014).
At present, the timescales over which cytoophidia are able to reversibly assemble in a physiologically relevant system are unknown. It is possible that enzyme re-activation by disassembly is rapid, however filament formation may require the diffusion of monomers with assembly being stochastic in nature. Therefore, cytoophidia formation may represent a long-term adaptation, which could explain why filament formation does not appear to necessarily correlate to cell cycle stage (Chen et al., Citation2011), throughout which nucleotide demand fluctuates rapidly, but can be regulated by cell cycle entry or exit (Aughey et al., Citation2014).
Possible functions of cytoophidia
Although the relationship between CTPS catalytic activity and filament formation has now been extensively characterized, several questions remain regarding the role of cytoophidia and other filament forming enzymes in cell biology (). In C. crescentus, CTPS filament formation has been shown to have a novel role in regulating the cell’s characteristic curved morphology through localization to the inner curved membrane where it is hypothesized to mediate a mechanical function in an analogous role to a cytoskeletal component such as actin (Ingerson-Mahar et al., Citation2010). However, it remains unclear whether changes in cell morphology are mediated by force generation through CTPS itself or through recruitment of other factors. Despite the highly conserved compartmentalization of CTPS across taxonomic domains, non-canonical roles for CTPS filaments have not yet been observed in eukaryotes. It has been hypothesized however, that cytoophidia may possess as yet unseen novel functions in eukaryotes (Liu, Citation2011). There are several lines of evidence indicating that this may be the case.
Figure 3. Schematic demonstrating hypothesized cellular functions of cytoophidia beyond regulation of enzymatic activity. (A) “Storage depot” downregulation or “Activator” upregulation of enzyme activity by filament assembly, as demonstrated by (Aughey et al., Citation2014; Barry et al., Citation2014; Noree et al., Citation2014) and (Chang et al., Citation2015; Strochlic et al., Citation2014), respectively. (B) Filament formation mediates structural roles analogous to cytoskeletal filaments as demonstrated in C. crescentus (Ingerson-Mahar et al., Citation2010). (C) The cytoophidium provides an intracellular scaffold for the sequestration of further cytoplasmic proteins. (D) Formation of intracellular filaments regulates traffic of CTPS between cellular compartments. (see colour version of this figure at www.informahealthcare.com/bmg)
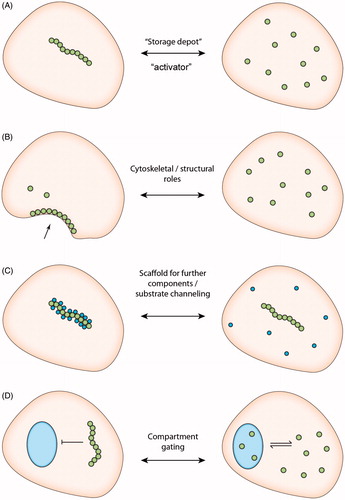
First, CTPS has been shown to colocalise with the de novo purine biosynthesis enzyme inosine-5′-monophosphate dehydrogenase (IMPDH) in human cells (Carcamo et al., Citation2011). IMPDH catalyzes the conversion of IMP to XMP, a rate limiting step in the production of guanine nucleotides, and is therefore a critical regulator of purine nucleotide metabolism (Hedstrom, Citation2009). It is not yet known whether IMPDH binds directly to CTPS or whether both proteins bind to a common third partner, however both CTPS and IMPDH filaments have been shown to exist independently of each other in vivo indicating that neither is necessary for the assembly of the other (Chang et al., Citation2015) (). However, the proximity between these two important enzymes indicates that there may be cross talk between the pyrimidine and purine de novo biosynthesis pathways. IMPDH has also been shown to have nucleic acid binding capability and it has been shown to move between the nucleus and cytoplasm to mediate a novel role as a transcription factor involved in cell cycle progression (Kozhevnikova et al., Citation2012; McLean et al., Citation2004). A novel hypothesis linking these two observations is that the compartmentalization of IMPDH via association with CTPS represents a more committed metabolic adaptation than the formation of IMPDH filaments alone. The generation of more stable, multi-component structures may act to restrict IMPDH to the cytoplasm by preventing diffusion, thereby inhibiting its transcription factor activity and cell cycle progression. Further components of the cytoophidium have not yet been reported, however “gaps” in the structure observed by confocal microscopy in Drosophila tissues have led to speculation that other components may be present (Liu, Citation2010).
