Abstract
Context: In its powdered form, turmeric [Curcuma longa L. (Zingiberaceae)], a spice of medical importance, is often adulterated lowering its quality.
Objective: The study sought to detect plant-based adulterants in traded turmeric powder using DNA barcoding.
Materials and methods: Accessions of Curcuma longa L., Curcuma zedoaria Rosc. (Zingiberaceae), and cassava starch served as reference samples. Three barcoding loci, namely ITS, rbcL, and matK, were used for PCR amplification of the reference samples and commercial samples representing 10 different companies. PCR success rate, sequencing efficiency, occurrence of SNPs, and BLAST analysis were used to assess the potential of the barcoding loci in authenticating the traded samples of turmeric.
Results: The PCR and sequencing success of the loci rbcL and ITS were found to be 100%, whereas matK showed no amplification. ITS proved to be the ideal locus because it showed greater variability than rbcL in discriminating the Curcuma species. The presence of C. zedoaria could be detected in one of the samples whereas cassava starch, wheat, barley, and rye in other two samples although the label claimed nothing other than turmeric powder in the samples.
Discussion and conclusion: Unlabeled materials in turmeric powder are considered as adulterants or fillers, added to increase the bulk weight and starch content of the commodity for economic gains. These adulterants pose potential health hazards to consumers who are allergic to these plants, lowering the product's medicinal value and belying the claim that the product is gluten free. The study proved DNA barcoding as an efficient tool for testing the integrity and the authenticity of commercial products of turmeric.
Introduction
Turmeric [Curcuma longa L. (Zingiberaceae)], already well known as a spice, a coloring agent for food, and a cosmetic, is becoming increasingly important as a medicinal herb for its anti-inflammatory, anticancerous, anti-oxidant, antimicrobial, and antiviral properties; as an antiseptic; and in the treatment of diabetes and Alzheimer's disease (Sasikumar, Citation2005). Turmeric has a history of 5000 years as a herb in the folk medicine as well as in Indian and Chinese systems of medicine.
India is the largest producer, consumer, and exporter of turmeric, which is traded mainly in the form of whole-dried rhizomes, as powder and in valued-added forms. In the powder form, which is the form mainly used in commerce and by the food, cosmetic and pharmaceutical industries, turmeric consists of particles approximately 0.2–0.25 mm (60–80 mesh). The powdered form has the highest share in exports, constituting about 42% of the world trade in turmeric.
Recent reports on the medicinal value of turmeric in treating a variety of ailments have further increased the demand for turmeric all over the world. Major importers of turmeric powder are USA, UAE, Saudi Arabia, UK, Australia, and Canada (Plotto, Citation2004).
Consumer preference for organic products has also spurred the demand for turmeric at the same time, as with other organic products, especially those that are sold in the powdered form, turmeric too is being adulterated, especially with filler materials that go visually undetected. Such filler materials add bulk, resulting in diluting the main product and thereby making it less effective, which, in turn, erodes consumer confidence. Adulteration is illegal as the consumption of adulterated products containing undeclared or unlabeled constituents may cause intoxication, allergies, or other undesirable side effects (Asensio et al., Citation2008; Marcus & Grollman, Citation2002). The American Spice Trade Association (ASTA) and the European Spice Association (ESA) have laid down stringent specifications to ensure the purity and the standards of spices. ASTA, for example, rejects a consignment of turmeric powder even if it contains as little as 0.5% of foreign matter (Plotto, Citation2004).
Another trend that encourages the search for efficient methods of detecting adulteration is the globalization of food trade, which also places a premium on the traceability of origin and a guarantee of the quality and authenticity of commercially available food products to ensure that they are safe for consumption (Barbuto et al., Citation2010). The process of tracing the origin of food products has been considered a proactive action to certify the origin and quality of products and to prevent fraudulent or deceptive labeling (European Commission, Citation2002). Therefore, detecting adulteration is a prerequisite to food safety also. Adulteration has been reported in commercially sold turmeric powder, adversely affecting the quality of the product and its trade (Dhanya et al., Citation2011; Sasikumar et al., Citation2005). Plant-based adulterants or fillers include wild species of Curcuma, namely C. zedoaria, starch from cheaper sources such as cassava, and sawdust (Dhanya & Sasikumar, Citation2010). Such plant-based adulterants and fillers are more difficult to detect whereas highly sensitive methods are already available to detect the synthetic adulterants in turmeric and other herbal spices.
