Abstract
Context: The Bowman-Birk inhibitors (BBIs) are currently investigated with renewed interest due to their therapeutic properties in cancer and other inflammatory disease treatment. The molecular mass of the BBI is a limitation, as sufficient amounts of the inhibitor do not reach the organs outside the gastrointestinal tract when administered orally.
Method: The anti-tryptic domain of HGI-III of horsegram (Dolichos biflorus) was cloned using the vector pET-20b (+) and expressed in E. coli BL21 (DE3) pLysS.
Results: Kinetic analysis of this anti-tryptic peptide (recombinant trypsin inhibitory domain (rTID)) reveals that it is a potent inhibitor of trypsin and human tryptase. The Ki (3.2 ± 0.17 × 10−8 M) establishes a very high affinity to bovine trypsin. rTID inhibited human lung tryptase (IC50 3.78 ± 0.23 × 10−7 M). The rTID is resistant to the digestive enzymes found in humans and animals.
Conclusion: These properties propagate further research on the use of rTID as a therapeutic for cancer and other related inflammatory diseases.
Introduction
Proteinase inhibitors, a promising class of therapeutic agents, play a vital role in regulating physiological proteases, which evoke critical pathologic conditions. The natural protein-proteinase inhibitors abundant in plant storage tissue such as seeds and tubers constitute a part of the plant defence system. The Bowman-Birk inhibitors (BBIs), bifunctional inhibitors have been extensively studied and their mechanism of inhibition very well establishedCitation1. The entire multigene BBI family and its genome organisation in common bean have been studiedCitation2. BBIs distinguish themselves from other protease inhibitors by their small size (6000–9000 Da), seven disulfide bridges and two symmetrical homology domains comprising the protease binding sites leading to the typical bow-tie motifCitation3. The inhibitory activity is vested in the closed nonapeptide loop with residue P1 directing specificity and β branched Thr (P2) strongly enhancing activityCitation4. Odani and IkenakaCitation5 bisected the BBI in such a way that each half retained its individual inhibitory properties. The smallest naturally occurring protease inhibitor that mimics BBI by combining size reduction, yet retaining biological activity is the sunflower trypsin inhibitorCitation6. The BBI-protease interactions are extensively reviewedCitation7,Citation8 and several three dimensional structures solvedCitation9–16.
Considerable scientific interest in BBIs is connected with their anticarcinogenicCitation17 and radioprotective effects on normal tissues. The possible mechanisms for these activities have been ascribed and linked to protease inhibitionCitation18. BBI concentrate (BBIC) a soybean extract enriched in BBI exhibits potent anti-inflammatory effect by inhibiting several human proteases associated with inflammation-mediating cellsCitation19. BBIC was effectively used in the remission of active ulcerative colitis without any apparent toxicity in humansCitation20. Mung bean BBI has been shown to inhibit the NS3 protease integral to the virus life cycle that causes Dengue hemorrhagic feverCitation21. An expressed lentil BBI exhibited antiproliferative properties in human colon cancer cellsCitation22. The anti-carcinogenic activity of Vigna unguiculata BBI was ascribed to apoptosis and permeabilisation of the lysosome membraneCitation23. The cancer-therapeutic property of BBIs has most often been attributed to the intrinsic ability to inhibit serine proteasesCitation24. Lanza et al.Citation25 showed that the Medicago scutellata trypsin inhibitor exhibits anticarcinogenic effects on cisplastin-induced cytotoxic breast and cervical cancer cells in the absence of any antichymotryptic activity suggesting that antichymotryptic activity is not a strict requirement for antitumoral activityCitation17. Considering the positive therapeutic benefits of BBIs and their relative stability at cooking temperatureCitation26 and the acidic digestive system in humansCitation27, BBI levels have been determined in commercial foodsCitation28.
