Abstract
Anti-apoptotic proteins such as BCL-2, BCL-XL and MCL-1 bind with pro-apoptotic proteins to induce apoptosis mechanism. BCL-2 family proteins are key regulators of apoptosis process. Over expression of these anti-apoptotic proteins lead to several cancers by preventing apoptosis. A number of studies revealed that ginseng derivatives reduce tumor growth. Ginseng, the most valuable medicinal herb found in eastern Asia belongs to Araliaceae family. In this study, docking simulations were performed for anti-apoptotic proteins with several ginsenosides from Panax ginseng. Our finding shows ginsenosides Rf, Rg1, Rg3 and Rh2 have more binding affinity with BCL-2, BCL-XL and MCL-1 and other ginsenosides also interact with each anti-apoptotic proteins. Therefore, ginseng derivatives represent a novel class of potent inhibitors and could be used for cancer chemotherapy.
Introduction
Apoptosis (programmed cell death) play a vital role in cell differentiation and in the elimination of cells that sustain genetic damage or that undergo uncontrolled cellular proliferationCitation1. BCL-2 family proteins are important integral membrane proteins located mainly on the outer membrane of mitochondria, which play a critical role in regulating and executing apoptosisCitation2. BCL2 family proteins can be divided into two classes, one involved in the promotion (pro-apoptotic) and another involved in the inhibition (anti-apoptotic) of apoptosis. The fine balance between the concentrations of pro-apoptotic and anti-apoptotic BCL-2 proteins is maintained by homeostasis; any perturbations to this balance may lead to disease including restenosis, stroke, heart failure, HIV infection, and cancerCitation3,Citation4. The anti-apoptotic proteins such as BCL-2, BCL-XL, and MCL1 contain four conserved BH (BCL 2 homology) homology domains such as BH1, BH2, BH3, and BH4, while pro-apoptotic proteins contain only BH3 domainsCitation2,Citation5. The BH3 region is responsible for mediating the interactions with anti-apoptotic proteins and is related to the ability of a protein to promote programmed cell deathCitation6. The structures of BCL-27 and BCL-XL8 are composed of eight α-helices with a hydrophobic groove on the protein surface containing all the four (BH1, BH2, BH3 and BH4) conserved domains. MCL -1 shows structural similarity with BCL-2 and BCL-XL except the absence of BH4 domain at the N-terminalCitation8. After receiving the appropriate signals, pro-apoptotic proteins bind to anti-apoptotic proteins via BH3 domains on their surfaces. When cytochrome c is released from the mitochondrial inner membrane space, the apoptosis process is initiatedCitation9. Overexpression of anti-apoptotic BCL-2 family proteins, prevents the release of cytochrome c from mitochondria, and is responsible for many types of human cancers like breast and prostate cancerCitation10,Citation11. Previous studies have revealed that ginseng has preventive activity against the development of cancer. Administration of ginseng extract has been demonstrated to increase resistance and decrease effects of colon, lung, liver, pancreas, pharynx, skin, stomach, ovarian, uterine cervix, mammary gland, and kidney cancerCitation12,Citation13. Korean Panax ginseng C.A. Meyer, commonly used as herbal medicine has numerous pharmacological activitiesCitation14 and belongs to the Araliaceae family found in eastern Asia. The biologically active constituents found in Panax ginseng include ginsenosides, polysaccharides, peptides, poly acetylene alcohols, and fatty acidsCitation15,Citation16. In addition, the main active compounds of Panax ginseng are ginsenosides, which are triterpene saponins. Triterpenes constitute one of the most important classes of natural products. These compounds also exhibit a wide range of structural diversity and biological activity and are one of the economically important natural medicines. There are 38 known ginsenosides and the structures of these ginsenosides include a four-ring rigid steroid skeleton with a modified side chainCitation17. These ginsenosides are reported to have various pharmacological effects, including immunomodulatory, anti-stress, anti-hyperglycemic, anti-inflammatory, anti-allergic, anti-atherosclerotic, anti-hypertensive, anti-oxidant, anti-diabetic, and anti-cancer effects, as well as effects on the cardiovascular system and central nervous systemCitation18–20. The pharmacological effects of ginseng have been demonstrated in many experimental studies. Based on those studies, ginseng compounds have been considered as potential agents for cancer chemotherapy.