Protein-based methods such as immunological assays, electrophoretic, and chromatographic techniques such as TLC and HPLC require fresh samples. Also, the results of these methods are less reliable because the composition of the same product from different manufacturers varies substantially. With highly processed items, the methods are not sensitive enough. Routine DNA-based methods such as RAPD, SCAR, and SSR/SNPs suffer from the lack of uniformity and inadequate standardization (Galimberti et al., Citation2013).
These problems have prompted the development of broadly accessible commercial tools such as DNA barcoding to detect product substitution and to authenticate commercial products (Newmaster et al., Citation2013). DNA barcoding uses short stretches of standardized gene sequences to identify plant materials. The variability of the gene sequences is analyzed to determine a unique “barcode” for each species (Hebert et al., Citation2003). The technique is currently used for detecting plant-based adulteration in many commodities including medicinal plants, commercial tea packets, olive oil, and medicinal spices (Gismondi et al., Citation2013; Kumar et al., Citation2011; Mattia et al., Citation2010; Stoeckle et al., Citation2011; Srirama et al., Citation2010; Vijayan & Tsou, Citation2010; Yuan et al., Citation2011). A recent study (Parvathy et al., Citation2014) has shown the sensitivity of this technique to detect adulteration as low as 0.5% in market samples of black pepper. Other studies have also attested the efficacy of barcoding in authenticating some socio-economically important herbs (Lockley & Bardsley, Citation2000; Mafra et al., Citation2008; Newmaster et al., Citation2013; Parvathy et al., Citation2014; Stoeckle et al., Citation2011; Wallace et al., Citation2012).
The present paper describes the use of DNA barcoding to detect adulterants including fillers in samples of turmeric powder sold in the market and to authenticate the validity of the labels affixed to these samples. The barcodes rbcL, matK, and ITS were selected based on earlier reports of their effectiveness. rbcL and matK were suggested as the ideal loci by the Consortium for Barcode of Life (CBOL) and ITS has shown greater variability even among closely related species (Newmaster et al., Citation2013; Tripathi et al., Citation2013).
Materials and methods
Study design
Whole fresh rhizomes of five accessions of C. longa (IC 143, 119, 126, 360, and 361) and four accessions of C. zedoaria (IC 465, 760, 765, and 1517) were obtained from the germplasm collection maintained at the Indian Institute of Spices Research, Kozhikode, Kerala, India, and five samples of cassava were obtained from the local market at the time of the study. The barcodes generated from these samples served as reference barcodes for determining the authenticity of commercially available turmeric powder. The reference samples for other species were selected based on earlier reports of commonly used adulterants in commercially available turmeric powder.
The authenticity of 10 popular brands (labeled MS 1–MS 10 to denote the 10 market samples) of turmeric powder procured locally from Kozhikode was tested. Each of the 10 brands represented a different firm.
DNA barcoding
Total genomic DNA was isolated in triplicate from the above-mentioned samples using the protocol of Remya et al. (Citation2004). An absorbance ratio (A260/A280) of 1.7–1.8 indicated insignificant levels of contaminating proteins and polysaccharides in the isolated DNA. The quality of DNA was confirmed on 0.8% agarose gel.
The three barcoding loci (ITS, rbcL, and matK) were used for PCR amplification using universal primers procured from Integrated DNA Technologies (IDT), Coralville, IA (). The reaction mix contained 10–50 ng of DNA, 1 × assay buffer with 1.5 mM MgCl2, 1 mM dNTP, 1 pmol µL−1 each of the forward, and reverse primers, and 1 U Taq DNA polymerase (Genei, Bangalore, India). The reaction was performed in an Eppendorf thermal cycler (Eppendorf, Hamburg, Germany) under conditions optimized for each locus ().
Table 1. Primers used for PCR amplification.
Table 2. PCR reaction conditions for the barcoding loci.
The amplified products were resolved using 1% agarose gel (containing 0.5 μg mL−1 ethidium bromide) in 1 × Tris-borate-ethylene diamine tetra acetic acid buffer (TBE) with 100 bp ladder (Fermentas, Vilnius, Lithuania) as a size marker. The gels were documented using the Syngene gel documentation system. PCR products of the expected size were extracted using QIA quick PCR purification kit (QIAGEN, Munich, Germany). The purified products were custom-sequenced at SciGenom Labs (Kochi, Kerala, India).
