Abstract
The catalytically inactive isoforms of carbonic anhydrase (CAs) are known as CA-related proteins (CARPs) VIII, X, and XI. They have highly conserved amino acid sequences. These proteins are predominantly expressed in human and mouse brain, however, their precise roles are poorly known. CARP VIII is functionally associated with motor coordination in human and mouse. CARP X is more highly expressed in the pineal gland during night compared to the day time, suggesting a function for wake/sleep patterns. Phylogeny shows that CARP XI has emerged from CARP X. It is only found in tetrapods and is highly expressed in the central nervous system (CNS) of humans and is also associated with several cancers. Detailed analysis of CARPs is in progress in our laboratory to understand their role in normal physiology. We present a review of literature on CARPs and present some novel data on CARPs obtained in our laboratory.
Introduction
The α-carbonic anhydrase (α-CA) gene family consists of 16 isoforms 13 of which are catalytically active and perform the reversible hydration of CO2. The three isoforms which are catalytically inactive lack one or more of the three histidine residues required for the co-ordination of the zinc atom in the active site, necessary for the catalytic activity of the enzymeCitation1. Due to high sequence and structural similarity with the active α-CAs, they are known as CA-related proteins (CARPs). The three mammalian CARPs are named as CARP VIII, CARP X and CARP XI in the order of discovery within the CA family. The existence of CARPs was reported two decades after the first discoveries of the active CA isozymes. In early 1990s, Kato reported CARP VIII based on the expression pattern Car8 cDNAs in the mouse brainCitation2. Since then several expression studies have been performed on CARP VIII in both mouse and humanCitation3–6. The association of CARP VIII with ataxia and neurodegeneration in humans has been studied recentlyCitation7,Citation8, and the crystal structure of human CARP VIII has been reportedCitation7–9. After the discovery of the first CARP VIII gene in 1990, many more cDNAs have been reported in mice and humans ()Citation10. So far, only few reports have been published on CARP X and XI mainly related to their expression in human and miceCitation5,Citation6. Previous studies suggest that CARP XI is associated with gastrointestinal tumorsCitation11. We have recently performed a phylogenetic analysis of all CARPs in the whole animal kingdom and expression analysis in several mouse tissuesCitation12. However, the exact biological functions of CARPs still remain poorly definedCitation13.
Table 1. Molecular properties of human and mouse CARPs.
In the present review, we report the most relevant previous literature on CARPs as well as the data on phylogeny and expression that have emerged recently in our lab. We also present the most recent data on CARP expression in normal and pathological conditions based on the microarray studies available at BioGPS and MediSapiens.
CARP VIII
Among the members of the catalytically inactive CA isoforms, CARP VIII was the first member to be reported based on its expression pattern in mouse brainCitation2,Citation10. After the first discovery several studies have been reported on CARP VIII, which have been mainly related to its expression pattern in both mice and humansCitation3,Citation4,Citation6. These studies include determining the role of CARP VIII in development of cancer, the involvement in neurodegeneration both in mouse and human, structural basis of its inactivity, and its role in other pathological conditionsCitation8,Citation9,Citation14–18. In our laboratory, we have performed the phylogenetic analysis to study the evolutionary history across the species and detailed expression analysis in several mouse tissues at mRNA and protein levelCitation12. Extensive information on CARP VIII has been published in a recent reviewCitation19.
Expression of CARP VIII protein and its mRNA in tissues
The expression of CA8 gene has been studied using northern hybridization. It was shown that there are two transcripts (2.1 and 1.6 kb) for Car8 gene in mice, and it was present in the cerebellum but absent in other parts of the brain, possibly reflecting two different polyadenylation sitesCitation3,Citation6. In-situ hybridization showed that Car8 gene is localized in the Purkinje cells in contrast to Car7 which is only expressed in glia cellsCitation4. Immunohistochemistry showed that CARP VIII is expressed in both adult and fetal brains. Cellular distribution of CARP VIII in the adult and fetal brain specimens is shown in .Citation3,Citation6 Although CARP VIII is predominantly expressed in the cerebellar Purkinje cells, its expression has also been demonstrated in several other human and mouse tissues using reverse transcription-PCR (RT-PCR) and quantitative PCR methods ()Citation3,Citation12,Citation14.