The non-receptor tyrosine kinase dACK (Drosophila orthologue of the mammalian enzyme Ack1 – activated cdc42-associated kinase 1) was also observed to colocalise with CTPS, and was shown to be necessary for normal cytoophidia morphology (Strochlic et al., Citation2014). The authors also showed that dAck co-localizes with cytoophidia in the human MCF7 cell line after DON treatment, suggesting that co-localization of dAck and CTPS in cytoophidia is evolutionarily conserved (Strochlic et al., Citation2014). These data provide further support for the idea that cytoophidia may act as a hub for the regulation of multiple interrelated biochemical activities. However, this observation is yet to be confirmed by independent studies, and it is unclear whether dACK is able to spontaneously polymerize in vitro. A study in budding yeast suggests that direct phosphorylation of CTPS does not play a significant role in cytoophidium assembly, indicating that the effect of dACK on filament formation is likely to be indirect (Noree et al., Citation2014).
CTPS activity is thought to be restricted to the cytoplasm. However, observations have indicated that cytoophidia are present within the nuclei of various human cultured cell types, leading to speculation that CTPS may have a novel role in the nucleus, separate from its canonical enzyme activity (Gou et al., Citation2014). Furthermore, IMPDH filament formation has also been demonstrated to occur within the nuclear envelope (Juda et al., Citation2014). Although nucleotide biosynthesis is thought to predominantly occur in the cytoplasm, there is evidence that some enzymes of pyrimidine biosynthesis may also have activity in the nucleus. For example a small fraction of the multifunctional enzyme, carbamoyl phosphate synthetase, aspartate transcarbamylase, and dihydroorotase (CAD) has been shown to have nuclear localization in mammalian cells which is responsive to cell cycle phase (Carrey et al., Citation2002, Chaparian & Evans, Citation1988, Sigoillot et al., Citation2003). Therefore, the observation that CTPS may be present in the nucleus under certain conditions is not unprecedented. The functions of cytoophidia in the nucleus are currently unclear; however, it could facilitate transcription through regulation of nucleotide production for incorporation into RNA in situ. The observation that CTPS may localize to different membrane bound cellular compartments invites the speculation that filament formation could act to prevent the movement of CTPS into or out of the nucleus as required. The filament forming property of CTPS, may therefore represent a novel mechanism to spatially regulate enzyme activity in addition to its role in downregulating catalysis.
Structural roles for filament-forming proteins have not been widely demonstrated. However, it is conceivable that even if individual filaments do not contribute roles that are directly analogous to those of cytoskeletal proteins, the assembly of filamentous macrostructures may reflect a wider re-organization of the cytoplasm in response to metabolic or physiological changes. It has been demonstrated that several metabolic enzymes reversibly assemble into macrostructures as an adaptive metabolic process, and large-scale screens have indicated that this process may be widespread (Petrovska et al., Citation2014, Noree et al., Citation2014). The transition of metabolic enzymes from diffuse to filamentous may result in changes in the mechanical properties of the cell, simply by altering the viscosity of the cytoplasm, thereby helping the cell to adapt to altered environmental conditions.
Regulation of cytoophidium assembly
That CTPS polymerization is regulated as adaptive metabolic response, dependant on environmental nutrient availability, is a feature that has been widely demonstrated. However, it has also been demonstrated that the reversible compartmentalization of CTPS occurs during various physiological processes, during which it is thought to facilitate the transition between cellular metabolic states necessary for co-ordinating proliferation or other energy-dependant cellular processes (Aughey et al., Citation2014). A notable example of reversible compartmentalization of CTPS in normal development is during neurogenesis. In Drosophila, a population of neural stem cells (neuroblasts) in the central nervous system (CNS) undergo a defined period of programed quiescence during larval development (Chell & Brand, Citation2010). These cells were shown to have prominent cytoophidia, which were seen to disassemble upon reentry of the cell cycle. Knockdown of the serine/threonine kinase AKT, which is necessary for initiation of neuroblast proliferation (Chell & Brand, Citation2010), was sufficient to prevent the dissociation of filaments. These data indicate that cytoophidia assembly plays a role in the regulation of normal developmental processes, as well as adaptive metabolism, and that filament formation is likely to be dependent on signaling from extrinsic cellular factors aside from direct allosteric regulation by nucleotide concentrations.