The purified PCR products were ligated to a pGEM-T vector system (Promega, Madison, WI) and transformed into an E. coli strain JM109 made competent using the competent cells preparation kit (Genei, Bangalore, India). The recombinants were identified using blue-white screening on LB plates supplemented with X-gal, IPTG, and ampicillin (Fermentas, Vilnius, Lithuania). Colony PCR was also done to screen the recombinants. The reaction mix contained 1 × assay buffer, 0.2 mM dNTPs, 1.5 mM MgCl2, and 5 pmol each of the M13 forward and reverse primers (IDT) with 1 U Taq polymerase in a total reaction volume of 25 µL. Thermal cycling conditions were as follows: 94 °C for 3 min followed by 30 cycles of denaturation at 94 °C for 1 min, primer annealing at 48 °C for 1 min, primer extension at 72 °C for 1 min, and final extension at 72 °C for 5 min. The amplified products were resolved on 1.5% agarose gel, and the positive transformants were identified based on the size of the insert. Plasmid DNA from the clones was isolated using the QIAprep Spin Miniprep Kit (QIAGEN, Munich, Germany). The recombinant plasmids were bi-directionally sequenced at SciGenom Labs (Kochi, Kerala, India).
Construction of the reference library
Consensus sequences and contig generation were accomplished by a DNA baser software package (SAS Inc., Cary, NC). Initial screening of the sequences was carried out using BLAST algorithm (Atschul et al., Citation1997) to identify the closest matching sequences in the nucleotide database of GenBank. The sequences were aligned using Clustal W (Larkin et al., Citation2007) and trimmed and edited using BioEdit (ver.7.2.5) (Agilent Technologies Inc., Santa Clara, CA) (Hall, Citation1999). Data were analyzed using MEGA ver. 5 (MEGA, Germantown, TN) (Tamura et al., Citation2011).
The sequences generated for the reference samples for rbcL and ITS were used for identifying the adulterants in the commercial samples. The reference samples were collected from different geographical regions and were thus rich in diversity and variability, enabling correct detection of SNPs in different species of Curcuma. The authenticity of all the sequences was also cross-checked with the available sequences in the National Centre for Biotechnology Information (NCBI, Bethesda, MD) and the Barcode of Life Data (BOLD, Ontario, Canada) system.
Analysis of commercial samples
The initial screening of the sequences generated from the commercial samples was carried out using BLAST and against the reference sequences. Cloned sequences that showed maximum sequence identity with other species (adulterants) rather than with C. longa were analyzed along with the reference sequences by BioEdit (Agilent Technologies Inc., Santa Clara, CA) and MEGA (MEGA, Germantown, TN) to detect the SNPs, and the multiple sequence alignment (MSA) tool was used for comparing the sequences against the maximum identity sequence.
Results
Barcode success
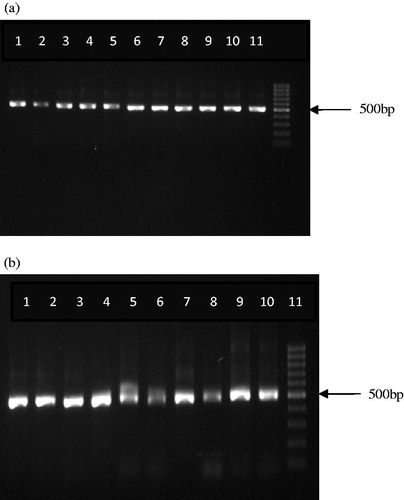
DNA of high molecular weight was isolated from all the genuine and market samples of turmeric. Amplification and sequencing efficiency of rbcL and ITS were found to be 100% whereas the success rate of PCR amplification with two different primer sets of matK was 0% despite using different concentrations of the DNA and less stringent conditions. rbcL locus yielded a sequence length of 600 bp whereas ITS yielded a sequence length of 500 bp for all the samples analyzed ( and ).
Figure 1. (a) Amplification of rbcL locus in C. longa and C. zedoaria (lanes 1–5 – C. longa acc. no. 119, 143, 126, 360, and 161; lanes 6–9 – C. zedoaria acc. no. 760, 465, 1517, and 765, and lane 10 – 100 bp ladder). (b) Amplification of rbcL locus in cassava starch and market samples of turmeric (lanes 1 – 100 bp ladder; lane 2 – cassava starch; lanes 3–12 – market sample (MS) 1 – 10; lane 13 – 100 bp ladder).