Only about half of the BBI administered orally is taken up into the blood stream via the intestinal epithelial cells or crosses the intestinal lumen and distributed throughout the bodyCitation29. The molecular mass of BBIs (6000–9000 Da) is a limitation, as sufficient amounts do not reach the organs outside the gastrointestinal tract when administered orally, leading to a decreased bioavailability. Engineering BBI to achieve size reduction while retaining either the antitryptic or antichymotryptic or both by synthetic and combinatorial chemistry are based on the structural scaffold of the nine residue cyclised loop of the BBICitation30–33. An important feature of legume BBIs is the well defined interaction with the cognate protease. Hydrophilic bridges and conformational rigidity are characteristic of the narrow specificity of the antitryptic domain of BBIsCitation12. The aim of this study is to genetically engineer and express the conformationally stable antitryptic domain of a BBI from horsegram (Dolichos biflorus). HGI-III, a 76 amino acid polypeptide is the major BBI present in the horsegram seed with potent anti-tryptic activityCitation34. Accordingly the anti-tryptic domain (36 amino acid residue), a disulfide cyclised loop of molecular mass ~4240 Da was cloned and expressed in E. coli BL21 (DE3) pLysS. Structural and kinetic analysis of this anti-tryptic peptide against bovine trypsin and human tryptase are reported.
Materials and methods
Materials
Bacterial strains and vectors
Bacterial strain E. coli DH5α was from Invitrogen Corporation, Carlsbad,CA; BL21 (DE3) pLysS strain and the vector pET20b(+) were from Novagen, Merck Specialities Private Ltd., Mumbai, India.
Chemicals
Tryptone-type 1, yeast extract, ampicillin sodium salt and isopropyl thiogalacto pyranoside (IPTG) were from Hi-Media Laboratories, Mumbai, India, Gel extraction kit and Taq DNA polymerase were from Qiagen, GmbH, Hilden, Germany. Pfu DNA polymerase was from Fermentas Life Sciences, GmbH, Hilden, Germany. Other restriction enzymes were purchased from New England BioLabs Inc., Ipswich, MA. Bovine serum albumin (BSA), bovine pancreatic trypsin (2 × crystallized, type III, EC 3.4.21.4), L-1-tosylamido-2-phenylalanine chloromethyl ketone-treated trypsin, N-α-benzoyl-dl-arginine-p-nitroanilide (BAPNA) and tryptase from human lung (EC 3.4.21.59) were from Sigma-Aldrich, St. Louis, MO. Sephadex G-50 was from Pharmacia LKB, Uppsala, Sweden.
Oligonucleotides were synthesised by Sigma-Genosys, Bangalore India. Molecular weight markers for sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) were obtained from Bangalore Genei, Bangalore, India. All other chemicals used were of highest purity.
Methods
Cloning and construction of recombinant plasmids for total prokaryotic expression of trypsin inhibitory domain (TID)
The TID of horsegram HGI-III was amplified from the plasmid pRSET-rHGICitation35. Primers designed to obtain the open reading frame of 108 bp were HGI-F (5′GATCATCATCAGTCAACTGATGAG3′) and HGIT-R (5′CTCGAGTTACCTAACGTCCGTACAGCGGCA3′). The thermocycling conditions used were initial denaturation at 94°C, 5 min; followed by 25 cycles of 94°C, 30 s; 60°C, 30 s; 72°C, 20 s; in a GeneAmp® PCR system 9700 (Applied Biosystems, Foster City, CA). The amplified product was separated by agarose gel electrophoresis and purified by using a gel extraction kit. The purified amplicon with a stop codon was blunt end ligated into the EcoRV site of pET20b to generate the plasmid pET20b-recombinant TID (rTID). Chemically competent E. coli DH5α cells were transformed and clones harbouring pET20b-rTID were identified by gel shift assay and HincII digestion. The DNA sequence was confirmed by dideoxy sequencing using the Big Dye R terminator version 3.1 cycle sequencing kit (Applied Biosystems).
Expression of rTID
E. coli BL21 (DE3) pLysS transformed with pET20b-rTID, was grown overnight at 37°C in Luria-Bertani medium (10 mL) supplemented with ampicillin (100 µg/mL). Cells (1 mL) were diluted into 250 mL 2YT medium supplemented with ampicillin (100 µg/mL) and incubated at 37°C in an orbital shaker (180 rpm) until the optical density at 600 nm reached 1.75 ± 0.1. IPTG was added to a final concentration of 0.3 mM and the culture further grown at 37°C for 4 h in an orbital shaker. The cells were harvested by centrifugation at 7650g for 15 min at 4°C. The pellet was lysed in 35 mL 0.1 M Tris-HCl pH 8.2 by ultrasonication (Vibracell™, Sonics and Materials Inc., New Town, CT). The cell free extract (CFE) containing the rTID was further purified.