Table 2. Distribution of CARP VIII protein and its mRNA in brain and other tissues.
Expression of CA8 gene has been studied in a large number of microarray experiments. From the data that have been compiled at the BioGPS online portal we can observe that the expression is predominantly seen in the cerebellum, but significant levels of CA8 expression have also been reported in several other tissues ()Citation20,Citation21. Similarly, the expression of CA8 gene has been studied in normal human tissues, cancerous tissues and in other pathological conditions and the data has been reported in MediSapiens portal at www.medisapiens.comCitation21. The expression patterns in the MediSapiens collection show upregulation of CA8 mRNA in several cancers and other diseases (), suggesting a role for CARP VIII in physiological and pathophysiological functions in humans. Recently, expression analysis of CARP VIII was done in zebrafish, both at mRNA and protein level. The expression was found to be similar to expression pattern of humans and mice.
Figure 1. Expression pattern of CA8 gene in human analyzed by microarray method. The expression profile figure is adapted from BioGPS (http://biogps.gnf.org, accessed July 2012). The figures shows expression of CA8 gene in different human tissues samples and expression is significantly high in cerebellum compared to other tissuesCitation20.
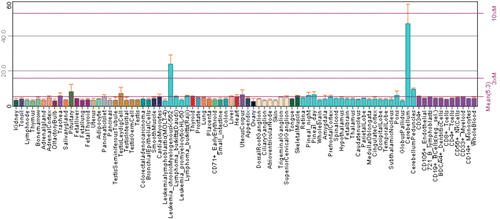
Figure 2. Expression of CA8 gene in normal and pathological conditions in human. Microarray data of the mRNA expression levels of human genes in normal and pathological tissues from MediSapiens. (http://www.medisapiens.com, accessed February 2012)Citation21.
Bioinformatic studies on CARP VIII
Comparative sequence and structural analysis of human CARP VIII and CA II has been reported recently to study the conservation pattern of surface amino acids residues between the “classical” active isozyme, CA II and inactive CARP VIIICitation19. The amino acid residue conservation patterns of CARP VIII and CA II were calculated from multiple sequence alignment (MSA) of the proteins from 11 comparable species and visualized on protein structures of CAII and CARP VIII. The majority of the highly conserved residues were in the active site of the CA II protein core, whereas the protein surface showed more variability. CARP VIII showed very high conservation even on the protein surface and remarkably, one face of CARP VIII protein is very highly conserved. Although the present information does not allow reliable functional correlations, it is tempting to speculate that this face may represent an interaction surface with other proteins which needs to be studied further. Few of the conserved surface residues of CARP VIII are ones shared with CA II.
CARP VIII sequences have been discovered in many deuterostome invertebrates, such as molluscs and chordates, but not in protostomes, such as insectsCitation12. In consequence, the phylogenetic origin of CA8 is more ancient than the origins of CA4, CA5, CA6, CA9, CA12, and CA14, which were formed during the radiation of jawed vertebrates, or CA1, CA2, CA3, and CA13, which are specific to the tetrapod lineage.
CARP VIII in ataxia and mental retardation
The evidence for the involvement of CA8 gene in neurodegeneration and ataxia came from waddles (wdl) mice, which are characterized by wobbly side-to-side ataxic movement throughout their life spanCitation18. Studies on wdl mice revealed a 19-base pair (bp) deletion in Car8 gene. This gene defect is responsible for wdl phenotype based on the evidence that (1) Car8 gene is located within the genetic region of the wdl, (2) Car8 deletion was the only defect detected among genes and expressed sequence tags (ESTs) within the wdl locus from wdl mice, and (3) CARP VIII protein was completely absent in wdl mice. Genetic studies indicated that wdl were autosomal recessive mutantsCitation18. Linkage mapping at The Jackson Laboratory placed the wdl mutation in close proximity to the wd locus on mouse chromosome 4. Interestingly, the waddler (wd) mice, which were discovered in 1959 but are now extinct, produced a similar phenotype to the wdl miceCitation22.
CARP VIII research recently gained a lot of new interest when the first human CA8 gene defect was reported in an Iraqi familyCitation8. The affected members of the family had mild mental retardation and congenital ataxia accompanied by quadrupedal gait.
