Abstract
Human swine influenza A [H1N1], also referred to as “swine flu,” is highly transmissible. The emergence of new strains will continue to pose challenges to public health and the scientific communities will have to prepare to detect them for appropriate treatment. Most sophisticated methods include immunofluorescence staining and antigen subtyping based on hemagglutination inhibition (HI). Another standard method is RT-PCR targeting hemagglutinin and neuraminidase genes. The recent availability of rapid, reliable, and easy-to-perform tests for detecting influenza virus infections has introduced rapid viral diagnosis. This review thus summarizes the current information on the present diagnostic methods for influenza virus H1N1.
Introduction: H1N1 etiology and epidemiology
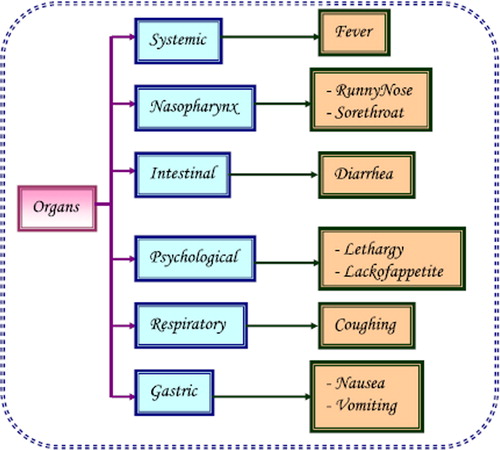
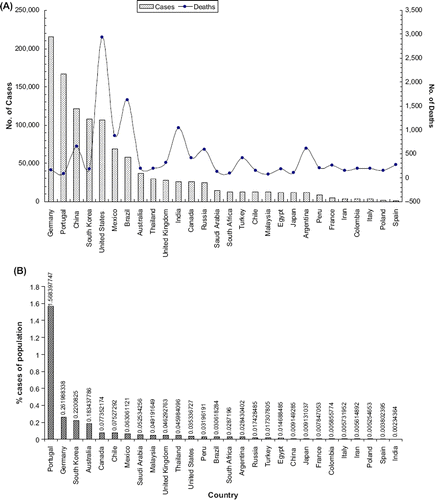
Pandemic H1N1 was first detected in the United States in April 2009. On June 11, 2009, the World Health Organization raised the worldwide pandemic alert level to phase 6. Epidemiological data indicate that the current outbreak of influenza-like respiratory illness started in Mexico, in February 2009. In early April, public health authorities began investigating high numbers of influenza-like illness and informed the Pan American Health Organization of a possible outbreak (Centers for Disease Control and Prevention Citation2009). Influenza A viruses are single stranded RNA viruses of negative sense with an eight segmented genome that belongs to the family Orthomyxoviridae. The viral haemagglutinin (HA) and neuraminidase (NA) proteins are envelope glycoproteins and are the key antigens against which humoral immune responses are directed. The HA protein has an important role in expressing high pathogenicity in many animal species and mediates the fusion of the viral and host endosomal membranes. They are used for the subtyping of influenza A viruses into 16 HA and 9 NA subtypes (Peiris et al. Citation2009). For instance, part of the process that allows influenza viruses to invade cells is the cleavage of the viral hemagglutinin protein by any one of several human proteases. In mild and a virulent virus, the structure of the hemagglutinin means that it can only be cleaved by proteases found in the throat and lungs, so these viruses cannot infect other tissues. However, in highly virulent strains, such as H5N1, the hemagglutinin can be cleaved by a wide variety of proteases, allowing the virus to spread throughout the body. The viral hemagglutinin protein is responsible for determining both the species that a strain can infect and where in the human respiratory tract a strain of influenza will bind. Strains that are easily transmitted between people have hemagglutinin proteins that bind to receptors in the upper part of the respiratory tract, such as in the nose, throat, and mouth. In contrast, the highly-lethal H5N1 strain binds to receptors that are mostly found deep in the lungs (United States Centers for Disease Control and Prevention Citation2009). This difference in the site of infection may be part of the reason why the H5N1 strain causes severe viral pneumonia in the lungs, but is not easily transmitted by people coughing and sneezing. Common symptoms of the flu such as fever, headaches, and fatigue are the result of the huge amounts of proinflammatory cytokines and chemokines (such as interferon or tumor necrosis factor) produced from influenza-infected cells (). In contrast to the rhinovirus that causes the common cold, influenza does cause tissue damage, so symptoms are not entirely due to the inflammatory response (Louie et al. Citation2009, Rosenberg et al. Citation2009, Jain et al. Citation2009). This massive immune response might produce a life-threatening cytokine storm. This effect has been proposed to be the cause of the unusual lethality of the H5N1 avian influenza. However, another possibility is that these large amounts of cytokines are just a result of the