Developmentally regulated changes in CTPS compartmentalization have also been proposed to occur in several other physiological contexts. The ovarian follicle cell epithelium in Drosophila has proved to be a useful system in which to study the regulation of cytoophidia formation (). This is due to the highly reliable distribution of filaments in these cells, as well as the overall genetic tractability and nutritional sensitivity of the tissue (Liu, Citation2010, Noree et al., Citation2010, Chen et al., Citation2011, Azzam & Liu, Citation2013, Strochlic et al., Citation2014, Wang et al., Citation2015, Aughey et al., Citation2016). Cytoophidia are easily visualized in follicle cells throughout most of oogenesis, then undergo a rapid disassembly in time with a period of increased anabolic activity during the chorion gene amplification stage of oogenesis (Aughey et al., Citation2016). In Drosophila follicle cells, the occurrence of cytoophidia correlates with the expression of the oncogene Myc. Knockdown of Myc results in loss of cytoophidia in follicle cells, while overexpressing Myc can induce cytoophidium formation (Aughey et al., Citation2016). It has been proposed that filament dissociation is required to facilitate this metabolic change, although this is yet to be conclusively demonstrated.
Cytoophidia have been shown to be regulated by a number of cellular processes in the Drosophila ovarian follicle cells. Knockdown of the ubiquitin E3 ligase, Cbl, has been shown to be required for filament formation without affecting CTPS protein levels. It remains unclear whether ubiquitination of CTPS directly by Cbl is required for cytoophidia formation, or whether filament formation is dependent on an indirect effect of Cbl action (Wang et al., Citation2015). Cytoophidia have also been shown to be regulated in these cells by basic helix-loop-helix transcription factor and oncogene, Myc, which is highly expressed in metabolically active and proliferating tissues.
Further evidence that cytoophidia may be regulated by the action of extrinsic factors comes from the observation that treatment of yeast cells with the broadly active kinase inhibitor staurosporine is sufficient to cause dramatic upregulation of filament formation (Noree et al., Citation2010). However, it is not clear whether this is a direct consequence of CTPS phosphorylation, or due to the metabolic perturbation which is undoubtedly induced by inhibiting multiple kinases. Further study will be required to determine whether post-translational modifications play any role in promoting or preventing CTPS polymerization into cytoophidia.
Widespread prevalence of filament-forming enzymes
CTP synthase (CTPS) remains one of the best characterized examples of compartmentalization of a metabolic enzyme through polymerization; however, there is evidence to suggest that this feature may be widespread in the modulation of protein catalytic activity. One of the earliest enzymes to be identified as subject to regulation by the assembly of monomers into higher-order structures is acetyl-CoA carboxylase (ACC). Purified ACC from animal tissues was shown by electron microscopy to reversibly form filamentous structures upon incubation with its allosteric activator, citrate (Kleinschmidt et al., Citation1969). Conversely, incubation with malonyl-CoA or Mg2+ ions, both inhibitors of ACC, caused rapid depolymerization of citrate-induced filaments (Beaty & Lane, Citation1983a,Citationb). For a long time, the polymerized form of ACC has been detectable as tiny subcellular filaments by electron microscopy (Kleinschmidt et al., Citation1969; Meredith & Lane, Citation1978). A study reported that Acc1p (the yeast ortholog of ACC) has diffused distribution under normal growth conditions, while prolonged starvation can drive Acc1p to form rod-like structures in budding yeast (Suresh et al., Citation2015). From these data, it has been inferred that the formation of higher order structures is necessary to positively regulate the enzymatic activity of ACC. Furthermore, it has been speculated that these structures may possess a further, possibly structural, role in fatty acid synthesis, however no evidence has yet been presented in support of this idea (Beaty & Lane, Citation1983b; Kim et al., Citation2010). The fact that the polymerization of ACC appears to have the opposite effect on its enzymatic activity to that of CTPS indicates that although there are broad similarities between these two filaments, the mechanisms of their assembly and nature of monomer subunits may differ greatly. Following the discovery of the regulation of ACC by filament assembly, several other enzymes with wide ranging functions have been shown to assemble into filamentous structures ().
Table 1. Filament-forming proteins and their regulatorsa.
In a yeast protein localization screen in which CTPS filaments were identified, eight further proteins localizing to four distinct intracellular structures were also observed including the metabolic enzymes glutamine synthase and GDP-mannose pyrophosphorylase (Noree et al., Citation2010). Although these structures are morphologically similar in appearance, it is not possible to tell whether they regulate enzyme activity by a common mechanism. However, it has been shown that glutamine synthase filaments appear to be regulated in a similar manner to CTPS, acting to mediate cellular glutamine homeostasis with their assembly being highly responsive to cellular metabolic state (Petrovska et al., Citation2014). This indicates that the “storage depot” model of enzyme filament formation may be more widespread than previously realized.