A number of chemical compounds were identified and designed based on computer modeling (In silico method) to understand the interactions among BCL2 family proteins. These molecules were tested in animals and cells based on a trial-and-error method. Based on the experimental evidence, we utilized various ginsenoside compounds, and BCL-2, BCL-XL and MCL-1 proteins to perform docking simulation. Further docked complexes were confirmed by molecular dynamics and calculated interaction energy. In addition, structural studies were performed to understand the interaction of three anti-apoptotic proteins with ginsenosides at the molecular level.
Material and methods
Protein preparation
For docking studies three-dimensional structures of anti-apoptotic proteins such as BCL-2, BCL-XL, and MCL-1 were retrieved from the RCSB protein data bankCitation13,Citation21 with their respective PDB id’s 2W3LCitation22, 2YXJCitation23, and 3KZ0Citation24. These protein structures were determined using X-ray diffraction methods. For docking simulations, the hetero atoms were removed, and H-atoms were added into all protein structures using the automated docking tool, AutoDockCitation25–27. Further active sites were analyzed for each protein using web-based online tool, Q site finderCitation28. These active sites were chosen as the most favorable binding residues for our docking simulation.
Ligand preparation
The structures of ginsenosides Rb1, Rb2, Rc, Rd, Re, Rf, Rg1, Rg2, Rg3, Rh1, Rh2, and Ro, was retrieved from the Pubchem compound database from the National Center for Biotechnology InformationCitation29,Citation30 with their respective id’s; Rb1: 432524, Rb2: 44584746, Rc: 44593677, Rd: 25203001, Re: 441921, Rf: 6441008, Rg1: 432116, Rg2: 6441009, Rg3: 159727, Rh1: 12855920, Rh2: 404133, and Ro: 11815492. These ginsenosides contain similar backbones with modified side chains. In addition, anti-apoptotic known inhibitors such as ABT-737 (11228183), ABT-702 (3965212) and obatoclax (4905081) were used as control for our study. The downloaded structures were converted into pdb format using freely available open source tool, Open BabelCitation31.
Sequence alignments
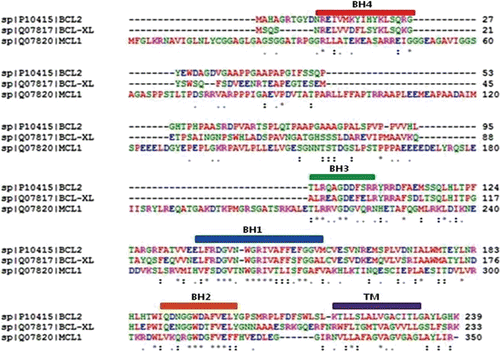
The anti-apoptotic protein sequences were retrieved from Swiss-Prot with the following accession numbers: BCL-2 (P10415), BCL-XL (Q07817), and MCL-1 (Q07820). Using these protein sequences, multiple sequence alignments were performed using the program ClustalWCitation32 to identify sequential alignments of structural motifs. The results of multiple sequence alignments are described in , including the conserved homology domain regions of each protein.
Molecular docking
Docking studies were performed with three anti-apoptotic proteins and twelve ginsenosides using the automated docking tool, AutoDock 4.2.3. This tool operates using a Lamarckian genetic algorithm (LGA)Citation33. The active sites were input and a grid parameter file for each protein was generated by fixing the number of grid points on the x, y, and z axes to 60 × 60 × 60, though the size changed depending on the ginsenosides size. AutoDock grids were calculated for regularly spaced points at intervals of 0.375 Å contained within a cube based on the BCL-2, BCL-XL, and MCL-1 active sites. The population size was set to 150 and the individuals were initialized randomly. The maximum number of energy evaluations was set to 500,000, and the maximum number of generations was 1000. Other docking parameters were set to the software’s default values. The Lamarckian genetic algorithm was chosen to determine the best conformers in fifty independent trials of each ginsenosides. The AutoGrid 4.2.3 and AutoDock 4.2.3 programs were used to produce grid maps and to obtain results.