massive levels of viral replication produced by these strains, and the immune response does not itself contribute to the disease (Kumar et al. Citation2009, Perez-Padilla et al. Citation2009, Chowell et al. Citation2009, Davies et al. Citation2009). The pandemic H1N1/09 virus, such as other influenza A viruses, is believed to be transmitted from infected individuals through air by coughs or sneezes, creating aerosols containing the virus (Branksto et al. Citation2007, Tellier Citation2006). Influenza can also be transmitted by saliva, nasal secretions, feces, and blood. Infections occur through contact with these body fluids or with contaminated surfaces. Uncomplicated human influenza virus infection causes transient tracheo-bronchitis, corresponding with predominant virus attachment to tracheal and bronchial epithelial cells. The main complication is the extension of viral infection to the alveoli, often with secondary bacterial super infection, resulting in severe pneumonia. Complications in extra-respiratory tissues such as encephalopathy, myocarditis, and myopathy occur occasionally (Kuiken and Taubenberger Citation2008). The majority of cases of H1N1 have been mild influenza illness, mostly associated with fever. Some patients have gastrointestinal symptoms including diarrhea. Only a small percentage of confirmed cases had been hospitalized. Underlying conditions such as asthma or other lung diseases, diabetes, morbid obesity, auto-immune disorders, immunosuppressive therapy, neurological or cardiovascular disorders or pregnancy are predisposing factors for hospitalization. In Mexico, where the largest numbers of fatalities have been seen, severe pneumonia with multifocal infiltrates and rapid progression to acute respiratory distress syndrome and multi-organ failure has been reported (Peiris et al. Citation2009). The worldwide status of swine flu was reported and deaths due to it are shown in . The most encouraging news for clinicians is that swine flu is treatable at all ages. The advantage of taking antiviral treatment within 48 hours of the onset of symptoms is that it may make the illness milder and it may also prevent serious complications of influenza infection (Taubenberger and Morens Citation2008, Fitzgerald Citation2009).
Discussion
Reassortment of different strains
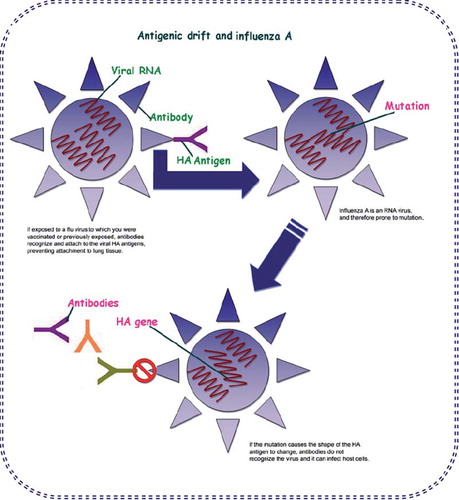
Like all influenza viruses, swine flu viruses change constantly. Pigs can be infected by avian influenza and human influenza viruses as well as swine influenza viruses. When influenza viruses from different species infect pigs, the viruses can reassort (i.e. swap genes) and new viruses that are a mix of swine, human and/or avian influenza viruses can emerge (). A new influenza virus strain emerges either from antigenic drift or from antigenic shift. Antigenic drift causes small mutations which may or may not lead to a new influenza viral strain but antigenic shift causes the emergence of new influenza viral strains.
Influenza pandemic plan strategy
A pandemic is generally defined as a novel infection that spreads globally. On that definition, novel H1N1 is already “pandemic.” The best solution to prevent a pandemic is stopping any virus from spreading in the first place. At the peak of an H1N1 pandemic, assessment of dischargeability of patients is critical and hospitals will have to be prepared to accept patients in exceptionally high number. Hospital staffs will be required to handle very large patient censuses. Emergency physicians, hospitality, critical care specialists, and infectious disease specialist will be called on to play leading roles. The first rapid diagnosis of influenza by detection of viral antigen was performed in 1956 (Liu Citation1956). The method was immunofluorescence and it rapidly grew in popularity. Now, much specific, high quality, commercially available reagents can reliably and rapidly detect viral antigens in exfoliated nasopharyngeal cells collected by nasal swab, aspirate, or wash.