An expanded screening has been carried out by our group recently. We identified 23 proteins which can form filamentous structures (Shen et al., Citation2016). We confirmed all the nine proteins identified in the Wilhelm study (Noree et al., Citation2010) and four septin proteins available in that collection. These enzymes are clustered in metabolic pathways. It will be interesting to see how many of them reside in the same structures. These screening studies could still be an underestimate of the number of filament-forming proteins since the assembly of filaments is sensitive to various culture conditions. Further study will be required to determine the full extent of enzymes which are regulated by filament formation.
A common feature that seems to be shared by many enzyme filaments is the ability to reversibly dissociate in response to metabolite changes in the cellular environment. Often these changes are seen in response to small molecules known to be allosteric regulators, pharmacological inhibitors or substrates (for example regulation of cytoophidia formation by CTP end-product inhibition, or ACC by malonyl-CoA). By this mechanism, these filaments seem to be clearly regulating their enzymatic activity in response to metabolic demand. However, a small number of enzyme filaments have been identified that do not have biosynthetic capacity resulting in the production of small molecules that are able to directly affect their polymerization. An example of this is the protein kinases Mps1 and Polo. These proteins were shown to form intracellular filaments upon exposure to hypoxic conditions (McLean et al., Citation2004). It is unclear whether these filaments are responding directly or indirectly to changes in the cellular environment in hypoxia. Intriguingly, Mps1/Polo filaments were shown to be sensitive to collagenase, indicating that they may be dependent on a collagen-like backbone for their formation.
Filament-forming enzymes and the cytoskeleton
It has been hypothesized that the ability of metabolic enzymes to polymerize into intracellular filaments helped to drive the evolution of the cytoskeleton (Barry & Gitai, Citation2011). Aside from obvious morphological and structural similarities of observed enzyme polymers to cytoskeletal elements, there are several lines of evidence to support this idea. In C. crescentus CTPS filaments have been shown to be involved in regulating cell curvature in an apparently structural role analogous to that of an intermediate filament in eukaryotic cells (Ingerson-Mahar et al., Citation2010). Similar structural roles for CTPS have not yet been observed in eukaryotes, however their restricted distributions (e.g. in rat neuronal axons), indicates that this is possible.
Further evidence has been presented for an evolutionary relationship between cytoskeletal components and metabolic enzymes. Intriguingly, the glycolytic enzyme hexokinase has high structural similarity to actin (although shares little sequence homology), being categorized as an “actin fold” protein with a core conserved ATP binding site (Bork et al., Citation1992; Wilson & Schwab, Citation1996). It has been suggested that hexokinase and actin evolved from a common ancestor, with actin having gained the ability to polymerize in order to mediate enzymatic activity (Barry & Gitai, Citation2011). In this scenario the catalytic activity of the protein was subsequently lost, possibly due to redundancy or selective pressure, resulting in a protein with a purely structural role which remains regulated by nucleotide interactions as a consequence of its catalytic evolutionary past. Despite being a compelling hypothesis, it is also conceivable to imagine these events happening in the opposite order. That is, in a cellular precursor with limited autonomous metabolic control, a polymerizing structural component may have been regulated by interactions with small molecules in its immediate molecular environment. Catalytic activity may then have evolved as a consequence of these interactions with these small molecules. Subsequently, some of these proteins may have lost their cytoskeletal properties. Therefore, the opposite hypothesis can be put forward; that filament forming proteins helped to drive the evolution of catalysis.
These ideas about the evolutionary relationship between polymerizing enzymes and cytoskeletal components may help to explain how non-polymerizing protein have retained features in common with filament forming enzymes such as oligomerization, tendency to aggregate and allosteric regulation. Regardless of the order in which this process occurred, it is clear that there is a close relationship between filament forming enzymes and cytoskeletal proteins, which indicates that many more proteins than currently appreciated may harbor polymerizing tendencies that have as yet gone unnoticed.
Implications for disease biology
Enzymes involved in nucleotide metabolism have been considered promising targets for human cancer therapy due to their proven antiproliferative effects in animal models and clinical trials (Jordheim et al., Citation2013). CTPS activity was shown to be upregulated by as much as ten-fold in human hepatomas, suggesting that the increased rate of CTP synthesis may confer a selective advantage to these tumors (Kizaki et al., Citation1980). Mutations in conserved allosteric regulatory sites in CHO cells were shown to increase resistance to anti-proliferative cytotoxic nucleosides as well as increase intracellular CTP concentration and promote a higher rate of spontaneous mutations (Chu et al., Citation1984; Meuth et al., Citation1979; Whelan et al., Citation1993). Furthermore, CTPS inhibitors were identified in a large scale chemical screen as one of the most effective anti-proliferative drugs in a Drosophila metastatic tumor model (Willoughby et al., Citation2013).