Molecular dynamic simulation
MD simulation studies were carried out as previously describedCitation34. All molecular dynamic simulations were run with YASARA using AMBER 03 force fieldCitation35 under periodic-boundary conditions and with explicit solvent method. In this study, we have conducted MD simulations for fifteen structures, which includes three apo proteins (2W3L, 2YXJ, 3KZ0) and also twelve holo proteins that were obtained from our docking studies (each apo protein complexes with four different ligands namely ginsenosides Rf, Rg1, Rg3 and Rh2). A simulation cell was constructed around the model with a 7.9 Å cutoff for Lennard-Jones forces and the direct space portion of electrostatic forces, which were calculated using the Particle Mesh Ewald method. The pKa values of the ionizable groups in the model were predicted and assigned the protonation states based on pH 7.0. The cell was filled with water, and the AMBER 03 electrostatic potential was evaluated for all water molecules; the one with the lowest and highest potential were turned into sodium and chloride counter ions until the cell was neutral. A short steepest descent minimization of all atoms was performed to remove severe bumps in the protein. A start-up simulation was then carried out for 5 picoseconds (ps) using a multiple time step of 1 femtosecond (fs) for intramolecular forces and 2 fs for intermolecular forces, with all heavy protein atoms fixed such that the solvent molecules smoothly covered the protein surface. Simulated annealing minimizations were carried out at 298 K, and the velocities were scaled down every 10 steps for a total time of 5 ps in 500 steps. The simulations were carried out using the AMBER 03 force field at 298K and 0.9% NaCl for 3 nanoseconds (ns)Citation35,Citation36. The snapshots of the simulation were stored at every 25 ps for further analysis. In this study, docking and dynamic simulations were performed using an Intel(R) 2.93 GHz Xenon(R) CPU 5670 and a 64-bit RHEL5.5 Server.
Interaction energy calculation
To calculate total interaction energy for twelve complex structures with three different proteins were used Calculate Interaction Energy protocol encoded in Discovery Studio 2.537. The CHARMm force field was assigned on complexes before calculation and all other parameters used as their default parameters. The final trajectories of MD simulations were selected to calculate non-bonded interactions (van der Waals and Electrostatic) between two sets of atoms.
Results
Docking studies were performed on ginsenoside compounds and anti-apoptotic proteins. After docking, the ligands were ranked according to their protein-ligand affinity. AutoDock results were analyzed based on the interactions between anti-apoptotic proteins and ginsenosides and the binding energies of the complexes. The accuracies of the AutoDock results were confirmed by considering the lowest binding free energy and the hydrogen bonds between the anti-apoptotic proteins and ginsenosides. Ginsenosides demonstrated good affinity with all anti-apoptotic proteins.
Docked results with BCL-2
The docking results are ranked according to the binding energies with ginsenosides. The docking simulation of BCL-2 to ginsenosides Rf, Rg3, and Rh1 resulted in the formation of one hydrogen bond with binding energies of −1.7, −1.9, and −3.2 kcal/mol and hydrogen bond lengths of 1.968 Å, 2.239 Å, and 2.075 Å, respectively. Also, formation of two hydrogen bonds was noted with ginsenoside Rg1, resulting in an energy of −1.08 kcal/mol and hydrogen bond lengths of 2.062 Å and 2.202 Å. A similar observation was noted with ginsenoside Rh2, with energy of −4.28 kcal/mol and hydrogen bond lengths of 2.215 Å and 2.166 Å. Based on these results, ginsenosides Rg1 and Rh2 were found to have a good binding affinity with BCL-2 proteins, which also have strong interactions with ginsenosides Rf, Rg3, and Rh1. No hydrogen bond interaction was observed with ginsenosides Rb1, Rb2, Rc, Rd, Re, Rg2 and Ro. Each ginsenoside interaction with binding free energy (kcal/mol) and the residues involved in van der Waals interactions are described in Supplementary Table 1.
Table 1. The binding energies of BCL-2, BCL-XL, and MCL-1 and the experimentally known small molecules.