Trypsin and tryptase inhibitory assay
The amidase activity of trypsin and tryptase and their inhibition were assayed using BAPNA according to the method of Kakade et al.Citation36 One trypsin/tryptase inhibitory unit (TIU) is defined as the amount of inhibitor, which reduces the enzyme activity by one unit.
Purification of the pET20b-rTID
The CFE was applied to a Sephadex G-50 size exclusion chromatography (SEC) column (100 × 2 cm) pre-equilibrated with 0.025 M Tris-HCl pH 8.2 at a flow rate of 10 mL/h. Each fraction (2 mL) was assayed for protein and anti-tryptic activity. The fractions exhibiting trypsin inhibitor activity were pooled, dialysed against water and lyophilised.
The lyophilised fraction was dissolved in 0.1% trifluoracetic acid (TFA) and purified by reversed phase high-performance liquid chromatography (RP-HPLC) (Waters Symmetry Shield™ RP18 5 µm 4.6 × 150 mm) using a binary gradient of 0.1% TFA and 70% acetonitrile in water containing 0.05% TFA.
Molecular weight determination
The apparent molecular weight of purified rTID was determined by SDS-PAGECitation37. Ovalbumin (Mr 45 14;000), carbonic anhydrase (Mr 29 14;000), soybean trypsin inhibitor (Mr 20 14;000), lysozyme (Mr 14 14;500) and aprotinin (Mr 6500) were the marker proteins. The gel was stained with 0.25% coomassie brilliant blue R-250 and destained with 10% (v/v) acetic acid.
Electrospray-mass spectrometry (ESI-MS) was carried out in the ionisation mode on an Acquity SYNAPT HRMS (Waters Associate). The RP-HPLC purified rTID was suspended in 50% CH3CN containing 0.1% HCOOH and directly infused.
SEC measurements were performed using a BIOSEP-SEC-S 3000 (300 × 8 mm, exclusion limit: 700 kDa for globular proteins) column on a Waters Associate HPLC equipped with a binary gradient pumping system and Waters Model 1296 photodiode array detector set at 230 nm. The column was pre-equilibrated with 0.1 M Tris-HCl pH 8.0 at a flow rate of 1 mL per min and calibrated using a mixture of standard proteins, BSA (66 14;000 Da), ovalbumin (45 14;000 Da), carbonic anhydrase (29 14;000 Da), lysozyme (14 14;400 Da), and insulin β chain (3500 Da).
Inhibition studies
The effect of varying substrate concentration (BAPNA) on bovine trypsin in the presence of fixed concentrations of rTID was studied. The modes of inhibition and dissociation constants were evaluated from the double reciprocalCitation38 and Dixon plotCitation39 of the data.
Stability studies
The purified rTID was dissolved in 0.1 M Tris-HCl buffer pH 8.2 and incubated at 90 ± 1°C in a constant temperature water bath. Aliquots were removed at regular time intervals, immediately cooled on ice and assayed for residual trypsin inhibitor activity as described above.
Determining the IC50 value for tryptase
Different concentrations of rTID were assayed against a fixed concentration of human lung tryptase. The IC50 value was determined by plotting log of % residual activity against varying inhibitor concentrations. The Ki was also deduced from the IC50 value following the equation Ki = IC50/1+ ((S)/Km).
In vitro stability of pET20b-rTID and HGI-III to gastrointestinal proteases
The stability of rTID and HGI-III to gastrointestinal proteases like pepsin and pancreatin was studied. The inhibitors were incubated with pepsin (1 mg/ mL) for 2 h and pancreatin (1 mg/ mL) for 4 h. The reaction was stopped by heat inactivation of the proteases. The residual trypsin inhibitory activity was determined as described above.
Results
Design of the stable mini inhibitors
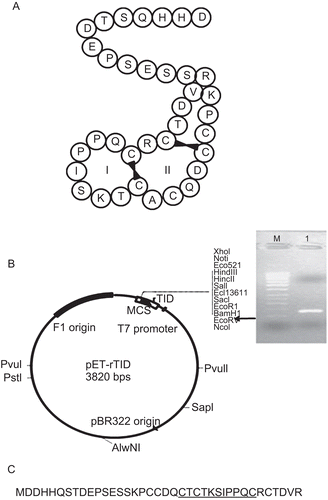
The poor bioavailability and high susceptibility to proteolysis are significant limitations of BBIs as efficient and potent cancer chemopreventive agents. These limitations are attributed due to its fairly large size. The aim of this study was to obtain a molecularly pruned inhibitor that was stable, yet exhibited trypsin inhibition. HGI-III similar to soybean BBI possesses two domains easily distinguishableCitation34. Primers were designed to amplify the TID of HGI-III comprising loops I and II, this included residues 1–36 of HGI-III. A ~108 bp amplicon obtained was blunt end ligated to obtain the plasmid pET20b-rTID (). The deduced amino acid sequence of the amplicon was commensurate with that of residue 1–36 of HGI-III in addition to that derived from the vector.