Diagnosis strategy of influenza A [H1N1]
A rapid diagnosis of infection with the swine influenza virus is essential to minimize spread of the condition and protect the patients from developing complications by treating with antivirals medicines. Upper respiratory specimens such as nasopharyngeal aspirates or nasopharyngeal, throat, and nose swabs are suitable for the detection of H1N1 (Dominguez et al. Citation1993). It is pertinent to establish molecular epidemiology study team in regions where high virulent influenza viruses are expected to emerge. Many tests are available in kits that allow the simultaneous detection of many respiratory viruses directly in the sample (Peiris et al. Citation2009, Waner et al. Citation1991, Todd et al. Citation1995). These reagents can differentiate influenza types A and B viruses, so virus typing is possible directly from the sample. The performance of more sensitive immunofluorescence assays requires laboratory expertise in the methodology and experienced technicians trained in immunofluorescence microscopy. Furthermore, the results may be affected by quality and the cellularity of the sample submitted for testing. Specimens with few cells may produce false-negative results leading to rejection of samples with few or no cells. Results are usually available the day after the sample is submitted, and costs can be contained by batching the samples daily (World Health Organization Citation2009, Centers for Disease Control and Prevention Citation2009). Most virology laboratories utilize conventional diagnostic methods for influenza detection including cell culture with immune-fluorescence (IF) staining and antigen subtyping based on hemagglutination inhibition (HI) with antisera raised against various reference strains (Kendal et al. Citation1982). These approaches are not only highly accurate and sensitive for viral detection but are also laborious and time consuming. Many laboratories also utilize commercially available immunoassays for rapid influenza detection. Although these commercial kits are fast and easy to use, they have been shown to be less sensitive and specific compared with traditional culture methods (Herrmann et al. Citation2001, Boivin et al. Citation2001, Kaiser et al. Citation1999). Previous studies have employed multiplex polymerase chain reaction (PCR) for the detection of human influenza viruses (Ellis et al. Citation1997, Zhang and Evans Citation1991, Wright et al. Citation1995, Stockton et al. Citation1998). These studies needed multistep approaches with random hexamers or multiple PCR amplifications with nested primer sets.
Laboratory diagnosis of influenza virus A [H1N1]
The virus when grown on culture helps one make a definitive specific diagnosis. The sensitivity and negative predictive value is also quite high at around 90% (Ginocchio et al. Citation2009). The virus grows on chick embryo as well as monkey kidney cell cultures within 48–72 hours of its inoculation (Ginocchio et al. Citation2009). The use of rapid diagnostic tests to detect antigens of the virus was compared with the standard RT-PCR in 65 patients and it was observed that the method had 60%–80% sensitivity (Balish et al. Citation2009). These findings indicate that although a positive test suggests a diagnosis of H1N1 influenza, a negative result does not rule out the same. Besides this, the test often requires a high virus concentration in the respiratory secretions, and if negative, its results are interpreted based on the clinical suspicion of illness (Balish et al. Citation2009). Real time PCR assays have been developed on the basis of publicly released hemagglutinin sequences of the currently circulating virus. The RNA extracted from nasopharyngeal aspirate samples is amplified and detected by this assay (Poon et al. Citation2009). Other samples which may be used include throat swabs and bronchial aspirates (World Health Organisation: Global alert and response Citation2009). The assay was seen to be highly specific for the swine origin H1N1 virus and was able to distinguish this from the seasonal H1N1 as well as non H1N1 organisms. These assays are rapid with results being available in a few hours (Poon et al. Citation2009). The comparison of antibody titer during the acute illness and 10–14 days later will help make a diagnosis too but the test is not used for diagnostic purposes and is useful primarily in retrospect (Dolin Citation2005, Perez-Padilla et al. Citation2009, Sebastian et al. Citation2009) ().
Table I. Summary of the different methods for detection of Influenza A (H1N1).
Viral culture
Isolation of pandemic H1N1 influenza A virus using culture is also diagnostic, but culture is usually too slow to help guide clinical management. A negative viral culture does not exclude pandemic H1N1 influenza A infection. However, viral culture is still the method of reference for the diagnosis of microorganisms.
