Abstract
A new series of pyrazolo[3,4-d]pyrimidine-6-one derivatives (2a–2j) were prepared by using the Biginelli multicomponent cyclocondensation of 3-methyl-1-phenyl-1H-pyrazol-5(4H)-one (1a), different aromatic aldehydes, and urea with a catalytic amount of HCl at reflux temperature. These compounds were characterized by IR, 1H NMR, 13C NMR, and Mass spectral data. In vitro antiamoebic activity was performed against HM1:IMSS strain of Entamoeba histolytica. The results showed that the compounds 2b, 2i, and 2j with IC50 values of 0.37 µM, 0.04 µM, and 0.06 µM, respectively, exhibited better antiamoebic activity than the standard drug metronidazole (IC50 = 1.33 µM). The toxicological studies of these compounds on human breast cancer MCF-7 cell line showed that the compounds 2b, 2i, and 2j exhibited >80% viability at the concentration range of 1.56–50 µM.
Introduction
Parasitic infections constitute one of the most widespread human health problems, and most of them occur through contaminated food or water. The human intestine is a major target of these ingested pathogenic microorganisms, resulting in severe infections, of which amoebiasis (potential life-threatening dysentery) is one. Amoebiasis is the second leading cause of death from a protozoan parasite, Entamoeba histolytica, and remains as a major health problem in Third World countries (Citation1). It affects more than 10% of the world’s population, and untreated infection may lead to severe complications including hepatic amoebiasis and intestinal tissue destruction (Citation2). Globally, amoebiasis accounts for 50 million clinical cases and is responsible for approximately 110,000 deaths annually (Citation3,Citation4). Metronidazole (MNZ) is known to be highly effective amoebicide and is considered to be the drug of choice for the treatment of amoebiasis, but recent studies have shown that this drug have several toxic effects such as genotoxicity, gastric mucus irritation, and spermatozoid damage (Citation5,Citation6). Furthermore, failures in the treatment of several intestinal protozoan parasites may result from drug resistant to parasites (Citation7,Citation8). According to the International Agency for Research on Cancer (IARC), MNZ is classified in the 2B group, that is, potentially carcinogenic to humans and proved to be carcinogenic to animals (Citation9). This prompted us to search for new antiamoebic agents.
Pyrimidines are of great importance in fundamental metabolism, being an integral part of DNA and RNA and found as core structure in a large verity of compounds that exhibited important biological activities such as anticancer, antimicrobial, antioxidant, and antiviral activities (Citation10–14). In the recent years, a lot of attention has been drawn by the pyrimidines derivatives due to their diverse range of activities, especially calcium channel blocker property (Citation15). Moreover, several alkaloids containing the dihydropyrimidine core unit that have been isolated from marine sources also showed interesting biological properties. In particular, the batzelladine alkaloids have been found to be potent HIV gp-120-CD4 inhibitors (Citation16,Citation17). According to the literature, pyrazolones are also used as starting materials for the synthesis of biologically active pyrazolo[3,4-d]pyrimidine derivatives (Citation18,Citation19).
In view of the number of pharmacological significances of pyrimidine derivatives and as a part of our continuous efforts toward the development of more potent amoebicidal agents, we herein report the synthesis, characterization, in vitro antiamoebic activity, and cytotoxicity of a new series of pyrazolo[3,4-d]pyrimidine-6-one derivatives (2b–2j), based on modification of the classical one-pot Biginelli reaction (Citation18,Citation20).
Materials and methods
All the required chemicals were purchased from Merck and Aldrich Chemical Company (USA). Precoated aluminum sheets (silica gel 60 F254, Merck Germany) were used for thin-layer chromatography (TLC) and spots were visualized under UV light. Elemental analysis was carried out on CHNS Elementar (Vario EL-III), and the results were within ±0.3% of the theoretical values. IR spectra were recorded on Perkin-Elmer model 1600 FT-IR RX1 spectrophotometer as KBr discs. 1H NMR and 13C NMR spectra were recorded on Bruker Spectrospin DPX 300 MHz and Bruker Spectrospin DPX 75 MHz spectrometer, respectively, using CDCl3 and DMSO-d6 as a solvent and trimethylsilane (TMS) as an internal standard. Splitting patterns are designated as follows; s, singlet; d, doublet; t, triplet; m, multiplet. Chemical shift values are given in ppm. The FAB mass spectra of the compounds were recorded on JEOL SX 102/DA-6000 mass spectrometer using Argon/Xenon 6KV (10 mA) as the FAB gas, and m-nitrobenzyl alcohol (NBA) was used as the matrix.
Preparation of 5-methyl-2-phenyl-2,4-dihydro-3H-pyrazol-3-one (1a)
A mixture of phenyl hydrazine (0.1 mol) and ethyl acetoacetate (0.1 mol) in 50 ml ethanol was heated under reflux for 5 h. After cooling, the reaction mixture was poured into 200 ml of ice water. The crude product was filtered, dried, and recrystallized with 95% ethanol.