Our newfound understanding of the regulation of CTPS activity through cytoophidia formation raises several questions regarding the role of CTPS in tumorigenesis. It has been demonstrated that increasing CTPS concentrations in vivo and in vitro lead to only modest increases in CTP production (Aughey et al., Citation2014; Barry et al., Citation2014). When mutations were made at the polymerization interface disrupting CTPS polymerization, the catalytic activity of the enzyme was shown to be slightly downregulated, however feedback inhibition was also disrupted, leading to an overall increase in CTP production in vivo (Barry et al., Citation2014). It is plausible that the upregulation of CTPS activity observed in tumor cells may be caused by mutations disrupting cytoophidia assembly. However, tumor cells have been reported to typically present a 3- to 4-fold increase in intracellular nucleotide pools (Traut, Citation1994), which is broadly in line with the theoretical limits imposed by models recently presented (Aughey et al., Citation2014; Barry et al., Citation2014). Therefore, the formation of cytoophidia may help to explain why higher nucleotide concentrations are not frequently observed and point to higher expression of CTPS as the primary mechanism for CTP pool increase.
CTP synthase (CTPS) is not the only enzyme for which a greater understanding of its polymerization may lead to insights into disease etiology or therapeutics. Specific point mutations in IMPDH are linked to the degenerative eye disease autosomal dominant retinitis pigmentosa (adRP). Interestingly, this disease leads to the specific degeneration of retinal cells despite the fact that IMPDH is broadly expressed and essential for purine metabolism. The same mutations implicated in adRP have also been shown to cause increased propensity for filament formation, indicating that this process may have a role in the pathology of the disease (Labesse et al., Citation2013).
A greater appreciation for the phenomenon of enzyme filament formation generally may lead to novel approaches to the design of inhibitors targeting a number of metabolic pathways. It is conceivable that therapeutics could be developed that specifically inhibit or promote polymerization. Greater understanding of the mechanisms by which enzyme filaments are regulated will be required to realize this goal. Furthermore, filament forming enzymes could potentially be used as novel biomarkers for disease. For example, autoantibodies against IMPDH filaments (also referred to as “rods and rings”/RR) have been observed to be enriched in patients being treated for hepatitis C (Calise et al., Citation2015).
Perspectives
Due to the closely interrelated properties of enzymatic activity and filament-formation displayed by enzymes such as CTPS, considerable difficulties may be encountered in the study of filament forming enzymes. A combination of genetic, biochemical, and microscopic approaches have been extensively applied to the investigation of filament-forming enzymes, all of which have certain advantages and shortcomings. Biochemical approaches have been essential for determining how enzyme dynamics are affected by filament formation in vitro. Furthermore, reconstitution of cytoophidia in test tubes, in combination with super-resolution microscopy, can be performed to determine cytoophidium behavior, dynamics and ultrastructure. When combined with genetic approaches, it may be possible to identify point mutations or truncations in which filament-forming propensities are disrupted, whilst enzymatic activity is retained. The phenotypic consequences of losing the filament-forming property can subsequently be assessed, without disrupting enzymatic activity. Unicellular organisms such as budding yeast and fission yeast are excellent models to determine the function of cytoophidia systematically. Series of point mutations can be generated and assessed for their effects on the cell cycle, cell size, organelle morphology or global metabolic profiling. Furthermore, genetic approaches in multicellular models such as Drosophila have been used to great effect to determine the physiological and cell-type specific effects of filament formation or disruption. Enzyme filaments such as cytoophidia are frequently detectable using standard light-microscopy methods in vivo, therefore changes in their distribution and appearance may be easily quantified in the context of a whole tissue. To acquire a full understanding of the role of filament formation in metabolic enzymes, a combination of these approaches is likely to be required.
The identification of multiple filament forming enzymes has opened up an exciting new chapter in our understanding of enzymatic regulation. The filament forming property of CTPS, in particular, has been the subject of intensive research by multiple groups over recent years; however, significant questions remain regarding the structure, composition and function of cytoophidia. Nevertheless, we have gained significant insights into how metabolic processes can be regulated by dynamic rearrangement of individual proteins. Further research will be required to understand the role of the ever-increasing complement of metabolic enzyme filaments in regulating catalysis and cellular homeostasis.
Disclosure statement
The authors declare no conflict of financial interest. The study of cytoophidia in the Liu lab was supported by UK Medical Research Council.