Rapid antigen tests
Clinicians may consider using rapid influenza antigen tests as part of their evaluation of patients suspected of having pandemic H1N1 influenza A, but results should be interpreted with caution (United States Centers for Disease Control and Prevention Citation2009, Crum-Cianflone et al. Citation2009). Certain rapid influenza antigen tests that are commercially available can distinguish between influenza A and B viruses, but cannot distinguish among different subtypes of influenza A (e.g., pandemic H1N1 influenza A versus seasonal H1N1 or H3N2 influenza A). Confirmation of pandemic H1N1 influenza A infection can only be made by real-time reverse-transcriptase (rRT)-PCR or culture. The sensitivity of rapid antigen testing for pandemic H1N1 influenza A virus infection has ranged from 10% to 70% compared with rRT-PCR (United States Centers for Disease Control and Prevention Citation2009, Crum-Cianflone et al. Citation2009, Drexler et al. Citation2009, United States Centers for Disease Control and Prevention Citation2009). Thus, a negative result does not rule out infection. The specificity of rapid antigen testing has generally been >95% (United States Centers for Disease Control and Prevention Citation2009), although in one study it was only 86% (Sabetta et al. Citation2009). Among 39 patients with pandemic H1N1 influenza A confirmed by rRT-PCR, 20 had a positive rapid antigen test using the QuickVue Influenza A + B (Quidel) assay (sensitivity 51%) (Faix et al. Citation2009). Twelve of 19 patients who had seasonal H1N1 influenza confirmed by rRT-PCR had a positive rapid antigen test (sensitivity 63%). In the same study, the specificity of rapid antigen testing was 99% for patients with either the pandemic strain or a seasonal strain of H1N1 influenza A of 21 critically ill patients with pandemic H1N1 influenza infection, upper respiratory tract specimens were positive in 17 of 21 patients (81%) using rRT-PCR, but in only 5 of 20 patients (25%) using rapid antigen testing (Blyth et al. Citation2009).
Immunofluorescent antibody testing
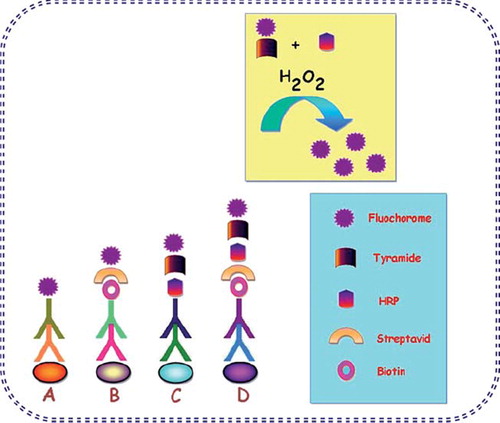
Primary, or direct, immunofluorescence uses a single antibody that is chemically linked to a fluorophore. The antibody recognizes the target molecule and binds to it, and the fluorophore it carries can be detected via microscopy (). This technique reduces the number of steps in the staining procedure making the process faster and can reduce background signal by avoiding some issues with antibody cross-reactivity or non-specificity. However, since the number of fluorescent molecules that can be bound to the primary antibody is limited, direct immunofluorescence is less sensitive than indirect immunofluorescence (Fritschy and Härtig Citation2001). Direct or indirect immunofluorescent antibody testing (DFA or IFA) can distinguish between influenza A and B, but does not distinguish among different influenza A subtypes (United States Centers for Disease Control and Prevention 2009, Riffelmann et al. Citation2005). In one study, among 42 samples that were positive for pandemic H1N1 influenza A by real-time reverse-transcriptase polymerase chain reaction (rRT-PCR), 39 (93%) were positive by direct fluorescent antibody testing (Harmon and Kendal Citation1989). However, in another study, among 21 critically ill patients with pandemic H1N1 influenza, lower respiratory tract specimens were positive in all patients when tested by rRT-PCR, but in only 5 of 20 patients (25%) tested by immunofluorescence (Whiley et al. Citation2009).
However, a negative DFA or IFA does not exclude pandemic H1N1 influenza A infection since larger studies are required to define the sensitivity to detect this virus.