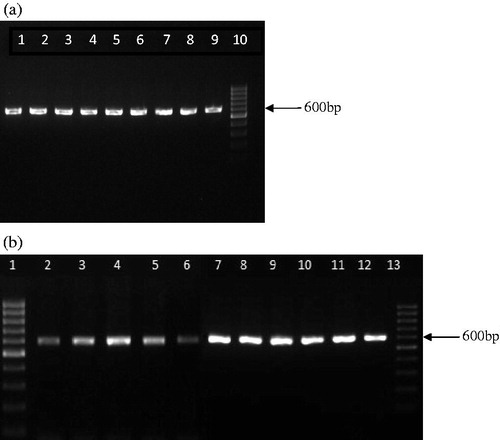
Figure 2. (a) Amplification of ITS locus in C. longa, C. zedoaria, and cassava starch (lanes 1–5 – C. longa acc. no. 119, 143, 126, 360, and 161; lanes 6–9 – C. zedoaria acc. no. 760, 465, 1517, and 765; lane 10 – cassava starch; lane 11 – 100 bp ladder). (b) Amplification of ITS locus in market samples of turmeric (lanes 1–10 – market sample (MS) 1–10; lane 11 – 100 bp ladder).
All the barcodes generated were of high quality and could be aligned bi-directionally. The quality of the sequences was confirmed by checking the peaks in the chromatogram and the background noise: the peaks were non-overlapping and broad, and there was little background noise. The sequences generated for genuine samples were 100% similar with those of the respective species in the reference library as well as in the BOLD or GenBank databases. The generated sequences were deposited in the NCBI database (KM226968–KM226977).
When the discriminatory powers of the loci were compared, ITS showed greater variability in the sequences with greater number of variable sites and SNPs (). ITS was able to discriminate between C. longa and its common adulterant, C. zedoaria. The low resolution of rbcL was expected given the insufficient nucleotide variation between the closely related species.
Table 3. SNPs that discriminates C. longa from C. zedoaria.
Authentication of powdered market samples
The market samples were tested using BLAST and SNP analysis. BLAST helps in identifying any unknown contaminant from a wider range of taxa, and SNP analysis is considered a valuable tool in genetic analysis of closely related species.
Of the 10 commercial samples, seven showed only one type of sequence (the barcode). All the five colonies picked and sequenced for each sample showed sequences that were identical to those of C. longa; none of the colonies showed any other barcode apart from the one that belonged to the authentic, or the reference sample, thus confirming that the product was indeed what the label claimed it to be.
Contamination or adulteration
The labels mention “turmeric” (C. longa) as the sole ingredient; therefore, if any sample yields a sequence other than that of C. longa, the sample is to be considered contaminated or adulterated. Since the label does not mention a filler, even that is to be considered a contaminant.
Thus any sample that generates more than one barcode is considered contaminated or adulterated. Of the 10 samples tested, one was found to be contaminated with species other than the labeled ingredient. This market sample generated sequences of C. zedoaria, a reported adulterant of C. longa. SNPs in ITS locus specific to C. zedoaria species were also found in this sample, confirming that the sample was adulterated with a cheaper and easily available substitute, namely C. zedoaria.
In another sample, some of the clones generated sequences that were identical (100% similarity) to those of Triticum aestivum L. (Poaceae) (wheat), Secale cereale L. (Poaceae) (rye), and 91% similar to those of Hordeum vulgare L. (Poaceae) (barley). Yet, another sample showed sequences that were virtually identical (99% similarity) to those of Manihot esculenta Crantz (Euphorbiaceae) (cassava), pointing to possible adulteration of turmeric powder with cassava starch. Overall, three samples were found to be adulterated with plant-based adulterants.
Discussion
One of the key objectives of this study was to find an appropriate locus to identify the adulterants in turmeric powder sold in the market. An ideal DNA barcode for this purpose should have universal priming sites, a high amplification rate, sequencing success, and enough variability to discriminate between samples (Vijayan & Tsou, Citation2010). We found ITS locus ideal: rbcL showed less variability and matK could not be amplified adequately. The low resolution of the plastid rbcL, which has been ascribed to its low mutation rate when compared with that of the nuclear ITS, was discussed by Newmaster et al. (Citation2013), who also observed that ITS has greater sequence variability, allowing to differentiate between closely related species. The efficiency of ITS over rbcL in differentiating tropical tree species of India was reported by Tripathi et al. (Citation2013). Our study confirmed these reports as ITS could distinguish between C. longa and C. zedoaria. The inefficiency of matK and its amplification problems have also been widely reported (Ford et al., Citation2009; Kress & Erickson, Citation2007; Mattia et al., Citation2010; Pettengill & Neel, Citation2010). Studies by Wallace et al. (Citation2012) on authenticating ginseng using ITS proved not only the efficiency of the locus in discriminating between the two species of ginseng but also the effectiveness of barcoding in detecting adulterants and fillers.