Docked results with BCL-XL
Analysis of the docking results suggests that BCL-XL has a binding affinity with ginsenosides Rg1, Rg2, Rg3, and Rd through the formation of one hydrogen bond with binding energies of −1.2, −1.05, −6.87, and −2.36 kcal/mol and hydrogen bond lengths of 2.06 Å, 2.218 Å, 2.068 Å, and 1.87 Å, respectively. Also, formation of two hydrogen bonds was observed with ginsenoside Rf with an energy of −3.83 kcal/mol and hydrogen bond distances of 2.127 Å, 2.18 Å, ginsenoside Rh2 with a binding energy of −0.31 kcal/mol and hydrogen bond distances of 2.228 Å, 1.183 Å, and ginsenoside Ro with a binding energy of −1.5 kcal/mol and hydrogen bond distances of 1.794 Å and 2.164 Å. The docked interaction results and residues involved in van der Waals forces are described in Supplementary Table 2. Ginsenosides Rb1, Rb2, Rc, Re, and Rh1 exhibited no binding affinity or hydrogen bond formation with BCL-XL. These interaction results show that BCL-XL interacts more highly with ginsenosides Rg1, Rg2, Rg3, Rd, Rf, Rh2, and Ro.
Table 2. Molecular interaction results of BCL-2, BCL-XL, and MCL-1 into Rg1, Rg3, Rh2, and Rf.
Docked results with MCL-1
The docked results showed (Supplementary Table 3) that ginsenoside compounds Rb1, Rb2, Rc, Rd, Re, Rg2, and Rh1 have no binding affinity with MCL-1 but the rest, ginsenosides Rg3, Ro, Rf, Rg1, and Rh2, found to have good binding affinity. The simulation results showed that ginsenosides Rg3 and Ro interacted with binding energies of −0.81 and −3.01 kcal/mol in the formation of two hydrogen bonds with lengths of 2.183 Å, 2.132 Å and 2.236 Å, 2.165 Å, respectively. In addition, like BCL-2 and BCL-XL, MCL-1 also formed a one hydrogen bond with ginsenosides Rf, Rg1, and Rh2 with hydrogen bond distances of −2.05 Å, −2.25 Å, and −3.25 Å and interaction energies of −2.05, −2.25, and −3.22 kcal/mol, respectively.
Table 3. Average energy interactions between anti-apoptotic proteins with ginsenosides complexes.
Structure Refinement and Stability Evaluation
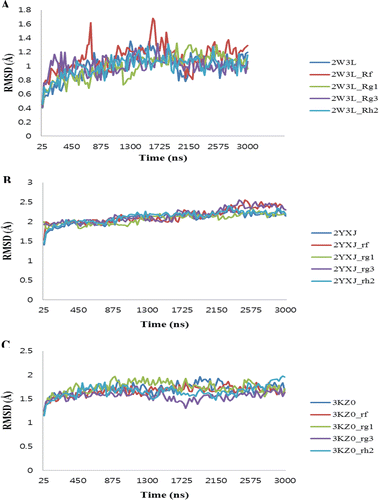
We have conducted the MD simulation for all apo proteins and also their complexes (see Methods) in order to assess the structural stability. shows the backbone RMSD plot for the protein Cα-atoms with reference to the initial structure and as a function of time. In case of 2W3L, 2YXJ and 3KZ0 complexes, the plot shows that all of the models reached an equilibrium state only after 1.1 ns, 0.95 ns and 0.4 ns of the simulation and remained constant until the end of the dynamics with RMSD values of 1.1, 2 and 1.54 Å, respectively. It is of worthwhile to note that these simulations are the longest explicit solvent MD simulations ever carried out on these protein families. Superimposition of the initial structure with the final refined structure revealed that the small structural rearrangement mainly takes place only in the active site. The final snapshots of all the structures were taken and subjected to energy minimization and were subsequently used for the binding energy calculations. The results of total interaction energy and non- bonded interaction energy values were listed in .