Yield 75%; Solid; m.p.: 123°C Anal. calc. for C10H10N2O: C 68.95, H 5.79, N 16.08%; found: C 68.88, H 5.73, N 18.03%; IR νmax (cm−1): 3044 (Ar-H), 2976 (C-H), 1709 (C=O), 1588 (C=N), 1340 (C-N); 1H NMR (CDCl3) δ(ppm): 7.86 (d, 2H, J = 7.8Hz, Ar-H), 7.40–7.35 (t, 2H, J = 7.8 Hz, Ar-H), 7.19 (t, 1H, J = 7.2Hz, Ar-H), 3.39 (s, 2H, -CH2 pyrazolone ring), 2.16 (s, 3H, CH3 pyrazolone ring), 13C NMR (CDCl3) δ(ppm): 170.48 (C=O), 147.1 (pyrazole), 139.5, 134.7, 131.7, 129.5, 124.5, (Ar-C), 13.6 (CH3 pyrazolopyrimidine ring); FAB-MS (m/z): [M++1] 175.19.
Synthesis of pyrazolo[3,4-d]pyrimidine-6-ones (2a–2j)
General procedures
A mixture of urea (0.01 mol), aromatic aldehyde (0.01 mol) and 3-methyl-1-phenyl-1H-pyrazol-5(4H)-one (0.01 mol) in absolute ethanol (30 ml) contain 2–3 drops of 37% HCl as catalyst was refluxed for 5 h. The crude product which precipitated on cooling was filtered and washed with cold ethanol and then recrystallized from appropriate solvent.
4, 5-Dihydro-3-methyl-1,4-diphenyl-1 H-pyrazolo[3,4-d]pyrimidin-6(7H)-one (2a)
Yield 35%; Solid; (Ethanol) mp: 167°C; Anal. calc. for C18H16N4O: C 71.04, H 5.30, N 18.41%; found: C 71.02, H 5.23, N 18.36%; IR νmax (cm−1): 3233 (N-H), 3064 (Ar-H), 2923 (C-H), 1689 (C=O); 1H NMR (DMSO-d6) δ(ppm): 8.22 (s, 1H, NH pyrimidine), 8.14 (s, 1H, NH pyrimidine), 7.37–7.08 (m, 10H, Ar-H), 5.13 (s, 1H, -CH pyrimidine), 2.48 (s, 3H, CH3 pyrazole ring); 13C NMR (DMSO-d6) δ(ppm): 162.5 (C=O), 151.8 (pyrazolopyrimidine ring), 147.1 (pyrazole ring), 139.5, 134.7, 131.7, 129.5, 124.5 (Ar-C), 117.8 (pyrazolopyrimidine ring), 50.1 (-CH pyrimidine), 13.6 (CH3 pyrazole ring); FAB-MS (m/z): [M++1] 305.44.
4-(4-Chlorophenyl)-4,5-dihydro-3-methyl-1-phenyl-1H-pyrazolo[3,4d]pyrimidin-6 (7H)-one (2b)
Yield 37%; Solid; (Ethanol) mp: 237°C; Anal. calc. for C18H15N4OCl: C 63.81, H 4.46, N 16.54%; found: C 63.72, H 4.39, N 16.48%; IR νmax (cm−1): 3255 (N-H), 3088 (Ar-H), 2846 (C-H), 1678 (C=O); 1H NMR (DMSO-d6) δ(ppm): 8.24 (s, 1H, NH pyrimidine), 8.01(s, 1H, NH pyrimidine), 7.65 (d, 2H, J = 7.8 Hz, Ar-H), 7.47–7.42 (t, 2H, J = 7.8 Hz, Ar-H), 7.33–7.21 (m, 5H, Ar-H) 4.99 (s, 1H, -CH pyrimidine), 2.48 (s, 3H, CH3 pyrazole ring); 13C NMR (DMSO-d6) δ(ppm): 161.8 (C=O), 152.4 (pyrazolopyrimidine ring), 148.8 (pyrazole ring), 139.5, 135.7, 130.2, 122.4 (Ar-C), 113.2 (pyrazolopyrimidine ring), 52.65 (-CH pyrimidine), 14.8 (CH3 pyrazole ring); FAB-MS (m/z): [M++1] 339.38.
4-(3-Chlorophenyl)-4,5-dihydro-3-methyl-1-phenyl-1H-pyrazolo[3,4-d]pyrimidin-6 (7H)-one (2c)
Yield 40%; Solid; (Ethanol:Methanol) mp: 133°C; Anal. calc. for C18H15N4OCl: C 63.81, H 4.46, N 16.54%; found: C 63.74, H 4.41, N 16.47 %; IR νmax (cm−1): 3257 (N-H), 3041 (Ar-H), 2853 (C-H), 1653 (C=O); 1H NMR (DMSO-d6) δ(ppm): 9.19 (s, 1H, NH pyrimidine), 8.22 (s, 1H, NH pyrimidine), 7.69 (d, 2H, J = 7.8 Hz, Ar-H), 7.49–7.19 (m, 7H, Ar-H), 4.97 (s, 1H, -CH pyrimidine), 2.48 (s, 3H, CH3 pyrazole ring); 13C NMR (DMSO-d6) δ(ppm): 162.3 (C=O), 151.7 (pyrazolopyrimidine ring), 147.6 (pyrazole ring), 135.7, 132.2, 131.1, 130.3, 122.4 (Ar-C), 118.4 (pyrazolopyrimidine ring), 50.97 (-CH pyrimidine), 14.04 (CH3 pyrazole ring); FAB-MS (m/z): [M++1] 339.21.