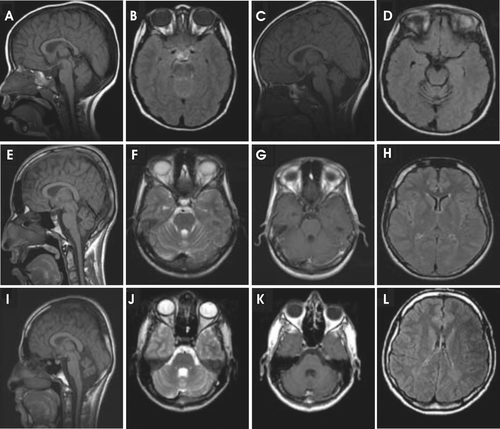
A very recent study reported a novel homozygous (G162R) variation in CA8 gene in members of a Saudi Arabian family. All the seven affected members showed cerebellar ataxia, mental retardation, and disequilibrium syndrome type 3. The patients had variable cerebellar ataxia and mild cognitive impairment without quadrupedal gait. The brain magnetic resonance imaging (MRI) showed loss in cerebellar volume and ill-defined peritrigonal white matter abnormalities ()Citation7. The fluorodeoxyglucose positron emission tomography (FDG PET) revealed hypometabolic cerebellar hemispheres, temporal lobes and mesial cortex. Mild cognitive impairment, a variable degree of cerebellar ataxia, absence of seizures and lack of dysmorphism were the features which these Saudi Arabian patients shared with the members of the Iraqi family reported by Türkmen et al.Citation8 However, the Saudi Arabian patients did not have quadrupedal gait similar to members of the Iraqi family, but it was suggested that this is probably due to environmental factors rather than genuine phenotypic variation of the diseaseCitation7. The brain MRI studies showed variable cerebellar volume and vermian volume loss, the latter progressing in a superior to inferior fashion, as well as peritrigonal white matter changes in all patients but the loss was more striking in older patients (). Functionally, the FDG PET scan provided evidence that the disease has a predilection for certain areas of the brain, in particular the temporal lobes and cerebellar hemispheres. At molecular level, initial copy number and chromosomal imbalances analyses showed no pathological changes. However, linkage analysis revealed a peak on chromosome 8q12.1–8q13.3 with a LOD score of 5.1, covering a linkage interval of 14.3 mb. Sequencing of 8 exons and exon–intron boundaries showed a novel homozygous variation (484G>A) in exon 4 of CA8 gene in all the affected members. Further analysis showed that the novel 484G>A variation to be in heterozygous state in the parents of the affected members. The novel 484G>A homozygous variation in the CA8 gene found in this family will lead to the substitution of a highly conserved glycine residue at position 162 by arginine (G162R). Homozygosity for the variation co-segregated with the disease phenotype and the substitution was not present in 200 unrelated healthy controls of same ethnic origin, indicating that G162R variation is very likely to be pathogenic. By using different prediction algorithms the mutation was predicted to be probably damaging. Moreover, a MSA from more than 20 different organisms including human indicated that the region around the site of the mutation was highly conserved. In addition, G162 localizes to the same region as I165, a residue previously implicated in the spatial constriction of the CO2 hydrophobic pocket of the proteinCitation9.
Figure 3. Reduction in cerebellar volume of Saudi Arabian patients with a mutation in CA8 gene (Gln162-Arg). The brain magnetic resonance imaging (MRI) of the (A–D) patient 1, (E–H) patient 2, and the (I–L) patient 3. Figures show the progressive cerebellar loss in patient 1 between the sagittal T1 (A and C) and axial flair images (B and D) in the scans performed at 4 (A and B) and 8 years of age (C, D), respectively. Sagittal T1 images (E and I) show reduction in cerebellar volume more prominent in patient 3 (I) than in patient 2 (E). The volume loss is not associated with parenchymal signal abnormality as evident in images F, G, J, and K. Small sella turcica and skull base fatty replacement are also shown (E and I). Axial flair images (H and L) demonstrate deep periventricular white matter abnormalities with more involvement seen in patient 3 (L). Modified from Kaya et al.Citation7
Both of these human phenotypes linked with CA8 gene defects are characterized by significant changes in brain function/development. In our laboratory, we aimed to create a novel animal model to test the phenotypic consequences caused by gene silencing of CA8. Our results have shown that knockdown of CA8 gene using antisense morpholinos in zebrafish larvae showed defects in swim pattern and histochemistry revealed reduction in cerebellar volume.