References
- An S, Kumar R, Sheets ED, Benkovic SJ. (2008). Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science 320:103–6.
- Aughey GN, Grice SJ, Liu JL. (2016). The interplay between Myc and CTP synthase in Drosophila. PLoS Genet 12:e1005867.
- Aughey GN, Grice SJ, Shen QJ, et al. (2014). Nucleotide synthesis is regulated by cytoophidium formation during neurodevelopment and adaptive metabolism. Biol Open 3:1045–56.
- Azzam G, Liu JL. (2013). Only one isoform of Drosophila melanogaster CTP synthase forms the cytoophidium. PLoS Genet 9:e1003256
- Barry RM, Bitbol AF, Lorestani A, et al. (2014). Large-scale filament formation inhibits the activity of CTP synthetase. eLife 3:e03638.
- Barry RM, Gitai Z. (2011). Self-assembling enzymes and the origins of the cytoskeleton. Curr Opin Microbiol 14:704–11.
- Beaty NB, Lane MD. (1983a). Kinetics of activation of acetyl-CoA carboxylase by citrate. Relationship to the rate of polymerization of the enzyme. J Biol Chem 258:13043–50.
- Beaty NB, Lane MD. (1983b). The polymerization of acetyl-CoA carboxylase. J Biol Chem 258:13051–5.
- Bork P, Sander C, Valencia A. (1992). An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc Natl Acad Sci USA 89:7290–4.
- Calise SJ, Keppeke GD, Andrade LE, Chan EK. (2015). Anti-rods/rings: a human model of drug-induced autoantibody generation. Front Immunol 6:41.
- Carcamo WC, Satoh M, Kasahara H, et al. (2011). Induction of cytoplasmic rods and rings structures by inhibition of the CTP and GTP synthetic pathway in mammalian cells. PLoS One 6:e29690.
- Carrey EA, Dietz C, Glubb DM, et al. (2002). Detection and location of the enzymes of de novo pyrimidine biosynthesis in mammalian spermatozoa. Reproduction 123:757–68.
- Chan CY, Zhao H, Pugh RJ, et al. (2015). Purinosome formation as a function of the cell cycle. Proc Natl Acad Sci USA 112:1368–73.
- Chang CC, Lin WC, Pai LM, et al. (2015). Cytoophidium assembly reflects upregulation of IMPDH activity. J Cell Sci 128:3550–5.
- Chaparian MG, Evans DR. (1988). Intracellular location of the multidomain protein CAD in mammalian cells. FASEB J 2:2982–9.
- Chell JM, Brand AH. (2010). Nutrition-responsive glia control exit of neural stem cells from quiescence. Cell 143:1161–73.
- Chen K, Zhang J, Tastan OY, et al. (2011). Glutamine analogs promote cytoophidium assembly in human and Drosophila cells. J Genet Genomics 38:391–402.
- Chu EH, Mclaren JD, Li IC, Lamb B. (1984). Pleiotropic mutants of Chinese hamster cells with altered cytidine 5'-triphosphate synthetase. Biochem Genet 22:701–15.
- French JB, Jones SA, Deng H, et al. (2016). Spatial colocalization and functional link of purinosomes with mitochondria. Science 351:733–7.
- Gilliland WD, Vietti DL, Schweppe NM, et al. (2009). Hypoxia transiently sequesters mps1 and polo to collagenase-sensitive filaments in Drosophila prometaphase oocytes. PLoS One 4:e7544.
- Gou KM, Chang CC, Shen QJ, et al. (2014). CTP synthase forms cytoophidia in the cytoplasm and nucleus. Exp Cell Res 323:242–53.
- Gunning BE. (1965). The fine structure of chloroplast stroma following aldehyde osmium-tetroxide fixation. J Cell Biol 24:79–93.
- Gunter J, Thomas E, Lengefeld N, et al. (2007). Charecterisation of inosine monphosphate dehydrogenase expression during retinal development: differences between isoforms and variants. Int J Biochem Cell Biol 40:1716–28.
- Hedstrom L. (2009). IMP dehydrogenase: structure, mechanism, and inhibition. Chem Rev 109:2903–28.
- Ingerson-Mahar M, Briegel A, Werner JN, et al. (2010). The metabolic enzyme CTP synthase forms cytoskeletal filaments. Nat Cell Biol 12:739–46.
- Ji Y, Gu J, Makhov AM, et al. (2006). Regulation of the interaction of inosine monophosphate dehydrogenase with mycophenolic Acid by GTP. J Biol Chem 281:206–12.