4, 5-Dihydro-3-methyl-1-phenyl- 4-p-tolyl-1H-pyrazolo [3,4-d] pyrimidin-6(7H)-one (2d)
Yield 45%; Solid; (Ethanol) m.p: 188°C; Anal. calc. for C19H18N4O: C 71.68, H 5.70, N 17.60%; found: C 71.59, H 5.62, N 17.54%; IR νmax (cm−1): 3250 (N-H), 3086 (Ar-H), 2857 (C-H), 1698 (C=O); 1H NMR (DMSO-d6) δ(ppm): 8.11 (s, 1H, NH pyrimidine), 8.01 (s, 1H, NH pyrimidine), 7.65 (d, 2H, J = 7.8 Hz, Ar-H), 7.49 (t, 2H, J = 7.5 Hz, Ar-H), 7.33–7.05 (m, 5H, Ar-H), 4.99 (s, 1H, -CH pyrimidine), 2.48 (s, 3H, CH3 pyrazole ring), 2.37 (s, 3H, CH3 phenyl); 13C NMR (DMSO-d6) δ(ppm): 162.6 (C=O), 155.3 (pyrazolopyrimidine ring), 148.3 (pyrazole ring), 140.5, 139.7, 132.4, 130.3, 124.3 (Ar-C), 113.4 (pyrazolopyrimidine ring), 50.07 (-CH pyrimidine), 21.5 (CH3 Aromatic), 14.1 (CH3 pyrazole ring); FAB-MS (m/z): [M++1] 319.54.
4,5-Dihydro-4-(4-isopropylphenyl)-3-methyl-1-phenyl-1H-pyrazolo[3,4-d]pyrimidin-6(7H)-one (2e)
Yield 48%; Solid; (Ethylacetate) m.p: 192°C; Anal. calc. for C21H22N4O: C 72.81, H 6.40, N 16.17%; found: C 72.76, H 6.33, N 16.19%; IR νmax (cm−1): 3293 (N-H), 3031 (Ar-H), 2921 (C-H), 1698 (C=O); 1H NMR (DMSO-d6) δ(ppm): 8.02 (s, 1H, NH pyrimidine), 7.97 (s, 1H, NH pyrimidine), 7.86 (d, 2H, J = 8.1 Hz, Ar-H), 7.79 (s, 1H, J = 8.1 Hz, Ar-H), 7.46–7.35 (m, 3H, Ar-H), 7.21–6.90 (m, 3H, Ar-H), 5.17 (s, 1H, -CH pyrimidine), 3.77 (m, 1H, -CH (CH3)2), 2.48 (s, 3H, CH3 pyrazole ring), 2.16 (s, 3H, CH3 isopropylphenyl), 2.11 (s, 3H, CH3 isopropylphenyl); 13C NMR (DMSO-d6) δ(ppm): 161.5 (C=O), 153.8 (pyrazolopyrimidine ring), 148.8 (pyrazole ring), 138.6, 135.3, 130.8, 123.4 (Ar-C), 118.8 (pyrazolopyrimidine ring), 51.47 (-CH pyrimidine), 13.8 (CH3 pyrazole ring); FAB-MS (m/z): [M++1] 347.56.
4,5-Dihydro-4-(3-hydroxyphenyl)-3-methyl-1-phenyl-1H-pyrazolo[3,4-d]pyrimidin-6(7H)-one (2f)
Yield 35%; Solid; (Ethanol) m.p: 176–177°C; Anal. calc. for C18H16N4O2: C 67.49, H 5.03, N 17.49%; found: C 67.43, H 5.05, N 17.42%; IR νmax (cm−1): 3408 (O-H), 3192 (N-H) 3071 (Ar-H), 2917 (C-H), 1665 (C=O); 1H NMR (DMSO-d6) δ(ppm): 10.2 (s, 1H, NH pyrimidine), 10.0 (s, 1H, Ar-OH), 9.74 (s, 1H, NH pyrimidine), 7.65–6.52 (m, 9H, Ar-H), 5.11 (s, 1H, -CH pyrimidine), 2.48 (s, 3H, CH3 pyrazole ring); 13C NMR (DMSO-d6) δ(ppm): 162.5 (C=O), 159.3 (C-OH), 154.9 (pyrazolopyrimidine ring), 147.8 (pyrazole ring), 139.3, 137.1, 135.42, 130.8, 122.4, (Ar-C), 112.4 (pyrazolopyrimidine ring), 52.44 (-CH pyrimidine), 13.9 (CH3 pyrazole ring); FAB-MS (m/z): [M++1] 321.11.