Structural changes in CARP VIII in disease
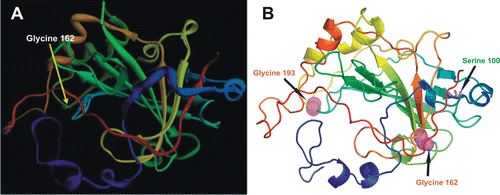
We have studied the three-dimensional location of the two known human CARP VIII mutations in the protein structure ( and ).
Figure 4. Locations of the CA8 gene variation in members of Iraqi and Saudi Arabian family. (A) The location of the variation (yellow arrow, Glycine 162 Arginine) on hypothetical 3-D structure of carbonic anhydrase-related protein VIII (CARP VIII) protein as shown with incorrect numbering in Kaya et al.Citation7 (B) The 3-D structure of human CARP VIII (PDB ID: 2W2J) showing the correct places of variations in the CARP VIII protein (substitution of amino acids) found in members of Iraqi (top right black arrow, Serine 100 Proline) and members of Saudi Arabian family (bottom right black arrow Glycine 162 Arginine), top left black arrow, the position of panel A correctly identified as Glycine 193.
The structural basis of misfolding of the S100P variant protein, which was found in some members of an Iraqi family, can be interpreted in the context of the crystal structure of the wild-type CARP VIII protein as shown in . The location of Ser 100 is at the end of beta strand 6Citation8,Citation9,Citation19. Substituting proline for serine will cause a rigid bend in the peptide backbone, which would likely preclude the sixth or the sixth and seventh (blue and green) loops from assuming the conformations they have in the wild-type protein. Having a relatively short and constrained loop between the strands 5 and 6 may amplify the folding problem in the S100P mutant protein even furtherCitation19.
In case of the other known disease causing variation, G162R, the original reportCitation7 shows an incorrect position of the variant residue (). The location labeled as G162 is in fact G193. The correct position of G162 is shown in . Glycine 162 is a fully buried residue, and it is obvious that substituting arginine in this position alters the structure severely. This agrees with prediction results of a pathogenic variationCitation8.
CARP X
CARP X was the second isoform in the order of discovery among CARPs after CARP VIII. Hewett and Tashian were the first to mention the existence of CARP X gene in 1996 based on many ESTCitation1. The ESTs showed significant homology to CA isozymes and lacked the histidine residues required for catalytic activity and were reported as CA10Citation1. A longer sequence of CA10 was discovered while screening for CCG repeats in cDNA libraries from human brain. Later, Okamoto et al.Citation24 obtained a full-length cDNA clone of 2720 bp and predicted that it encodes a protein of 328 amino acid residues (). The deduced amino acid sequence of the cDNA showed a similarity of 25–57% to other CA isozymes and the greatest identity to CARP XI. Two out of 3 histidine residues needed for binding to zinc were absent from CARP X, suggesting that this CA isozyme has no CA enzymatic activityCitation23. The cDNA presented a 987 bp open reading frame that encoded a 328 amino acid protein with a predicted molecular mass of 37.6 kDa. Similar to CARP XI, CARP X contained a hydrophobic N-terminal extension compared to CA isozymesCitation24. Interestingly, the CARP X sequence includes seven CCG repeats in the 5′-untranslated region followed by two CCG repeats at 16 bp downstream. Polymorphism of its repeat number was reported in normal human populationCitation25.
Expression of CARP X protein and its mRNA in tissues
The mRNA expression of CA10 was studied in a panel of human tissues by northern blot analysis, which showed a 2.8 kb transcript () that corresponded to a full-length cDNA in kidney and brain. No detectable signal was observed in the heart, lung, skeletal muscle, or pancreas. A shorter transcript signal was seen in the placentaCitation23. A number of CA10 ESTs from human placenta have been reportedCitation13. The expression of CA10 mRNA was also studied in the human pancreas, kidney, liver, salivary glands, brain, mammary glands, placenta, and testis by using RT-PCR. The amplification was seen in the kidney, salivary glands, and brainCitation23. The CA10 mRNA expression was also evaluated by RNA dot blotting on a large panel of normal human adult and fetal tissues. Weak but significant signals were observed in almost all parts of the CNS, including the amygdala, cerebellum, cerebral cortex, frontal lobe, hippocampus, medulla oblongata, occipital lobe, putamen, substantia nigra, temporal lobe, thalamus, nucleus accumbens and spinal cord. However, no detectable signal was seen in human fetal brainCitation23.