Figure 1. (A) Pictorial representation of the trypsin inhibitory domain (TID) of the bifunctional horsegram BBI (HGI-III). (B) Schematic representation of the expression cassette in pET20b vector to produce rTID protein. Inset: Agarose gel electrophoresis showing 108 bp amplicon of the TID domain. Lane 1: 50 bp DNA ladder; Lane 2: PCR amplicon. C: Translated protein sequence with the additional residues of the vector.
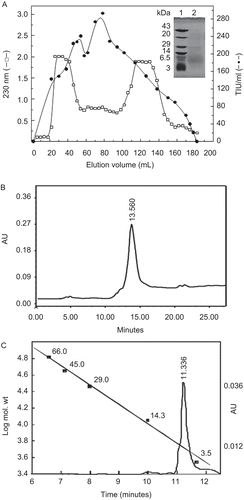
Purification and characterisation of rTID
rTID was expressed in E. coli BL21 (DE3) pLysS and solubilised by sonication. The trypsin inhibitory activity of the crude cell lysate was 4.7 ± 0.2 × 105 TIU/L culture. The inhibitor was partially purified by Sephadex G-50 SEC (). Further purification by RP-HPLC () afforded the rTID in an apparently homogenous form. The specific activity of the purified rTID was 4.6 ± 0.5 × 103 U/mg. The apparent molecular mass estimated by SDS-PAGE was ~4000 ± 1000 Da ( inset), which was in close agreement to the theoretical molecular mass (). The rTID contains in addition to residues 1–36 of HGI-III two additional residues M&D from the vector. ESI-tandem MS indicated that the molecular mass of rTID was 4299 Da.
Figure 2. (A) Purification profile of rTID. The E. coli cell lysate was loaded on a Sephadex G-50 column equilibrated with 25 mM Tris-HCl buffer (pH 8.2) at a flow rate 10 mL/h. (–•–) TIU/mL,(–□–) A 230. Inset: SDS-PAGE (15% T, 2.7% C) of rTID. Lane 1: Molecular weight marker; Lane 2: purified rTID. (B) The pooled trypsin inhibitor fraction of Sephadex G-50 purification step was lyophilised and repurified by RP-HPLC using a binary gradient. (C) The purified rTID was dissolved in 0.1 M Tris-HCl, pH 8.0 and loaded on to a BIOSEP-SEC-S 3000 column pre-equilibrated in the same buffer and eluted at 1 mL/min. Retention time vs log molecular weight standard proteins is also shown.
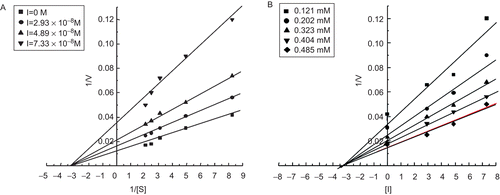
Inhibitory properties of rTID
The initial rates of trypsin reaction in the presence and absence of rTID followed Michaelis Menten kinetics (results not shown). The mode of rTID inhibition was evaluated from the double reciprocal plots of trypsin titrated with different concentrations of rTID. The results indicate that rTID is a non-competitive inhibitor (). The apparent Ki from Dixon plots of the same data was 3.2 ± 0.17 × 10−8 M () for trypsin. The reported Ki of seed HGI-III is 8.7 × 10−8 M for trypsin whereas the Ki of a refolded rHGI is 6.1 ± 0.13 × 10−8 MCitation35. The Ki thus establishes a very high affinity between trypsin and rTID, close in agreement with the Kis reported for other legume BBIs, but one order lower than the cyclic SFTICitation6.
Figure 3. (A) Lineweaver-Burk plot determining mode of inhibition of rTID. (B) Dixon plot for determining the equilibrium dissociation constant (Ki) of rTID. Data points are the average of three determinations.