4,5-Dihydro-4-(3,4-dimethoxyphenyl)-3-methyl-1-phenyl-1H-pyrazolo[3,4-d]pyrimidin-6 (7H)-one (2g)
Yield 30%; Solid; (Ethanol:Methanol) m.p: 183°C; Anal. calc. for C20H20N4O3: C 5.92, H 5.53, N 15.38%; found: C 5.87, H 5.47, N 15.33%; IR νmax (cm−1): 3263 (N-H), 3076 (Ar-H), 2853 (C-H), 1663 (C=O); 1H NMR (DMSO-d6) δ(ppm): 8.81 (s, 1H, NH pyrimidine), 8.04 (s, 1H, NH pyrimidine), 7.90 (d, 2H, J = 8.1 Hz, Ar-H), 7.90 (d, 2H, Ar-H), 7.69 (s, 1H, Ar-H), 7.43–7.11 (m, 3H, Ar-H), 5.10 (s, 1H, -CH pyrimidine), 3.86 (s, 6H, 2xOCH3), 2.48 (s, 3H, CH3 pyrazole ring); 13C NMR (DMSO-d6) δ(ppm): 161.9 (C=O), 152.9 (pyrazolopyrimidine ring), 147.2 (pyrazole ring), 140.7, 135.4, 131.6, 130.3, 122.4 (Ar-C), 116.3 (pyrazolopyrimidine ring), 52.08 (-CH pyrimidine), 14.64 (CH3 pyrazole ring); FAB-MS (m/z): [M++1] 365.17.
4,5-Dihydro-4-(1H-indol-3-yl)-3-methyl-1-phenyl-1H-pyrazolo[3,4-d]pyrimidin-6(7H)-one (2h)
Yield 45%; Solid; (Ethylacetate) m.p: 238°C; Anal. calc. for C20H17N5O: C 69.96, H 4.99, N 20.40%; found: C 69.91, H 4.93, N 20.32%; IR νmax (cm−1): 3249 (N-H), 3067 (Ar-H), 2891 (C-H), 1653 (C=O); 1H NMR (DMSO-d6) δ(ppm): 12.66 (s, 1H, NH, Indole), 9.80 (s, 1H, NH pyrimidine), 8.16 (s, 1H, NH pyrimidine), 7.97 (d, 2H, J = 7.8 Hz, Ar-H), 7.59–7.12 (m, 8H, Ar-H), 5.22 (s, 1H, -CH pyrimidine), 2.47 (s, 3H, CH3 pyrazole ring); 13C NMR (DMSO-d6) δ (ppm): 162.5 (C=O), 153.6 (pyrazolopyrimidine ring), 148.9 (pyrazole ring), 140.1, 134.7, 132.2, 131.6, 130.8, 123.4, (Ar-C), 115.3 (pyrazolopyrimidine ring), 50.34 (-CH pyrimidine), 13.84 (CH3 pyrazole ring); FAB-MS (m/z): [M++1] 344.25.
4-(2-Chloroquinoline-3-yl)-4,5-dihydro-3-methyl-1-phenyl-1H-pyrazolo[3,4-d] pyrimidine-6(7H)-one (2i)
Yield 28%; Solid; (Ethanol) m.p: 168°C; Anal. calc. for C21H16N5OCl: C 64.70, H 4.14, N 7.96%; found: C 64.63, H 4.08, N 7.87%; IR νmax (cm−1): 3298 (N-H), 3061 (Ar-H), 2955 (C-H), 1668 (C=O); 1H NMR (DMSO-d6) δ(ppm): 12.3 (s, 1H, NH pyrimidine), 10.0 (s, 1H, NH pyrimidine), 8.49 (s, 1H, quinoline), 7.97–7.17 (m, 9H, Ar-H & quinoline), 5.13 (s, 1H, -CH pyrimidine), 2.48 (s, 3H, CH3 pyrazole ring); 13C NMR (DMSO-d6) δ(ppm): 161.6 (C=O), 152.3 (pyrazolopyrimidine ring), 147.3 (pyrazole ring), 139.6, 133.7, 132.2, 131.6, 128.3, 121.4 (Ar-C), 115.3 (pyrazolopyrimidine ring), 52.80 (-CH pyrimidine), 14.67 (CH3 pyrazole ring); FAB-MS (m/z): [M++1] 390.37.