RT-PCR: a standard golden method for detection of H1N1
Accurate and rapid diagnosis of novel H1N1 infections is critical for minimizing further spread through timely implementation of antiviral treatment and public health based measures. Whiley et al. (Citation2009) have developed two sensitive and specific TaqMan-based reverse transcription PCRs (RT-PCR) for the detection of novel H1N1 targeting the hemagglutinin and neuraminidase genes. Carr et al. (Citation2009) applied a one-step real-time RT-PCR approach to target the matrix gene of the novel influenza virus and validated this assay against a panel of seasonal human influenza A, swine influenza, and avian influenza. This assay successfully recognizes clinical novel H1N1 specimens, which were confirmed by sequencing, and does not cross-react with other influenza subtypes and displayed a similar detection limit as universal influenza PCR assays (Stone et al. Citation2004). The current outbreak of novel influenza A (H1N1) virus continues to expand globally. During outbreaks of emerging infectious diseases, accurate and rapid diagnosis is critical for minimizing further spread through timely implementation of appropriate vaccines and antiviral treatment and prophylaxis where available, and other public health based, non-pharmaceutical measures. Real-time polymerase chain reaction (PCR) is more rapid and sensitive than traditional techniques including virus isolation by cell culture.
A limitation of PCR methods is that false-negative results may occur due to sequence variation in primer and probe targets and is particularly relevant for the detection of emerging viruses. Extraction step of RNA may also turn out to be the limitations of PCRs. In this study, Whiley et al. (Citation2009) developed two TaqMan-based reverse transcription PCR (RT-PCR) methods for the detection of novel influenza A (H1N1) virus targeting the HA and NA genes (Harmon and Kendal Citation1989).
Future implications: the challenges/solutions
There still remains a need for a rapid and sensitive penside diagnostic test to identify swine flu. A field-deployable biosensor is expected to provide a low-cost device for assessment of swine influenza infection. Such a biosensor is likely to be very worthwhile and can be applied to any pandemic disease to increase the rapidity and sensitivity of medical diagnosis. This model can be implemented quickly in response to detection in large patient volume.
Conclusions
The rapid and accurate laboratory diagnosis of influenza is necessary through a variety of laboratory modalities. The pandemic H1N1/09 influenza virus is now the dominant strain in most areas of the world. It is identified by HA and NA proteins whose combination of H1N1 has been of greatest concern. If the diagnosis of a viral illness is sought primarily and made quickly, then unnecessary antibiotic therapy can be avoided, or if started empirically, can be terminated and the duration of antibiotic use shortened. Rapid testing for influenza viruses also allows a hospital to identify and to isolate infected patients from other patients to reduce the spread of infection. Such a testing is done so far by viral culture, rapid antigen tests, and RT-PCR but all have limitations. An influenza biosensor may be a better option due to low cost, rapidity, and high sensitivity factors.
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
References
- Balish A, Warnes CM, Wu K, Barnes N, Emery S, Berman L, et al. 2009. Evaluation of rapid influenza diagnostic tests for detection of Novel Influenza A (H1N1) Virus — United States, 2009. MMWR. 58:826–829.
- Blyth CC, Iredell JR, Dwyer DE. 2009. Rapid-test sensitivity for novel swine-origin influenza a (H1N1) virus in humans. N Engl J Med. 361:2493.
- Boivin C, Hardy I, Kress A. 2001. Evaluation of a rapid optical immunoassay for influenza viruses (FLU OIA Test) in comparison with cell culture and reverse transcription-PCR. J Clin Microbiol. 39: 730–732.
- Branksto G, Gitterman L, Hirji Z, Lemieux C, Gardam M. 2007. Transmission of influenza A in human beings. Lancet Infect Dis. 7:257–265.
- Carr MJ, Gunson R, Maclean A, Coughlan S, Fitzgerald M, Scully M, et al. 2009.Development of a real-time RT-PCR for the detection of swine-lineage influenza A (H1N1) virus infections. J Clin Virol. 45:196–199.
- Centers for Disease Control and Prevention. 2009. Novel H1N1 Flu: Facts and Figures. (http://cdc.gov/h1n1flu/surveillanceqa.htm).
- Centers for Disease Control and Prevention. 2009. H1N1 flu (Swine Flu): a pandemic is declared. (www.cdc.gov/H1N1FLU).
- Chowell G, Bertozzi SM, Colchero MA, Lopez-Gatell H, Alpuche-Aranda C, Hernandez M, et al. 2009. Severe respiratory disease concurrent with the circulation of H1N1 influenza. N Engl J Med. 361:674–679.
- Crum-Cianflone NF, Blair PJ, Faix D, Arnold J, Echols S, Sherman SS, et al. 2009. Clinical and epidemiologic characteristics of an outbreak of novel H1N1 (swine origin) influenza A virus among United States military beneficiaries. Clin Infect Dis. 9:1801–1810.