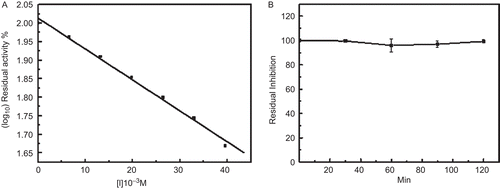
The inhibition of rTID towards human lung tryptase was also evaluated. Although BBIs do not commonly inhibit human tryptaseCitation18 the BBI-derived peptides are potent inhibitors of tryptaseCitation33. The purified rTID inhibited human lung tryptase with an IC50 value 3.78 ± 0.23 × 10−7 M () and the deduced Ki was 2.2 × 10−7 M. Preliminary results indicate that rTID inhibits trypsin like proteases in the colonal lumen of ulcer patients (data not shown).
Thermal stability of rTID
Pre-incubation of rTID for different time periods at 90°C had no effect on the trypsin inhibitor activity (). These results are in agreement to that reported for the legume seed BBIs and also for the seed inhibitor HGI-III. The remarkable stability of BBIs have been ascribed to the conserved array of seven disulfide bonds. In rTID two of these conserved disulfides are still intact, which plausibly explains the high heat stability.
In vitro stability of rTID to gastrointestinal proteases
rTID was resistant to the digestion of gastrointestinal proteases like pepsin and pancreatin or the combination of both. HGI-III, the seed inhibitor used as a control also showed similar results (). These results are in agreement to the other BBI proteins which reach the colon in their active form.
Discussion
Physiological proteinase inhibitors are required for the precise spatial and temporal regulation of proteinase activity. Naturally occurring protein-proteinase inhibitors like BBI therefore find use in therapeutic intervention to limit the activity of proteinases associated with disease. Following the recognition that legume BBIs are effective in suppressing carcinogenesis in several in vitro and in vivo modelsCitation11, there has been renewed scientific interest on these molecules. A goal in BBI research has been to reduce their size to minimal structural elements yet retaining inhibitor potency and specificity. The growing interest of protein mimicking coupled with use of natural scaffolds as templates and capacity for diversity in chemical synthesis has resulted in an enormous number of synthetic peptide mimicsCitation31,Citation33,Citation40,Citation41. Horsegram seed HGI-III consists of two similar domains each displaying heterologous anti-proteinase activityCitation42. The seed HGI-III inhibits trypsin with a Ki of 87 nM. A functional form of the inhibitor expressed in E. coli is equally active against bovine trypsin with a Ki of 61 ± 0.13 nMCitation35. The larger size of BBI inhibitor limits its bioavailability, rendering the requirement for higher doses when ingested orallyCitation17. Designing smaller versions of this inhibitor, comprising only the domain that displays either trypsin or chymotrypsin inhibitory activity would increase the bioavailability. We therefore designed and expressed rTID, a potent inhibitor of trypsin with a Ki of 3.2 ± 0.17 × 10−8 M, and of a much smaller size (~4200 Da). This inhibitor was not only smaller but was three fold more potent than the seed inhibitor. Very recently a similar study on the expression of a 33 residue peptide derived from mung bean trypsin inhibitor in E. coli as a glutathione-S-transferase fusion protein was reported and the equilibrium dissociation constant for trypsin was 2.12 ± 0.24 × 10−8 MCitation43, very close to that reported for rTID. However the anti-tryptase activity had not been studied. rTID not only inhibited trypsin but also inhibited human lung tryptase with a dissociation constant Ki = 2.2 × 10−7M. Chymase and tryptase are chymotrypsin and trypsin like proteases produced and stored in human mast cells and released upon degranulation as in inflammation. Tryptase is involved in inflammatory and allergic disorders, among them asthma, rhinitis, multiple sclerosis, psoriasis and rheumatoid arthritisCitation32. Soybean BBI is a highly effective inhibitor of chymase but not human tryptase. It is opined that the active site of tryptase is restricted in some manner so that it is not accessible to large protein inhibitors normally effective against trypsin-like proteasesCitation44. HGI-III which inhibits both trypsin and chymotrypsin did not inhibit tryptase. The tryptase inhibition by rTID is consistent with the reported structural constrains of tryptaseCitation44. The smaller size of rTID probably allows it to access the restricted active site of tryptase, which has an unusual tetrameric arrangement with a central pore flanked by active sites. Although tryptase is inhibited by the general synthetic inhibitors of trypsin like proteases viz di-isopropyl fluorophosphate, phenyl methyl sulfonyl fluoride, these compounds are unsuitable for human applications due to their toxicity and low stability. Preliminary data suggest that rTID inhibits trypsin like enzymes of human colon lumen of patients with ulcerative colitis. This result suggests a potential application as a therapeutic agent for ulcerative colitis. BBIC is efficacious with high trypsin and chymotrypsin inhibitor activity without any apparent toxicity for both achieving clinical response and induction of remission in patient with active ulcerative colitisCitation20. A leech derived tight binding tryptase inhibitor, (Ki = 1.4 nM) of 46 amino acids differs from BBI in its sizeCitation45. The N-terminal residue of the larger inhibitors such as bovine pancreatic trypsin inhibitor and ovomucoid inhibitor accord a steric clash with insertion in the restricted active site of tryptase. The observation that rTID inhibits tryptase at the nM level bolsters the proposition that insertion of rTID occurs after residue 174 of tryptase, and thereby inactivating it.