4-(2-Chloro-7-methylquinoline-3-yl)-4, 5-dihydro-3-methyl-1-phenyl-1 H-pyrazolo[3,4-d]pyrimidin-6(7H)-one (2j)
Yield 35%; Solid; (Ethanol) m.p: 194°C Anal. calc. for C22H18N5OCl: C 65.43, H 4.49, N 17.34%; found: C 65.38, H 4.43, N 17.27%; IR νmax (cm−1): 3296, (N-H) 3048 (Ar-H), 2977 (C-H), 1665 (C=O); 1H NMR (DMSO-d6) δ(ppm): 12.2 (s, 1H, NH pyrimidine), 10.1 (s, 1H, NH pyrimidine), 8.40 (s, 1H, quinoline), 8.13 (s, 1H, quinolne), 7.67 (d, 1H, J= 8.1 Hz, quinoline), 7.59–7.20 (m, 6H, Ar-H & quinoline), 5.10 (s, 1H, -CH pyrimidine), 2.48 (s, 3H, CH3 pyrazole ring), 2.38 (s, 3H, quinoline); 13C NMR (DMSO-d6) δ(ppm): 162.4 (C=O), 153.6 (pyrazolopyrimidine ring), 148.4 (pyrazole ring), 140.6, 133.7, 132.1, 131.9, 129.3, 123.1 (Ar-C), 114.3 (pyrazolopyrimidine ring), 50.13 (-CH pyrimidine), 14.12 (CH3 pyrazole ring); FAB-MS (m/z): [M++1] 404.42.
Pharmacology
All the pyrazolo[3, 4-d]pyrimidine-6-one derivatives (2a–2j) were screened in vitro against HM1:IMSS strain of E. histolytica by microdilution method (Citation21). All the experiments were carried out in triplicate at each concentration level and repeated thrice. Cytotoxicity of active compounds has been studied by MTT assay on breast cancer MCF-7 cell line. The results of biological activity and cytotoxicity are summarized in .
Table 1. In vitro antiamoebic activity of pyrazolo[3,4-d] pyrimidine-6-one derivatives (2a–2j) against HM1:IMSS strain of Entamoeba histolytica and cytotoxicity profile of compounds 2b, 2i, 2j and metronidazole.
In vitro antiamoebic assay
All the compounds (2a–2j) were screened in vitro for antiamoebic activity against HM1:IMSS strain of E. histolytica by microdilution method (Citation21). E. histolytica trophozoites were cultured in wells of 96-well microtiter plate by using Diamond TYIS-33 growth medium (Citation22). The test compounds (1 mg) were dissolved in 40 μl DMSO and the final solutions in each well contained 0.1% DMSO or less, which did not affect viability (Citation23,Citation24). The stock solutions of the compounds were prepared freshly before use at a concentration of 1 mg/ml. Twofold serial dilutions were made in the wells of 96-well microtiter plate (costar). Each test includes MNZ as a standard amoebicidal drug, control wells (culture medium plus amoebae) and a blank (culture medium only). All the experiments were carried out in triplicate at each concentration level and repeated thrice. The amoeba suspension was prepared from a confluent culture by pouring off the medium at 37°C and adding 5 ml of fresh medium, chilling the culture tube on ice to detach the organisms from the side of the flask. The number of amoeba/ml was estimated with a haemocytometer, using trypan blue exclusion to confirm the viability. The suspension was diluted to 105 organism/ml by adding fresh medium and 170 μl of this suspension was added to the test and control wells in the plate so that the wells were completely filled (total volume, 340 μl). An inoculum of 1.7 × 104 organisms/well was chosen so that confluent, but not excessive growth, took place in control wells. Plates were sealed and gassed for 10 min with nitrogen before incubation at 37°C for 72 h. After incubation, the growth of amoeba in the plate was checked with a low-power microscope. The culture medium was removed by inverting the plate and shaking gently. Plate was then immediately washed with sodium chloride solution (0.9%) at 37°C. This procedure was completed quickly and the plate was not allowed to cool to prevent the detachment of amoebae. The plate was allowed to dry at room temperature, and the amoebae were fixed with methanol and when dried, stained with (0.5%) aqueous eosin for 15 min. The stained plate was washed once with tap water, then twice with distilled water and allowed to dry. A 200-μl portion of 0.1N sodium hydroxide solution was added to each well to dissolve the protein and release the dye. The optical density of the resulting solution in each well was determined at 490 nm with a microplate reader. The percent inhibition of amoebal growth was calculated from the optical densities of the control and test wells and plotted against the logarithm of the dose of the drug tested. Linear regression analysis was used to determine the best fitting line from which the IC50 value was found. The IC50 values in μM are reported in .
MTT assay
MCF-7 cells were cultured and maintained as monolayer in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% of fetal calf serum (FCS), antibiotics: 100 IU/ml of penicillin and 100 μg/ml of streptomycin. All cells were cultured in flasks at 37°C in the 100%-humidity atmosphere and 5% of CO2 (Citation25). Only viable cells were used in the assay. Exponentially growing cells were plated at 1.2 × 104 cells per well into 96-well plates and incubated for 48 h before the addition of drugs. Stock solutions of compounds were initially dissolved in 20% (v/v) DMSO and further diluted with fresh complete medium. The growth-inhibitory effects of the compounds were measured using standard tetrazolium MTT assay. After 48 h of incubation at 37°C, the medium was removed and 25µl of MTT reagent (5 mg/ml) in serum-free medium was added to each well. The plates were incubated at 37°C for 4 h. At the end of the incubation period, the medium was removed and pure DMSO (100 µl) was added to each well. The metabolized MTT product dissolved in DMSO was quantified by reading the absorbance at 570 nm. All assays were performed in triplicate. Percent viability was defined as the relative absorbance of treated versus untreated control cells. Plates were analyzed in an ELISA plate reader (Labsystems Multiskan RC, Helsinki, Finland) at 570 nm with a reference wavelength of 655 nm.