The cellular distribution of CARP X has been studied in the brain using monoclonal antibodies for human CARP X, which cross-react with mouse homologues. The immunohistochemical analysis showed expression of CARP X protein in many parts of the brain as shown in the .Citation5,Citation6 In the humans, developmental expression of CARP X has been studied from day 84 to 222 of gestation and the expression was seen throughout the gestation period studied (). In our laboratory, we recently studied the expression of Car10 gene both at mRNA and protein levels in a panel of mouse tissues. The expression of CA10 mRNA was predominantly found in all parts of the brain and a faint signal was also found in the eye ( and ). Immunohistochemistry showed expression of CARP X protein in the brain, lung and liver (). Recent investigations have shown that CA10 is expressed in almost all the zebrafish tissues and the expression is highest in the brain. Although the exact functions in the developing brain are still uncertain, these findings suggest certain roles of CARPs in the early development or differentiation of neuroprogenitor cellsCitation5.
Table 3. Distribution of CARP X protein and its mRNA in brain and other tissues.
Figure 5. Expression profile of Car10 mRNA in murine tissues. The relative expression pattern of Car8 mRNAs in 20 different mouse tissues using RT-qPCR. The predominant expression of Car8 mRNA is seen central nervous system (CNS), expression being very high in cerebellum followed by parietal cortex and frontal cortex, very low level of expression was also seen in midbrain and eye (). Adopted from Aspatwar et al.Citation12
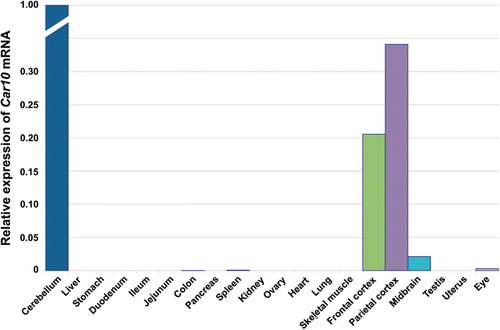
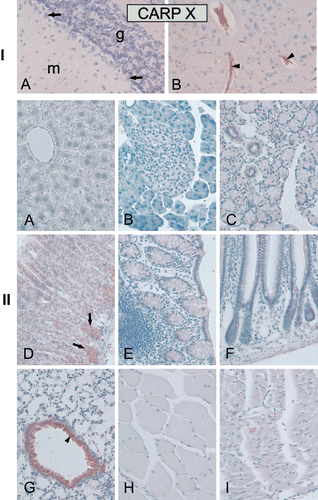
Figure 6. Immunohistochemical staining of carbonic anhydrase-related protein X (CARP X) protein in mouse tissues. Panel I. Expression of CARP X in the brain. (A) There was no expression in cerebellum. (B) The arrowheads in figure show CARP X-positive microcapillaries in the cerebrum. Panel II. Moderate CARP X expression was observed in the respiratory epithelium (arrowhead) of the lung (G). Moderate expression was also present in the gastric glands (arrows in panel D). The heart muscle cells occasionally showed extremely weak signals (I). The other tissues including the (A) liver, (B) pancreas, (C) submandibular gland, (E) colon, (F) small intestine, (H) skeletal muscle remained negative. Original magnifications are at ×20. Adopted from Aspatwar et al.Citation12
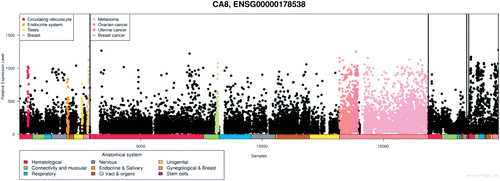
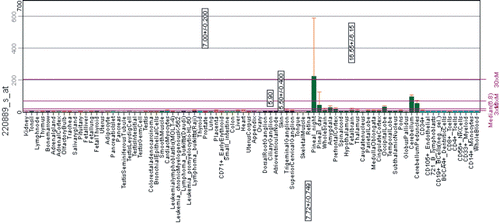
The data on human CA10 gene from BioGPS show expression in the cerebellum and also high levels in pineal gland in the night time but low levels in the day time (). Pineal gland is responsible for sleep and wake patterns and seasonal functions. The periodic expression pattern of CA10 mRNA would suggest its involvement in circadian rhythm in humans, but further studies are required to confirm its function in the pineal glandCitation20.