Discussion
In earlier days, chemical methods were used to identify anti-cancer agents; however, effective chemo-preventive agents were found in natural products. The commercially available ginseng roots are classified into fresh, white, and red ginseng; these have been officially approved and are used in more than 50 countries. Several experimental studies have been performed to evaluate the effects of ginseng in cancer therapy. Toxicological and clinical studies support the production and consumption of ginseng, which has been shown to reduce cancer risk by up to 50%Citation38. Previous investigations have shown that ginsenosides Rg3 inhibit the endothelial cell apoptosis through increased expression of BAX and decreased BCL-2 in prostate cancer cellsCitation39,Citation40. Ginsenoside Rh2 induces apoptosis and interacts with anti-apoptotic proteins in rat glioma C6Bu-1 cellsCitation41 as well as inhibits the growth of human ovarian cancer cells in nude miceCitation42. The mechanism through which ginsenosides regulate the apoptosis pathway is still unclear. In this study we analyzed the interaction between ginsenosides Rg1, Rg3, Rh2, and Rf with BCL-2, BCL-XL, and MCL-1 proteins.
Detailed simulation results of twelve ginsenosides with three anti-apoptotic proteins involved in the docking simulation were given in . Intermolecular energy (van der Walls energy, hydrogen bonding energy, desolvation energy, and electrostatic energy), internal energy, docking energy, binding energy and torsional free energy of ginsenosides are calculated using simulation results. During the docking interactionsTYR67, GLU95, ARG142, ARG12, THR137, and ASN131 were involved in the formation of hydrogen bonds with BCL-2. ALA142, GLU153, THR109, LYS16, LYS20 and LEU267, THR266, GLU288, HIS224 were involved in BCL-XL and MCL-1 binding respectively (Supplementary Table 1, 2, 3). The hydrogen bonds and their distances are shown . Docking experimentally known inhibitors with same proteins was done to confirm our simulation and interaction results were mentioned in . Moreover docking simulation between ginsenosides and known inhibitors with same protein showed similar results to those of original modes, suggesting Autodock was appropriate for docking studies. From the present study it can be suggested that ginsenosides Rg1, Rg3, Rh2, and Rf interacted with BCL-2, BCL-XL, and MCL-1 anti-apoptotic proteins. Thus the expression of BCL-2, BCL-XL, and MCL-1 proteins were decreased.
Figure 2. Illustration of docked complexes of BCL-2 (A), BCL-XL (B) and MCL-1 (C) with ginsenosides Rg1, Rg3, Rh2 and Rf.
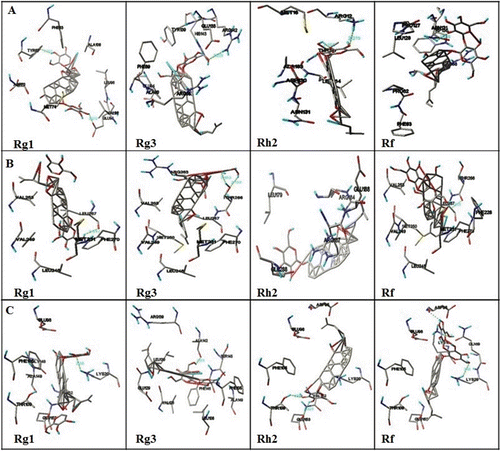
Figure 3. The RMSD of the twelve studied complexes to their initial structure obtained MD simulations. (A) 2W3L-ginsenosides, (B) 2YXJ-ginsenosides, (C) 3KZ0-ginsenosides.
The MD simulations were performed to obtain complex models in the state close to natural conditions. The MD simulation results show that binding mode of ligand-receptor interaction is nearly the same as in molecular docking. Also final MD simulation trajectory files confirmed that strong interactions between ligand and receptor (based on total interaction energy and non-bonded interaction). Our results suggest that hydrogen bonding and electrostatic interaction residues are potential inhibitor sites for all three anti-apoptotic proteins. Furthermore, these residues also bind with BH3 domains to down-regulate apoptosis for the possible prevention of cancer. Docking simulation and molecular dynamics results confirmed that ginsenosides compounds are potential ligands for anti-apoptotic proteins such as BCL-2, BCL-XL, and MCL-1.