Results and Discussion
Chemistry
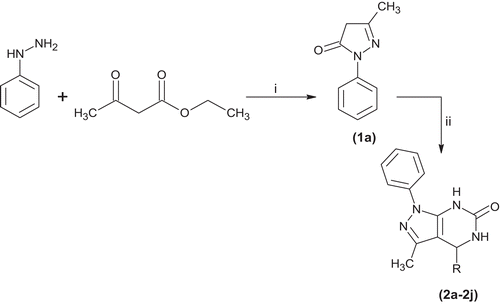
The synthesis of pyrazolo[3,4-d]pyrimidine-6-one derivatives (2a–2j) was performed in a manner as outlined in . The reaction of 3-methyl-1-phenyl-1H-pyrazol-5(4H)-one, urea and various aromatic aldehydes in absolute ethanol using HCl as a catalyst and refluxed for 5 h gave pyrazolo[3,4-d]pyrimidine-6-one derivatives (2a–2j). The compounds were stable in solid state and were soluble in methanol and DMSO. Melting points were recorded on KSW melting point apparatus and are uncorrected.
Scheme 1. General method for the synthesis of pyrimidine derivatives (2a–2j). Reagent and conditions: (i) C2H5OH, 78°C reflux 5–6 h, (ii) Urea, HCl (2–3 drops), different aromatic aldehydes, where R = aryl group of different aldehydes.
Synthesis
In the present work, Biginelli-type cyclocondensation reaction was studied for the preparation pyrazolo[3,4-d]pyrimidine-6-ones (2a–2j). The classical three-components urea, aromatic aldehyde, and 3-methyl-1-phenyl-1H-pyrazol-5(4H)-one were carried out in alcoholic solution containing a few drops of concentrated hydrochloric as catalyst, and refluxed for 5 h, as shown in Scheme 1. All the compounds (2a–2j) were recrystallized from appropriate solvents and the yield was in the range of 36–48%. All the compounds were highly soluble in methanol and DMSO and were characterized by IR, 1H and 13C NMR, and Mass spectra. The purity of the compounds was confirmed by elemental analysis and data were found in accordance with ±0.3%.
Characteristics IR bands provides significance indication for the formation of 5-methyl-2-phenyl-2,4-dihydro-3H-pyrazol-3-one (1a). The appearance of absorption bands at 1709, 1588, and 1340 cm−1 corresponding to the C=O, C=N, C-N stretching vibrations, confirmed the formation of cyclized product (1a). In cases of pyrazolo[3,4-d]pyrimidine-6-ones (2a–2j), a sharp band was observed for NH-stretching vibrations in the range 3233–3298 cm−1 showed the formation of pyrazolo[3,4-d]pyrimidine ring in all the compounds (2a–2j). The appearance of strong band in the range of 1653–1698 cm−1 due C=O stretching vibration also confirmed the formation of desired compounds (2a–2j) containing pyrazolo[3,4-d]pyrimidine ring.
Moreover the structure of 5-methyl-2-phenyl-2,4-dihydro-3H-pyrazol-3-one (1a) was further confirmed by 1H NMR spectra. The appearance of singlet for two protons at 3.39 due to CH2 group showed the formation of desired pyrazolon ring in the structure (1a). The appearance of singlet at 2.16 ppm corresponds to the methyl group of pyrazolon nucleus also confirmed the formation of cyclized structure (1a). The structure of 5-methyl-2-phenyl-2,4-dihydro-3H-pyrazol-3-one (1a) was further supported by 13C NMR spectra. The appearance of signal at 170 ppm due to (C=O) group of desire pyrazolon nucleus confirmed the formation of 5-methyl-2-phenyl-2,4-dihydro-3H-pyrazol-3-one.
In case of pyrazolo[3,4-d]pyrimidine-6-ones (2a–2j), NH protons were appeared in the range of 7.97–12.6 ppm only as a singlet due to poor resolution of its signal revealed the presence of pyrimidines nucleus in all the compounds. A sharp peak representing the methine proton of the pyrimidine was observed in the range of 4.97–5.22 ppm also confirming the formation of the pyrazolo[3,4-d]pyrimidine nucleus in all the synthesized compounds (2a–2j) (Citation18). The appearance of signals at 8.11–8.13 and 8.40–8.49 ppm were found in compounds (2i) and (2j) for chloroquinoline rings, a singlet at 10.0 ppm for OH proton was observed in compound (2f). For methyl group of pyrazol ring, a singlet at 2.47–2.48 ppm was found in all the compounds (2a–2j), whereas for compounds (2e) and (2j), the appearance of singlet for methyl group was found in the range of 2.11–2.38 ppm. A singlet at 3.86 ppm for two OCH3 protons was found in compound (2g).The structure of the compounds (2a–2j) was further supported by 13C NMR spectra. The appearance of signals in the range of 161.5–162.8 ppm and 50.1–52.8 ppm due to C=O and methine carbon atom (pyrimidine nucleus) confirmed the formation of pyrazolo[3,4-d]pyrimidine-6-ones (2a–2j) (). The signal due to aromatic and aliphatic carbons resonates at their usual positions and the data shown in the experimental section.