- Jordheim LP, Durantel D, Zoulim F, Dumontet C. (2013). Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases. Nat Rev Drug Discov 12:447–64.
- Juda P, Smigova J, Kovacik L, et al. (2014). Ultrastructure of cytoplasmic and nuclear inosine-5'-monophosphate dehydrogenase 2 “rods and rings” inclusions. J Histochem Cytochem 62:739–50.
- Kain J, He GG, Losick R. (2008). Polar localization and compartmentalization of ClpP proteases during growth and sporulation in Bacillus subtilis. J Bacteriol 190:6749–57.
- Kim CW, Moon YA, Park SW, et al. (2010). Induced polymerization of mammalian acetyl-CoA carboxylase by MIG12 provides a tertiary level of regulation of fatty acid synthesis. Proc Natl Acad Sci USA 107:9626–31.
- Kim SY, Kim YW, Hegerl R, et al. (2005). Novel type of enzyme multimerization enhances substrate affinity of oat beta-glucosidase. J Struct Biol 150:1–10.
- Kizaki H, Williams JC, Morris HP, Weber G. (1980). Increased cytidine 5'-triphosphate synthetase activity in rat and human tumors. Cancer Res 40:3921–7.
- Kleinschmidt AK, Moss J, Lane DM. (1969). Acetyl coenzyme A carboxylase: filamentous nature of the animal enzymes. Science 166:1276–8.
- Korennykh AV, Egea PF, Korostelev AA, et al. (2009). The unfolded protein response signals through high-order assembly of Ire1. Nature 457:687–93.
- Kozhevnikova EN, van der Knaap JA, Pindyurin AV, et al. (2012). Metabolic enzyme IMPDH is also a transcription factor regulated by cellular state. Mol Cell 47:133–9.
- Labesse G, Alexandre T, Vaupre L, et al. (2013). MgATP regulates allostery and fiber formation in IMPDHs. Structure 21:975–85.
- Landgraf D, Okumus B, Chien P, et al. (2012). Segregation of molecules at cell division reveals native protein localization. Nat Methods 9:480–2.
- Lauritsen I, Willemoes M, Jensen KF, et al. (2011). Structure of the dimeric form of CTP synthase from Sulfolobus solfataricus. Acta Crystallogr Sect F Struct Biol Cryst Commun 67:201–8.
- Levitzki A, Koshland DE, Jr. (1976). The role of negative cooperativity and half-of-the-sites reactivity in enzyme regulation. Curr Top Cell Regul 10:1–40.
- Li H, Korennykh AV, Behrman SL, Walter P. (2010). Mammalian endoplasmic reticulum stress sensor IRE1 signals by dynamic clustering. Proc Natl Acad Sci USA 107:16113–8.
- Liu JL. (2010). Intracellular compartmentation of CTP synthase in Drosophila. J Genet Genomics 37:281–96.
- Liu JL. (2011). The enigmatic cytoophidium: compartmentation of CTP synthase via filament formation. Bioessays 33:159–64.
- Long CW, Levitzki A, Koshland DE. JR. (1970). The subunit structure and subunit interactions of cytidine triphosphate synthetase. J Biol Chem 245:80–7.
- Massiere F, Badet-Denisot MA. (1998). The mechanism of glutamine-dependent amidotransferases. Cell Mol Life Sci 54:205–22.
- Mclean J, Hamaguchi N, Belenky P, et al. (2004). Inosine 5'-monophosphate dehydrogenease binds nucleic acids in vitro and in vivo. Biochem J 15:243–51.
- Meng Q, Turnbough CL, Jr, Switzer RL. (2004). Attenuation control of pyrG expression in Bacillus subtilis is mediated by CTP-sensitive reiterative transcription. Proc Natl Acad Sci USA 101:10943–8.
- Meredith MJ, Lane MD. (1978). Acetyl-CoA carboxylase. Evidence for polymeric filament to protomer transition in the intact avian liver cell. J Biol Chem 253:3381–3.
- Meuth M, L'Heureux-Huard N, Trudel M. (1979). Characterization of a mutator gene in Chinese hamster ovary cells. Proc Natl Acad Sci USA76:6505–9.
- Miller RE, Shelton E, Stadtman ER. (1974). Zinc-induced paracrystalline aggregation of glutamine synthetase. Arch Biochem Biophys 163:155–71.
- Narayanaswamy R, Levy M, Tsechansky M, et al. (2009). Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc Natl Acad Sci USA 106:10147–52.
- Noree C, Monfort E, Shiau AK, Wilhelm JE. (2014). Common regulatory control of CTP synthase enzyme activity and filament formation. Mol Biol Cell 25:2282–90.