The rTID like the legume BBIs is stable at cooking temperature and towards the digestive enzymes found in humans and animals. Soybean BBIs are reported to survive faecal fermentation and are active anticarcinogensCitation46. These properties propagate their use as therapeutics in the diet for treatment of cancer and other related inflammatory diseases. Human populations whose diet comprises of high concentration of BBI are reported to have lower incidences of colon, breast, prostrate and skin cancerCitation27. rTID consisting of the nine residue loop remains active and is a potent inhibitor of trypsin and human tryptase. The Ki values indicate that it is as potent as the horsegram seed inhibitor and other BBIs. The successful expression of rTID in conjunction with it being smaller and stable demonstrates the feasibility of producing a BBI-derived pharmaceutical for therapeutic interventions.
Table 1. In vitro stability of rTID to gastrointestinal proteases.
Acknowledgements
We are grateful to Dr. V. Prakash, Director, Central Food Technological Research Institute (CFTRI), Mysore and Dr. A. G. Appu Rao Head, Department of Protein Chemistry and Technology, for their advice and valuable suggestions. This investigation was supported by a grant from Department of Science and Technology, New Delhi, India (SR/SO/BB-47/2002). DGM acknowledges CSIR India, for the award of a Senior Research Fellowship.
Declaration of interest
The authors report no declarations of interest
References
- Steiner RF. The interaction of the Bowman-Birk inhibitor with trypsin and chymotrypsin. Eur J Biochem 1972;27:87–92.
- Galasso I, Piergiovanni AR, Lioi L, Campion B, Bollini R, Sparvoli F. Genome organization of Bowman–Birk inhibitor in common bean (Phaseolus vulgaris L.). Mol Breeding 2009;23:617–624.
- Chen P, Rose J, Love R, Wei CH, Wang BC. Reactive sites of an anticarcinogenic Bowman-Birk proteinase inhibitor are similar to other trypsin inhibitors. J Biol Chem 1992;267:1990–1994.
- Terada S, Sato K, Kato T, Izumiya N. Inhibitory properties of nonapeptide loop structures related to reactive sites of soybean Bowman-Birk inhibitor. FEBS Lett 1978;90:89–92.
- Odani S, Ikenaka T. Scission of soybean Bowman-Birk proteinase inhibitor into two small fragments having either trypsin or chymotrypsin inhibitory activity. J Biochem (Tokyo). 1973;74:697–715.
- Luckett S, Garcia RS, Barker JJ, Konarev AV, Shewry PR, Clarke AR et al. High-resolution structure of a potent, cyclic proteinase inhibitor from sunflower seeds. J Mol Biol 1999;290:525–533.
- Bode W, Huber R. Structural basis of the endoproteinase-protein inhibitor interaction. Biochim Biophys Acta 2000;1477:241–252.
- Qi RF, Song ZW, Chi CW. Structural features and molecular evolution of Bowman-Birk protease inhibitors and their potential application. Acta Biochim Biophys Sin (Shanghai) 2005;37:283–292.
- Barbosa JA, Silva LP, Teles RC, Esteves GF, Azevedo RB, Ventura MM et al. Crystal structure of the Bowman-Birk Inhibitor from Vigna unguiculata seeds in complex with beta-trypsin at 1.55 A resolution and its structural properties in association with proteinases. Biophys J 2007;92:1638–1650.