Conclusions
Ginseng has been used as a valuable oriental medicine for thousands of years. Recently, many studies have focused on the identification of cancer inhibitors from natural sources, and clinical trials with such chemicals have begun. Our study concludes that ginsenosides, particularly Rg1, Rg3, Rh2, and Rf will be effective compounds to control the overexpression of anti-apoptotic proteins such as BCL-2, BCL-XL, and MCL-1 demonstrating that these agents may be used in cancer treatment. Many other convincing studies also suggested that ginseng can be used in chemotherapy. Ginsenosides should be subjected to further experimental study in order to confirm these findings.
Supplementary Material
Download PDF (30.2 KB)Acknowledgment
The authors are very grateful for the help and cooperation from Dr. Balachandran Manavalan, Ajou University, South Korea.
Declaration of interest
This research was supported by the MKE (Ministry of Knowledge Economy), Korea, under the ITRC (Information Technology Research Center) support program supervised by the NIPA (National IT Industry Promotion Agency)’’ (NIPA-2011- (C1090-1121-0003)).
References
- Raff M. Cell suicide for beginners. Nature 1998;396:119–122.
- Reed JC. Mechanisms of apoptosis. Am J Pathol 2000;157:1415–1430.
- Wang D, Liao W, Arora PS. Enhanced metabolic stability and protein-binding properties of artificial alpha helices derived from a hydrogen-bond surrogate: application to Bcl-xL. Angew Chem Int Ed Engl 2005;44:6525–6529.
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000;100:57–70.
- Adams JM, Cory S. The Bcl-2 protein family: arbiters of cell survival. Science 1998;281:1322–1326.
- Chittenden T, Flemington C, Houghton AB, Ebb RG, Gallo GJ, Elangovan B et al. A conserved domain in Bak, distinct from BH1 and BH2, mediates cell death and protein binding functions. Embo J 1995;14:5589–5596.
- Zhai D, Ke N, Zhang H, Ladror U, Joseph M, Eichinger A et al. Characterization of the anti-apoptotic mechanism of Bcl-B. Biochem j 2003;376:229–236.
- Michels J, Johnson PW, Packham G. Mcl-1. Int J Biochem Cell Biol 2005;37:267–271.
- Petros AM, Olejniczak ET, Fesik SW. Structural biology of the Bcl-2 family of proteins. Biochim Biophys Acta 2004;1644:83–94.
- Fernández Y, Gu B, Martínez A, Torregrosa A, Sierra A. Inhibition of apoptosis in human breast cancer cells: role in tumor progression to the metastatic state. Int J Cancer 2002;101:317–326.
- Lebedeva I, Rando R, Ojwang J, Cossum P, Stein CA. Bcl-xL in prostate cancer cells: effects of overexpression and down-regulation on chemosensitivity. Cancer Res 2000;60:6052–6060.
- Kakizoe T. Asian studies of cancer chemoprevention: latest clinical results. Eur J Cancer 2000;36:1303–1309.
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H et al. The Protein Data Bank. Nucleic Acids Res 2000;28:235–242.
- Leung KW, Wong AS. Pharmacology of ginsenosides: a literature review. Chin Med 2010;5:20.
- Attele AS, Wu JA, Yuan CS. Ginseng pharmacology: multiple constituents and multiple actions. Biochem Pharmacol 1999;58:1685–1693.
- Sathiyamoorthy S, In JG, Gayathri S, Kim YJ, Yang DC. Generation and gene ontology based analysis of expressed sequence tags (EST) from a Panax ginseng C. A. Meyer roots. Mol Biol Rep 2010;37:3465–3472.
- Choi KT. Botanical characteristics, pharmacological effects and medicinal components of Korean Panax ginseng C A Meyer. Acta Pharmacol Sin 2008;29:1109–1118.
- Briskin DP. Medicinal plants and phytomedicines. Linking plant biochemistry and physiology to human health. Plant Physiol 2000;124:507–514.
- Vogler BK, Pittler MH, Ernst E. The efficacy of ginseng. A systematic review of randomised clinical trials. Eur J Clin Pharmacol 1999;55:567–575.
- Park J, Rhee D and Lee Y. Biological Activities and Chemistry of Saponins from Panax ginseng C. A. Meyer. Phytochemistry Reviews. 2005; 4: 159–75.