In the present study, ITS yielded SNPs specific to C. zedoaria. These SNPs recovered using four to five genuine specimens, in triplicate, proved useful in detecting adulteration with C. zedoaria in one of the 10 samples. These SNPs can be used in designing specific primers for developing diagnostic kits to detect adulteration of turmeric with C. zedoaria.
The reliability of DNA barcoding for detecting adulteration also depends on the presence of a reliable reference database library, which is lacking in the case of spices. This problem is one of the main limitations of barcoding in authenticating medicinal herbs and spices.
We checked the authenticity of market samples using barcode databases in GenBank and BOLD in addition to the reference database library made by us for the reported adulterants and fillers such as C. zedoaria and cassava starch. Cloning experiments along with BLAST analysis ensured that none of the sequences of any of the species present in a single product were missed. The barcode sequences generated in this study were deposited in the NCBI database as a contribution to construct a universal barcode library.
Because C. zedoaria is cheap and easily available, manufactures of turmeric powder may be tempted to mix it with C. longa. Curcuma zedoaria has been reported to be toxic (Lakshmi et al., Citation2011) and if mixed with turmeric powder, it lowers the medicinal value of turmeric thereby eroding consumer confidence and constituting a health hazard. Other adulterants that we found, namely cassava starch, wheat, rye, and barley, may have been added deliberately – despite the label's claim that the packet contains nothing but turmeric powder – to increase bulk so as to obtain higher profits.
Purity of a natural product is the cornerstone of its perceived biological efficacy. Although authenticating value-added herbal products such as powdered spices is a daunting task in the face of unethical practices including substitution, adulteration, use of fillers, and mislabeling, DNA barcoding may prove handy to meet these challenges in the near future.
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Acknowledgements
We are grateful to the Ministry of Food Processing Industries (MOFPI), Government of India, for the financial support. We also acknowledge Director, Indian Institute of Spices Research (IISR), Kozhikode, Kerala, for providing necessary facilities.
References
- Asensio L, Gonzalez I, Garcya T, Martyn R. (2008). Determination of food authenticity by enzyme-linked immunosorbent assay (ELISA). Food Control 19:1–8
- Atschul SF, Madden TL, Schaffer AA, et al. (1997). Gapped BLAST and PSI BLAST: A new generation of protein database search programs. Nucleic Acids Res 25:3389–402
- Barbuto M, Galimberti A, Ferri EE, et al. (2010). DNA barcoding reveals fraudulent substitutions in shark seafood products: The Italian case of “palombo” (Mustelus spp). Food Res Int 43:376–81
- Cuenoud P, Savolainen V, Chatrou LW, et al. (2002). Molecular phylogenetics of caryophyllales based on nuclear 18S rDNA and plastid rbcL, atpβ, and matK DNA sequences. Am J Bot 89:132–44
- Dhanya K, Sasikumar B. (2010). Molecular marker based adulteration detection in traded food and agricultural commodities of plant origin with special reference to spices. Curr Trends Biotechnol Pharm 4:454–89
- Dhanya K, Syamkumar S, Siju S, Sasikumar B. (2011). Sequence characterised amplified region markers: A reliable tool for adulterant detection in traded turmeric powder. Food Res Int 44:2889–95
- European Commission (2002). Regulation (EC) No. 178/2002 of the European Parliament and of the Council of 28 January 2002 laying down the general principles and requirements of food law, establishing the European Food Safety Authority and laying down procedures in matters of food safety. OJ L 31:1–24
- Ford CS, Ayres KL, Toomey N, et al. (2009). Selection of candidate coding DNA barcoding regions for use on land plants. Bot J Linn Soc 159:1–11
- Galimberti A, De Mattia FA, Losa, Bruni I. (2013). DNA barcoding as a new tool for food traceability. Food Res Int 50:55–63