Figure 7. Expression pattern of CA10 gene in healthy human tissues analyzed by microarray method. The expression profile figure is adapted from BioGPS (http://biogps.gnf.org, accessed July 2012). The figures shows expression of CA10 gene in different human tissues samples and expression is markedly high in the pineal gland during the night compared to day time. Some expression can also be seen in cerebellum and other parts of the brain.Citation20
Bioinformatic analyses of CARP X
The annotation and sequence analysis performed in our laboratory identified 31 full length sequences which included a CARP X sequence from a chordate, Branchiostoma floridae. MSA and analysis of sequences showed 70–100% amino acid identity among vertebrate sequences. The sequence alignment between invertebrates and vertebrates showed 40%-45% amino acid identity. Comparative sequence analysis of active CAs was also done and sequence conservation was found to be higher in all the CARPs than in any of the active CAsCitation12.
The phylogenetic analysis showed that CARP X sequences form three groups. Sequences in group one consisted of sequences from mammals, frogs and lizard, and the other two smaller groups contained sequences from fishes. The phylogenetic analysis showed that there has been a duplication of the ancestral vertebrate CA10 gene in the fish lineage producing the genes CA10a and CA10bCitation12.
A study of invertebrate CACitation26 showed that CARP X-like proteins are present even in nematodes and insects. This means that CARP X is the most ancient CA isoform in present-day animals.
CARP X in disease conditions
The expression of CARP X was studied using monoclonal antibodies for human CARP X in two unique pathological conditions. In one case, the expression was studied in acute disseminated encephalomyelitis, a disease in which there is focal demyelinization in the human brain. In a demyelinized lesion of a patient’s brain, the axons were clearly shown by luxol fast blue stain, but CARP X expression was selectively lostCitation5. The other is the shiverer demyelinated model mouse. In this mutant mouse, exons 3 to 7 of the myelin basic protein gene are deleted and thus myelinization is left incompleteCitation27. In the brain of the shiverer mouse, CARP X expression is almost eliminated from the myelin sheathCitation5. In cultured tumor cells, immunohistochemical analysis showed that CARP X is localized in the cytoplasm. The antibody to CARP X clearly demonstrated linear structures and failed to stain demyelinated regions of brain sections from a patient with acute disseminated encephalomyelitis. These findings indicated that CARP X was expressed in the cytoplasm of the myelin sheathCitation5. The expression pattern in the myelin sheath of the normal brain and the loss of expression in demyelinated areas suggested that CARP X is involved in myelin sheath organizationCitation5.
Expansion mutations in trinucleotide repeats, such as CCG, have been proposed to account for genetic susceptibility to a number of neuropsychiatric disorders. It has been reported earlier that CARP X has a CCG repeat in the 5′-untranslated region with a repeat length polymorphism. For instance, large expansion of 5′-untranslated CCG repeats is responsible for the fragile X A (FMR1 gene) and fragile X E (FMR2 gene) syndromes. Because CARP X is expressed relatively specifically in the myelin sheath, it is possible that CA10 is a causative gene for demyelination disorders, and the presence of CCG repeats would provide one mechanism for a dysfunction of CA10Citation25.
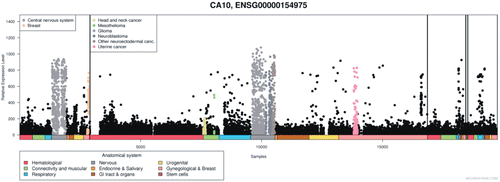
The expression pattern of human CA10 gene from nearly 20,000 microarrays (from www.medisapiens.com) showed upregulation of CA10 mRNA in several cancers (), suggesting that it could be involved in the development of cancers and in other pathological conditionsCitation21.