- Davies A, Jones D, Bailey M, Beca J, Bellomo R, Blackwell N, et al. 2009. Australia and New Zealand Extracoporeal membrane oxygenation (ANZ ECMO) influenza invesigators. JAMA. 302:1888–1895.
- Dolin R. 2005. Influenza. In: Kasper DL, Braunwald E, Fauci AS, Hauser SL, Longo DL, Jameson LJ. Eds. Harrisson's Principles of Internal Medicine. New York: McGraw Hill, p 1066.
- Dominguez EA, Taber LH, Couch RB. 1993. Comparison of rapid diagnostic techniques for respiratory syncytial and influenza A virus respiratory infections in young children. J Clin Microbiol. 31:2286–2290.
- Drexler JF, Helmer A, Kirberg H, Reber U, Panning M, Müller M, et al. 2009. Poor clinical sensitivity of rapid antigen test for influenza A pandemic (H1N1) 2009 virus. Emerg Infect Dis. 15:1662–1664.
- Ellis JS, Fleming DM, Zambon MC. 1997. Multiplex reverse transcription-PCR for surveillance of influenza A and B viruses in England and Wales in 1995-1996. J Clin Microbiol. 35:2076–2082.
- Faix DJ, Sherman SS, Waterman SH. 2009. Rapid test sensitivity for novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 361:728–729.
- Fitzgerald DA. 2009. Human swine influenza A [H1N1]: practical advice for clinicians early in the pandemic. Paedi Resp Rev. 10:154–158.
- Fritschy J-M, Härtig W. 2001. Immunofluorescence. In: Delves P. Ed. Indirect Immunofluorescence of Cultured Cells 2001. eLS. Wiley, USA.
- Ginocchio CC, Zhang F, Manji R, Arora S, Bornfreund M, Falk L, et al. 2009. Evaluation of multiple test methods for the detection of the novel 2009 influenza A (H1N1) during the New York City outbreak. J Clin Virol. 45:191–195.
- Harmon MW, Kendal AP. 1989. Influenza viruses. In: Schmidt NJ, Emmons RW, Eds. Diagnostic procedures for viral rickettsial and chlamydial infections. 6th ed. Washington DC: p. 631–668.
- Herrmann B, Larsson C, Zweygberg BW. 2001. Simultaneous detection and typing of influenza viruses A and B by a nested reverse transcription-PCR: comparison to virus isolation and antigen detection by immunofluorescence and optical immunoassay (FLU OIA). J Clin Microbiol. 39:134–138.
- Jain S, Kamimoto L, Bramley AM, Schmitz AM, Benoit SR, Louie J, et al. 2009. Hospitalized patients with 2009 H1N1 influenza in the United States. N Engl J Med. 361:1935–1944.
- Kaiser L, Briones MS, Hayden FG. 1999. Performance of virus isolation and Directigen Flu A to detect influenza A virus in experimental human infection. J Clin Virol. 14:191–194.
- Kendal AP, Pereura MS, Skehel J. 1982. Concepts and Procedures for Laboratory Based Influenza Surveillance. Geneva, Switzerland: World Health Organization.
- Kuiken T, Taubenberger JK. 2008. Pathology of human influenza revisited. Vaccine. 26:D59–D66.
- Kumar A, Zarychanski R, Pinto R, Cook DJ, Marshall J, Lacroix J, et al. 2009. Critically ill patients with 2009 influenza A(H1N1) infection in Canada. JAMA. 302:1872–1879.
- Liu C. 1956. Rapid diagnosis of human influenza infection from nasal smears by means of fluorescein-labeled antibody. Proc Soc Exp Biol Med. 92:883–887.
- Louie JK, Meileen A, Kathleen W, Cynthia J, Shilpa G, Robert S, et al. 2009. Factors associated with death or Hospitalization due to pandemic influenza A(H1N1) infection in California. JAMA. 302:1896–1902.
- Peiris JSM, Poon LM, Guan YJ. 2009. Emergence of novel swine-origin influenza A virus (S-OIV) H1N1 virus in humans. Clin Virol. 45: 169–173.
- Perez-Padilla R, Rosa-Zamboni D, Leon SP, Hernandez M, Quiñones-Falconi F, Bautista E, et al. 2009. INER working group on influenza. Pneumonia and respiratory failure from swine-origin influenza A (H1N1) in Mexico. N Engl J Med. 361:680–689.