- Gismondi A, Fanali F, Labarga JMM, et al. (2013). Crocus sativus L. genomics and different DNA barcode applications. Plant Syst Evol 299:1859–63
- Hall TA. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acid Symp Ser 41:95–8
- Hebert PDN, Cywinska A, Ball SL, de Waard JR. (2003). Biological identification through DNA barcodes. Proc R Soc Lond 270:313–21
- Kress WJ, Erickson DL. (2007). A two-locus global DNA barcode for land plants: The coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE 2:1–10
- Kumar S, Kahlon T, Chaudhary S. (2011). A rapid screening for adulterants in olive oil using DNA barcodes. Food Chem 127:1335–41
- Lakshmi S, Padmaja G, Remani P. (2011). Antitumour effects of isocurcumenol isolated from Curcuma zedoaria rhizomes on human and murine cancer cells. Int J Med Chem [Online] Available from: http://www.hindawi.com/journals/ijmc/2011/253962/ [last accessed 12 Jun 2014]
- Larkin MA, Blackshields G, Brown NP, et al. (2007). Clustal W & Clustal X version 2.0. Bioinformatics 23:2947–8
- Lockley AK, Bardsley RG. (2000). DNA-based methods for food authentication. Trends Food Sci Tech 11:67–77
- Mafra I, Ferreira IMPVO, Oliveira MBPPO. (2008). Food authentication by PCR-based methods. Eur Food Res Technol 227:649–65
- Marcus DM, Grollman AP. (2002). Botanical medicines – The need for new regulations. N Engl J Med 347:2073–6
- Mattia DF, Bruni I, Galimberti A, et al. (2010). A comparative study of different DNA barcoding markers for the identification of some members of Lamiacaea. Food Res Int 44:693–702
- Newmaster SG, Grguic M, Shanmughanadhan D, Ramalingam S. (2013). DNA barcoding detects contamination and substitution in North American herbal products. BMC Med 11:222–35
- Parvathy VA, Swetha VP, Sheeja TE, et al. (2014). DNA barcoding to detect chilli adulteration in traded black pepper powder. Food Biotechnol 28:25–40
- Pettengill JB, Neel MC. (2010). An evaluation of candidate plant DNA barcodes and assignment methods in diagnosing 29 species in the genus Agalinis (Orobanchaceae). Am J Bot 97:1392–406
- Plotto A. (2004). Turmeric-post harvest operations. In: Mazaud F, Rottger A, Steffel K, eds. INPhO Post Harvest Compendium. Rome, Italy: Food and Agriculture Organizations of United Nations. FAO, 2–8
- Remya R, Syamkumar S, Sasikumar B. (2004). Isolation and amplification of DNA from turmeric powder. Br Food J 106:673–8
- Sasikumar B. (2005). Genetic resources of Curcuma: Diversity, characterization and utilization. Plant Genet Res 3:130–51
- Sasikumar B, Syamkumar S, Remya R, John Zachariah T. (2005). PCR based detection of adulteration in the market samples of turmeric powder. Food Biotechnol 18:299–306
- Srirama R, Senthilkumar U, Sreejayan N, et al. (2010). Assessing species admixtures in raw drug trade of Phyllanthus, a hepato-protective plant using molecular tools. J Ethanopharmacol 130:208–15
- Stoeckle MY, Gamble CC, Kirpekar R, et al. (2011). Commercial teas highlight plant DNA barcode identification successes and obstacles. Sci Rep 1:42–9
- Tamura K, Peterson D, Stecher G, et al. (2011). MEGA 5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–9
- Tripathi AM, Tyagi A, Kumar A, et al. (2013). The internal transcribed spacer (ITS) region and trnH-psbA are suitable candidate loci for DNA barcoding of tropical tree species of India. PLoS ONE [Online]. Available from: http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0057934 [last accessed 12 Jun 2014]
- Urbatsch LE, Baldwin BG, Donoghue MJ. (2000). Phylogeny of the coneflowers and relatives (Heliantheae: Asteraceae) based on nuclear rDNA internal transcribed spacer (ITS) sequences and chloroplast DNA restriction site data. Syst Bot 25:539–65
- Vijayan K, Tsou CH. (2010). DNA barcoding in plants: Taxonomy in a new perspective. Curr Sci 99:1530–41
- Wallace LJ, Boilard SMAL, Eagle SHC, et al. (2012). DNA barcodes for everyday life: Routine authentication of natural health products. Food Res Int 49:446–52
- Yuan M, Wei MFU, Cheng XJ. (2011). Identification of species within Tetrastigma (Miq.) Planch. (Vitaceae) based on DNA barcoding techniques. J Syst Evol 49:237–45