Antiamoebic activity
Preliminary experiments were carried out to determine the in vitro antiamoebic activity of all the compounds (2a–2j) by microdilution method using HM1:IMSS strain of E. histolytica. The results are summarized in . The data are present in terms of percent growth inhibition relative to untreated controls and plotted as probit values as a function of drug concentration. The antiamoebic effect was compared with the most widely used antiamoebic medication MNZ which had a 50% inhibitory concentration (IC50) of 1.33 μM in our experiments. The results showed that the synthesized pyrazolo[3,4-d]pyrimidine-6-one derivatives (2a–2j) and their structural activity relationship (SAR) established on the basis of substituted and unsubstituted aromatic ring attached with the methine carbon atom of pyrimidine nucleus. The pyrazolo[3,4-d]pyrimidine-6-one derivatives with substituted and unsubstituted phenyl ring (2a–2h) and quinoline ring (2i, 2j) attached with the pyrimidine ring showed IC50 for antiamoebic activity in the range of 0.04–14.3 μM. Therefore out of ten compounds screened in vitro antiamoebic activity, three compounds (2b, 2i, and 2j) were found to be more active against E. histolytica as compared with reference drug MNZ (IC50 = 1.33 μM). Among these three compounds, two compounds 2i (IC50 = 0.04 µM) and 2j (IC50 = 0.06 µM) substituted by chloroquinoline rings showed excellent antiamoebic activity and the rest one compounds 2b (IC50 = 0.37 µM) substituted by hydroxyl phenyl showed moderatively active than the standard drug MNZ.
Cytotoxicity profile
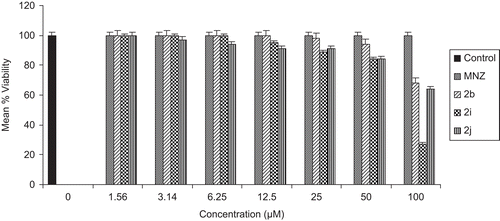
To examine the effect of antiamoebic compounds 2b, 2i, 2j and MNZ on cell proliferation, we ensured their cytotoxicity on human breast cancer MCF-7 cell line. A subconfluent population of MCF-7 cells was treated with increasing concentrations of compounds, and the number of viable cells was measured after 48 h by MTT cell viability assay. The concentration range for all the compounds 2b, 2i, 2j and MNZ was 1.56–100 µM. depicts the compounds 2b, 2i, 2j and MNZ exhibited >80% viability at the concentration range of 1.56–50 µM. On increasing the concentration range up to 100 µM, only compound 2i (viability 27%) showed cytotoxicity against the human breast cancer MCF-7 cell line. The cytotoxicity IC50 values along with the standard deviation values of 2b, 2i, 2j and MNZ are given in .
Conclusion
A new series of pyrazolo[3,4-d]pyrimidine-6-one derivatives (2a–2j) was prepared by simple one-pot Biginelli method. Compounds 2b, 2i, and 2j with IC50 values of 0.37 µM, 0.04 µM, and 0.06 µM, respectively, exhibited higher antiamoebic activity than the standard drug MNZ (IC50 = 1.33 µM). Cytotoxicity studies were performed on human breast cancer MCF-7 cell line and results showed that the compounds 2b, 2i, 2j and MNZ offered remarkable viability (>80% at 50 μM).
Declaration of interest
This work was supported by Council of Scientific and Industrial Research [grant no. 01(2278)/08/EMR-II New Delhi, India]. F.H. is thankful to the University Grant commission (UGC), India for the financial support.
References
- Stanley SL Jr. Amoebiasis. Lancet 2003;361:1025–1034.
- Marion S, Guillén N. Genomic and proteomic approaches highlight phagocytosis of living and apoptotic human cells by the parasite Entamoeba histolytica. Int J Parasitol 2006;36:131–139.
- Ackers JP, Mirelman D. Progress in research on Entamoeba histolytica pathogenesis. Curr Opin Microbiol 2006;9:367–373.
- Stanley SL. Pathophysiology of amoebiasis. Trends Parasitol 2001;17:280–285.
- el-Nahas AF, el-Ashmawy IM. Reproductive and cytogenetic toxicity of metronidazole in male mice. Basic Clin Pharmacol Toxicol 2004;94:226–231.
- Purohit V, Basu AK. Mutagenicity of nitroaromatic compounds. Chem Res Toxicol 2000;13:673–692.
- Abboud P, Lemée V, Gargala G, Brasseur P, Ballet JJ, Borsa-Lebas F et al. Successful treatment of metronidazole- and albendazole-resistant giardiasis with nitazoxanide in a patient with acquired immunodeficiency syndrome. Clin Infect Dis 2001;32:1792–1794.
- Petri WA Jr. Therapy of intestinal protozoa. Trends Parasitol 2003;19:523–526.