- Capaldi S, Perduca M, Faggion B, Carrizo ME, Tava A, Ragona L et al. Crystal structure of the anticarcinogenic Bowman-Birk inhibitor from snail medic (Medicago scutellata) seeds complexed with bovine trypsin. J Struct Biol 2007;158:71–79.
- Catalano M, Ragona L, Molinari H, Tava A, Zetta L. Anticarcinogenic Bowman Birk inhibitor isolated from snail medic seeds (Medicago scutellata): solution structure and analysis of self-association behavior. Biochemistry 2003;42:2836–2846.
- Koepke J, Ermler U, Warkentin E, Wenzl G, Flecker P. Crystal structure of cancer chemopreventive Bowman-Birk inhibitor in ternary complex with bovine trypsin at 2.3 A resolution. Structural basis of Janus-faced serine protease inhibitor specificity. J Mol Biol 2000;298:477–491.
- Li Y, Huang Q, Zhang S, Liu S, Chi C, Tang Y. Studies on an artificial trypsin inhibitor peptide derived from the mung bean trypsin inhibitor: chemical synthesis, refolding, and crystallographic analysis of its complex with trypsin. J Biochem 1994;116:18–25.
- Lin G, Bode W, Huber R, Chi C, Engh RA. The 0.25-nm X-ray structure of the Bowman-Birk-type inhibitor from mung bean in ternary complex with porcine trypsin. Eur J Biochem 1993;212:549–555.
- Park EY, Kim JA, Kim HW, Kim YS, Song HK. Crystal structure of the Bowman-Birk inhibitor from barley seeds in ternary complex with porcine trypsin. J Mol Biol 2004;343:173–186.
- Tsunogae Y, Tanaka I, Yamane T, Kikkawa J, Ashida T, Ishikawa C et al. Structure of the trypsin-binding domain of Bowman-Birk type protease inhibitor and its interaction with trypsin. J Biochem 1986;100:1637–1646.
- Kennedy AR. The Bowman-Birk inhibitor from soybeans as an anticarcinogenic agent. Am J Clin Nutr 1998;68:1406S–1412S.
- Ware JH, Wan XS, Rubin H, Schechter NM, Kennedy AR. Soybean Bowman-Birk protease inhibitor is a highly effective inhibitor of human mast cell chymase. Arch Biochem Biophys 1997;344:133–138.
- Larionova NI, Gladysheva IP, Tikhonova TV, Kazanskaia NF. Inhibition of cathepsin G and elastase from human granulocytes by multiple forms of the Bowman-Birk type of soy inhibitor. Biokhimiia 1993;58:1437–1444.
- Lichtenstein GR, Deren JJ, Katz S, Lewis JD, Kennedy AR, Ware JH. Bowman-Birk inhibitor concentrate: a novel therapeutic agent for patients with active ulcerative colitis. Dig Dis Sci 2008;53:175–180.
- Murthy HM, Judge K, DeLucas L, Padmanabhan R. Crystal structure of Dengue virus NS3 protease in complex with a Bowman-Birk inhibitor: implications for flaviviral polyprotein processing and drug design. J Mol Biol 2000;301:759–767.
- Caccialupi P, Ceci LR, Siciliano RA, Pignone D, Clemente A, Sonnante G. Bowman-Birk Inhibitors in Lentil: Heterologous Expression, Functional Characterisation and anti-proliferative Properties in Human Colon Cancer Cells. Food Chemistry 2010;120:1058–1066.
- Joanitti GA, Azevedo RB, Freitas SM. Apoptosis and lysosome membrane permeabilization induction on breast cancer cells by an anticarcinogenic Bowman-Birk protease inhibitor from Vigna unguiculata seeds. Cancer Lett 2010;293:73–81.
- Clemente A, Moreno FJ, Marín-Manzano Mdel C, Jiménez E, Domoney C. The cytotoxic effect of Bowman-Birk isoinhibitors, IBB1 and IBBD2, from soybean (Glycine max) on HT29 human colorectal cancer cells is related to their intrinsic ability to inhibit serine proteases. Mol Nutr Food Res 2010;54:396–405.
- Lanza A, Tava A, Catalano M, Ragona L, Singuaroli I, Robustelli della Cuna FS et al. Effects of the Medicago scutellata trypsin inhibitor (MsTI) on cisplatin-induced cytotoxicity in human breast and cervical cancer cells. Anticancer Res 2004;24:227–233.