- Perez-Padilla R, Zamboni DR, Ponce de Leon S, Hernandez M, Quiñones-Falconi F, Bautista E, et al. 2009. Pneumonia and respiratory failure from swine-origin influenza A (H1N1) in Mexico. NEJM. 361:680–689.
- Poon LLM, Chan KH, Smith GJ, Leung CSW, Guan Y, Yuen KY, et al. 2009. Molecular detection of a Novel Human Influenza (H1N1) of pandemic potential by conventional and real-time quantitative RT-PCR assays. Clin Chem. 55:1555–1558.
- Riffelmann M, Wirsing von König CH, Caro V, Guiso N. 2005. Nucleic acid amplification tests for diagnosis of Bordetella infections. J Clin Microbiol. 43:4925–4929.
- Rosenberg M, Tram C, Kuper A, Daneman N. 2009. Rash associated with pandemic (H1N1) influenza. CMAJ. 182:E146.
- Sabetta JR, Smardin J, Burns L, Barry K. 2009. Performance of rapid influenza diagnostic tests during two school outbreaks of 2009 pandemic influenza A (H1N1) virus infection – Connecticut, 2009. MMWR. 58:1029–1032.
- Sebastian MR, Lodha R, Kabra SK. 2009. Swine origin influenza (swine flu). Indian J Pediatr. 76:833–841.
- Stockton J, Ellis JS, Saville M, Clewley JP, Zambon MC. 1998. Multiplex PCR for typing and subtyping influenza and respiratory syncytial viruses. J Clin Microbiol. 36:2990–2995.
- Stone B, Burrows J, Schepetiuk S, Higgins G, Hampson A, Shaw R, et al. 2004. Rapid detection and simultaneous subtype differentiation of influenza A viruses by real time PCR. J Virol Methods. 117:103–112.
- Taubenberger JK, Morens DM. 2008. The pathology of influenza virus infections. Rev Pathol Mech Dis. 3:499–522.
- Tellier R. 2006. Review of aerosol transmission of influenza A virus. Emerg Infect Dis. 12:1657–1662.
- Todd SJ, Minnich L, Waner JL. 1995. Comparison of rapid immunofluorescence procedure with TestPack RSV and Directigen FLU-A for diagnosis of respiratory syncytial virus and influenza A virus. J Clin Microbiol. 33:1650–1651.
- United States Centers for Disease Control and Prevention. 2009. Updated interim recommendations for the use of antiviral medications in the treatment. and prevention of influenza for the 2009–2010 (www.cdc.gov/h1n1flu/recommendations.htm).
- United States Centers for Disease Control and Prevention. 2009. Interim Recommendations for Clinical Use of Influenza Diagnostic Tests During the 2009-10 Influenza Season. http://www.cdc.gov/h1n1flu/guidance/diagnostic/tests.htm(Accessed September 30, 2009).
- United States Centers for Disease Control and Prevention. 2009. Interim guidance for the detection of novel influenza A virus using rapid influenza diagnostic tests. (http://www.cdc.gov/h1n1flu/guidance/rapid/testing.htm).
- Waner JL, Todd SJ, Shalaby H. 1991. Comparison of Directigen FLU-A with viral isolation and direct immunofluorescence for the rapid detection and identification of influenza A virus. J Clin Microbiol. 29:479–482.
- Whiley DM, Bialasiewicz S, Bletchly C, Faux CE, Harrower B, Gould AR, Lambert SB, Nimmo GR, Nissen MD, Sloots TP. 2009. Detection of novel influenza A(H1N1) virus by real-time RT-PCR. J Clin Virol. 45:203–204.
- World Health Organization. 2009. Laboratory-confirmed cases of pandemic (H1N1) 2009 as officially reported to WHO by States Parties to the IHR (2005) as on 6 August 2009. (http://www.who.int/csr/don/2009/08/12/en/index.html).
- World Health Organisation: Global alert and response. 2009. Clinical management of human infection with new influenza A (H1N1) virus: initial guidance. (http://www.who.int/csr/resources/publications/swineflu/clinical/management/en/index.html).
- Wright KE, Wilson G, Novosad D, Dimock C, Tan D, Weber JM. 1995. Typing and subtyping of influenza viruses in clinical samples by PCR. J Clin Microbiol. 33:1180–1184.
- Zhang W, Evans DH. 1991. Detection and identification of human influenza viruses by polymerase chain reaction. J Virol Methods 33:165–189.