- Noree C, Sato BK, Broyer RM, Wilhelm JE. (2010). Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster. J Cell Biol 190:541–51.
- O'Connell JD, Zhao A, Ellington AD, Marcotte EM. (2012). Dynamic reorganization of metabolic enzymes into intracellular bodies. Annu Rev Cell Dev Biol 28:89–111.
- Petrovska I, Nuske E, Munder MC, et al. (2014). Filament formation by metabolic enzymes is a specific adaptation to an advanced state of cellular starvation. eLife 3:e02409.
- Robertson JG. (1995). Determination of subunit dissociation constants in native and inactivated CTP synthetase by sedimentation equilibrium. Biochemistry 34:7533–41.
- Shen QJ, Kassim H, Huang Y, et al. (2016). Filamentation of metabolic enzymes in Saccharomyces cerevisiae. J Genet Genomics, in press. [Epub ahead of print]. doi:10.1016/j.jgg.2016.03.008.
- Sigoillot FD, Berkowski JA, Sigoillot SM, et al. (2003). Cell cycle-dependent regulation of pyrimidine biosynthesis. J Biol Chem 278:3403–9.
- Simmons LA, Grossman AD, Walker GC. (2008). Clp and Lon proteases occupy distinct subcellular positions in Bacillus subtilis. J Bacteriol 190:6758–68.
- Strochlic TI, Stavrides KP, Thomas SV, et al. (2014). Ack kinase regulates CTP synthase filaments during Drosophila oogenesis. EMBO Rep 15:1184–91.
- Suresh HG, Da Silveira Dos Santos AX, Kukulski W, et al. (2015). Prolonged starvation drives reversible sequestration of lipid biosynthetic enzymes and organelle reorganization in Saccharomyces cerevisiae. Mol Biol Cell 26:1601–15.
- Tastan OY, Liu JL. (2015). CTP synthase is required for optic lobe homeostasis in Drosophila. J Genet Genomics 42:261–74.
- Thuku RN, Weber BW, Varsani A, Sewell BT. (2007). Post-translational cleavage of recombinantly expressed nitrilase from Rhodococcus rhodochrous J1 yields a stable, active helical form. FEBS J 274:2099–108.
- Traut TW. (1994). Physiological concentrations of purines and pyrimidines. Mol Cell Biochem 140:1–22.
- Wang PY, Lin WC, Tsai YC, et al. (2015). Regulation of CTP synthase filament formation during DNA endoreplication in Drosophila. Genetics 201:1511–23.
- Weng ML, Zalkin H. (1987). Structural role for a conserved region in the CTP synthetase glutamine amide transfer domain. J Bacteriol 169:3023–8.
- Werner JN, Chen EY, Guberman JM, et al. (2009). Quantitative genome-scale analysis of protein localization in an asymmetric bacterium. Proc Natl Acad Sci USA 106:7858–63.
- Whelan J, Phear G, Yamauchi M, Meuth M. (1993). Clustered base substitutions in CTP synthetase conferring drug resistance in Chinese hamster ovary cells. Nat Genet 3:317–22.
- Willoughby LF, Schlosser T, Manning SA, et al. (2013). An in vivo large-scale chemical screening platform using Drosophila for anti-cancer drug discovery. Dis Model Mech 6:521–9.
- Wilson JE, Schwab DA. (1996). Functional interaction of hexokinase with ATP requires participation by both small and large lobes of the enzyme: implications for other proteins using the actin fold as a nucleotide binding motif. FASEB J 10:799–801.
- Zalkin H. (1993). The amidotransferases. Adv Enzymol Relat Areas Mol Biol 66:203–309.
- Zalkin H, Smith JL. (1998). Enzymes utilizing glutamine as an amide donor. Adv Enzymol Relat Areas Mol Biol 72:87–144.
- Zeiri L, Reisler E. (1978). Uncoupling of the catalytic activity and the polymerization of beef liver glutamate dehydrogenase. J Mol Biol 124:291–5.
- Zhang J, Hulme L, Liu JL. (2014). Asymmetric inheritance of cytoophidia in Schizosaccharomyces pombe. Biol Open 3:1092–7.
- Zhao A, Tsechansky M, Ellington AD, Marcotte EM. (2014). Revisiting and revising the purinosome. Mol Biosyst 10:369–74.
- Zhao H, French JB, Fang Y, Benkovic SJ. (2013). The purinosome, a multi-protein complex involved in the de novo biosynthesis of purines in humans. Chem Commun (Camb) 49:4444–52.