- IARC monographs on the evaluation of the carcinogenic risk of chemicals to humans; International Agency for Research on Cancer: Lyon, France, 1987; Supplement 7, pp. 250–251.
- Gangjee A, Yu J, Kisliuk RL, Haile WH, Sobrero G, McGuire JJ. Design, synthesis, and biological activities of classical N-[4-[2-(2-amino-4-ethylpyrrolo[2,3-d]pyrimidin-5-yl)ethyl]benzoyl]-l-glutamic acid and its 6-methyl derivative as potential dual inhibitors of thymidylate synthase and dihydrofolate reductase and as potential antitumor agents. J Med Chem 2003;46:591–600.
- Naimi E, Zhou A, Khalili P, Wiebe LI, Balzarini J, De Clercq E et al. Synthesis of 3′- and 5′-nitrooxy pyrimidine nucleoside nitrate esters: “nitric oxide donor” agents for evaluation as anticancer and antiviral agents. j Med Chem 2003;46:995–1004.
- Gangjee A, Adair OO, Queener SF. Synthesis and biological evaluation of 2,4-diamino-6-(arylaminomethyl)pyrido[2,3-d]pyrimidines as inhibitors of Pneumocystis carinii and Toxoplasma gondii dihydrofolate reductase and as antiopportunistic infection and antitumor agents. J Med Chem 2003;46:5074–5082.
- Ismaili L, Nadaradjane A, Nicod L, Guyon C, Xicluna A, Robert J-F, Refouvelet B. Synthesis and antioxidant activity evaluation of new hexahydropyrimido[5,4-c]quinoline-2,5-diones and 2-thioxohexahydropyrimido[5,4-c]quinoline-5-ones obtained by Biginelli reaction in two steps. Eur J Med Chem 2008;43:1270–1275.
- Rostom AFS, Fahmy TYH, Saudi NSM. Synthesis and in vitro anti-HIV screening of certain 2-(benzoxazol-2-ylamino)-3H-4-oxopyrimidines. Sci Pharm 2003;71:57–74.
- Zorkun IS, Saraç S, Çelebi S, Erol K. Synthesis of 4-aryl-3,4-dihydropyrimidin-2(1H)-thione derivatives as potential calcium channel blockers. Bioorg Med Chem 2006;14:8582–8589.
- Patil AD, Kumar NV, Kokke WC, Bean, MF, Freyer AJ, Brossi CD, Mai S, Truneh A, Faulkner DJ, Carte B, Breen AL, Hertzberg RP, Johnson RK, Westly JW, Potts BCM. Novel Alkaloids from the Sponge Batzella sp.: Inhibitors of HIV gpl20-Human CD4Binding. J Org Chem 1995;60:1182–1188.
- Snider BB, Chen J, Patil AD, Freyer A. Synthesis of the Tricyclic Portions of Batzelladines A, B and D. Revision of the Stereoehemistry of Batzelladines A and D. Tetrahed Lett 1996;37:6977–6980.
- Trivedi AR, Siddiqui AB, Shah VH. Design, synthesis, characterization and antitubercular activity of some 2-heterocycle-substituted phenothiazines. Arkivoc 2008;(ii):210–217.
- Akbari JD, Tala SD, Dhaduk MF, Joshi HS, Mehta KB, Pathak SJ. Synthesis of Some New Pyrazolo[3,4-d]pyrimidines and Thiazolo [4,5 d]pyrimidines and Evaluation of Their Antimicrobial Activities. Phosph Sul Silicon 2008;183:1471–1477.
- Hassani Z, Islami MR, Kalantari M. An efficient one-pot synthesis of octahydroquinazolinone derivatives using catalytic amount of H2SO4 in water. Bioorg Med Chem Lett 2006;16:4479–4482.
- Wright CW, O’Neill MJ, Phillipson JD, Warhurst DC. Use of microdilution to assess in vitro antiamoebic activities of Brucea javanica fruits, Simarouba amara stem, and a number of quassinoids. Antimicrob Agents Chemother 1988;32:1725–1729.
- Diamond LS, Harlow DR, Cunnick CC. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans R Soc Trop Med Hyg 1978;72:431–432.
- Gillin FD, Reiner DS, Suffness M. Bruceantin, a potent amoebicide from a plant, Brucea antidysenterica. Antimicrob Agents Chemother 1982;22:342–345.
- Keene AT, Harris A, Phillipson JD, Warhurst DC. In vitro amoebicidal testing of natural products; part I. Methodology. Planta Med 1986;52:278–285.
- Gümüs F, Algül O, Eren G, Eroglu H, Diril N, Gür S et al. Synthesis, cytotoxic activity on MCF-7 cell line and mutagenic activity of platinum(II) complexes with 2-substituted benzimidazole ligands. Eur J Med Chem 2003;38:473–480.
![Figure 1. Pyrazolo[3,4-d]pyrimidine-6- one derivatives (2a–2j).](/cms/asset/4c133d0e-ceb5-422f-963f-c2620094acdc/ienz_a_528414_f0002_b.gif)