- Birk Y. The Bowman-Birk inhibitor. Trypsin- and chymotrypsin-inhibitor from soybeans. Int J Pept Protein Res 1985;25:113–131.
- Yavelow J, Finlay TH, Kennedy AR, Troll W. Bowman-Birk soybean protease inhibitor as an anticarcinogen. Cancer Res 1983;43:2454s–2459s.
- Blanca HL, Hsieh CC, Lumen BO. Lunasin and Bowman-Birk protease inhibitor (BBI) in US commercial soy foods. Food Chem 2009;115:574–580.
- Billings PC, St Clair WH, Maki PA, Kennedy AR. Distribution of the Bowman Birk protease inhibitor in mice following oral administration. Cancer Lett 1992;62:191–197.
- Fernandez JH, Mello MO, Galgaro L, Tanaka AS, Silva-Filho MC, Neshich G. Proteinase inhibition using small Bowman-Birk-type structures. Genet Mol Res 2007;6:846–858.
- Jaulent AsM Leatherbarrow, RJ. Design, synthesis and analysis of novel bicyclic and bifunctional protease inhibitors. Protein Eng Des Sel 2004;17:681–687.
- Scarpi D, McBride JD, Leatherbarrow RJ. Inhibition of human beta-tryptase by Bowman-Birk inhibitor derived peptides: creation of a new tri-functional inhibitor. Bioorg Med Chem 2004;12:6045–6052.
- Zablotna E, Jaskiewicz A, Legowska A, Miecznikowska H, Lesner A, Rolka K. Design of serine proteinase inhibitors by combinatorial chemistry using trypsin inhibitor SFTI-1 as a starting structure. J Pept Sci 2007;13:749–755.
- Sreerama YN, Das JR, Rao DR, Gowda LR. Double-headed trypsin/chymotrypsin inhibitors from horse gram (Dolichos biflorus): purification, molecular and kinetic properties. J Food Biochem 1997;21:461–477.
- Muricken DG, Gowda LR. Functional expression of Horsegram (Dolichos biflorus) Bowman-Birk inhibitor and its self association. Biochim Biophys Acta 2010;1804:1413–1423.
- Kakade ML, Simons N, Liener IE. An evolution of natural vs. synthetic substrate for measuring the antitryptic activity of soybean samples. Cereal Chem 1969;46:518–526.
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970;227:680–685.
- Lineweaver H, Burk D. The determination of enzyme dissociation constants. J Am Chem Soc 1934;56:658–666.
- Dixon M. The determination of enzyme inhibitor constants. Biochem J 1953;55:170–171.
- Fear G, Komarnytsky S, Raskin I. Protease inhibitors and their peptidomimetic derivatives as potential drugs. Pharmacol Ther 2007;113:354–368.
- McBride JD, Leatherbarrow RJ. Synthetic peptide mimics of the Bowman-Birk inhibitor protein. Curr Med Chem 2001;8:909–917.
- Prakash B, Selvaraj S, Murthy MR, Sreerama YN, Rao DR, Gowda LR. Analysis of the amino acid sequences of plant Bowman-Birk inhibitors. J Mol Evol 1996;42:560–569.
- Qi RF, Liu ZX, Xu SQ, Zhang L, Shao XX, Chi CW. Small peptides derived from the Lys active fragment of the mung bean trypsin inhibitor are fully active against trypsin. Febs J 2010;277:224–232.
- Stubbs MT, Morenweiser R, Stürzebecher J, Bauer M, Bode W, Huber R et al. The three-dimensional structure of recombinant leech-derived tryptase inhibitor in complex with trypsin. Implications for the structure of human mast cell tryptase and its inhibition. J Biol Chem 1997;272:19931–19937.
- Sommerhoff CP, Söllner C, Mentele R, Piechottka GP, Auerswald EA, Fritz H. A Kazal-type inhibitor of human mast cell tryptase: isolation from the medical leech Hirudo medicinalis, characterization, and sequence analysis. Biol Chem Hoppe-Seyler 1994;375:685–694.
- Marín-Manzano MC, Ruiz R, Jiménez E, Rubio LA, Clemente A. Anti-carcinogenic soyabean Bowman-Birk inhibitors survive faecal fermentation in their active form and do not affect the microbiota composition in vitro. Br J Nutr 2009;101:967–971.