- Deshpande N, Addess KJ, Bluhm WF, Merino-Ott JC, Townsend-Merino W, Zhang Q et al. The RCSB Protein Data Bank: a redesigned query system and relational database based on the mmCIF schema. Nucleic Acids Res 2005;33:D233–D237.
- Porter J, Payne A, de Candole B, Ford D, Hutchinson B, Trevitt G et al. Tetrahydroisoquinoline amide substituted phenyl pyrazoles as selective Bcl-2 inhibitors. Bioorg Med Chem Lett 2009;19:230–233.
- Lee EF, Czabotar PE, Smith BJ, Deshayes K, Zobel K, Colman PM et al. Crystal structure of ABT-737 complexed with Bcl-xL: implications for selectivity of antagonists of the Bcl-2 family. Cell Death Differ 2007;14:1711–1713.
- Dutta S, Gullá S, Chen TS, Fire E, Grant RA, Keating AE. Determinants of BH3 binding specificity for Mcl-1 versus Bcl-xL. J Mol Biol 2010;398:747–762.
- Goodsell DS, Morris GM, Olson AJ. Automated docking of flexible ligands: applications of AutoDock. J Mol Recognit 1996;9:1–5.
- Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J Mol Biol 1997;267:727–748.
- Rarey M, Kramer B, Lengauer T, Klebe G. A fast flexible docking method using an incremental construction algorithm. J Mol Biol 1996;261:470–489.
- Laurie AT, Jackson RM. Q-SiteFinder: an energy-based method for the prediction of protein-ligand binding sites. Bioinformatics 2005;21:1908–1916.
- Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Bryant SH. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res 2009;37:W623–W633.
- Wang Y, Bolton E, Dracheva S, Karapetyan K, Shoemaker BA, Suzek TO et al. An overview of the PubChem BioAssay resource. Nucleic Acids Res 2010;38:D255–D266.
- Geldenhuys WJ, Gaasch KE, Watson M, Allen DD, Van der Schyf CJ. Optimizing the use of open-source software applications in drug discovery. Drug Discov Today 2006;11:127–132.
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 1994;22:4673–4680.
- Morris GM, Goodsell DS, Halliday RS, et al. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. Journal of Computational Chemistry. 1998; 19: 1639–62.
- Manavalan B, Basith S, Choi YM, Lee G, Choi S. Structure-function relationship of cytoplasmic and nuclear I?B proteins: an in silico analysis. Plos one 2010;5:e15782.
- Krieger E, Darden T, Nabuurs SB, Finkelstein A, Vriend G. Making optimal use of empirical energy functions: force-field parameterization in crystal space. Proteins 2004;57:678–683.
- Ponder JW, Case DA. Force fields for protein simulations. Adv Protein Chem 2003;66:27–85.
- Accelrys. Accelrys Discovery Studio 2.5, San Diego, CA. 2009.
- Yun TK, Choi SY. Non-organ specific cancer prevention of ginseng: a prospective study in Korea. Int J Epidemiol 1998;27:359–364.
- Kim HS, Lee EH, Ko SR, Choi KJ, Park JH, Im DS. Effects of ginsenosides Rg3 and Rh2 on the proliferation of prostate cancer cells. Arch Pharm Res 2004;27:429–435.
- Min JK, Kim JH, Cho YL, Maeng YS, Lee SJ, Pyun BJ et al. 20(S)-Ginsenoside Rg3 prevents endothelial cell apoptosis via inhibition of a mitochondrial caspase pathway. Biochem Biophys Res Commun 2006;349:987–994.
- Kim YS, Jin SH, Lee YH, Kim SI, Park JD. Ginsenoside Rh2 induces apoptosis independently of Bcl-2, Bcl-xL, or Bax in C6Bu-1 cells. Arch Pharm Res 1999;22:448–453.
- Nakata H, Kikuchi Y, Tode T, Hirata J, Kita T, Ishii K et al. Inhibitory effects of ginsenoside Rh2 on tumor growth in nude mice bearing human ovarian cancer cells. Jpn J Cancer Res 1998;89:733–740.