Figure 8. Expression of CA10 gene in normal and pathological conditions in human: Microarray data of the mRNA expression levels of human genes in normal and pathological tissues from MediSapiens. (http://www.medisapiens.com, accessed February 2012)Citation21.
CARP XI
Among the three CARPs, the CARP XI was the last to be identified. It was discovered accidentally during the construction of a physical map for the cone-rod retinal dystrophy, CORD2 (an inherited blinding condition). The authors identified an anonymous EST D19S799E that mapped close to the distal flanking polymorphic marker D19S412 and reported it as CARP-2 on human chromosome 19qCitation28. Simultaneously, CARP XI was also identified by two other groupsCitation29,Citation30. There were several differences in the reported cDNA sequences, which included amino acid substitutions: 23 Gly(GGT) to Ala(GCT), 24 Asn(AAC) to His(CAC), and 75 Leu(CTG) to Val (GTG) between the sequence submitted by Fujikawa-Adachi et al.Citation29 and the CARP-2 sequence of Bellingham et al.Citation27 and the difference of one amino acid (280 Ile(ATC) to Met(ATG) between the sequence of Fujikawa-Adachi et al. and Lovejoy et al.Citation29,Citation30
A full-length cDNA clone of the CA11 transcript was sequenced, containing 1475 bp and predicted to encode a 328-amino acid polypeptide with a molecular mass of 36.2 kDa ()Citation29. The cDNA included an open reading frame that encoded a 328-residue protein and untranslated sequences of 369 bp at 5′ and 119 bp at 3′ ends. The 3′ untranslated region includes a putative polyadenylation cleavage signal (ATTAAA). The deduced amino acid sequence of CARP XI showed an overall similarity of 42–53% to the active site residues of the other active CA isozymes, however, it lacked three zinc-binding histidine residues, making it catalytically inactiveCitation30. Northern blot analysis demonstrated strong expression of an approximately 1.5 kb transcript in the human brain, particularly in the cerebellum, cerebral cortex and putamenCitation30.
Lovejoy et al.Citation28 observed a strongly predicted signal peptide in the N-terminus of CARP XI, which suggests that CARP XI is a secreted protein.
Expression of CARP XI protein and its mRNA in tissues
CARP XI mRNA expression was studied by northern blot analysis in a panel of human tissues, which had eight vital organs including heart, brain, placenta, lung, liver, skeletal muscle, kidney and pancreas. A strong signal for an ~1.5 kb transcript for CARP XI was detected only in the brain. However, expression analysis with RT-PCR showed presence of CA11 mRNA in the pancreas, liver, kidney, salivary gland and spinal cord as shown in Citation28. The expression was also studied by Bellingham et al. by northern blot analysis in 23 human tissues and the mRNA was found to be abundantly expressed in the brain, moderately in the spinal cord and thyroid, and weakly in the skeletal muscle, kidney, pancreas, ovary, small intestine, colon, peripheral blood leukocyte, stomach, lymph node and adrenal glandCitation28.
Table 4. Distribution of CARP XI protein and its mRNA in brain and other tissues.
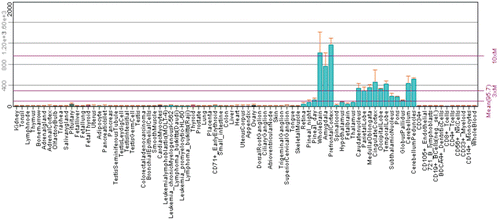
The expression of CARP XI protein was studied in different parts of the adult human brain and human fetal brain at five gestational periods (days 84, 95, 121, 141 and 222) using monoclonal antibodies for human CARP XI protein. The expression pattern of CARP XI protein is shown in Citation5. In our laboratory, we have performed expression analysis of Car11 mRNA and its protein product in a set of mouse tissues using RT-qPCR and immunohistochemistry. The Car11 mRNA was found to be predominantly expressed in the CNS and in many other tissues as seen in and and Citation12. The latest expression profile of CA11 gene is also available at the BioGPS portal, which shows predominant expression of CA11 gene in the CNS ()Citation20. Although the exact functions of CARP XI still remain uncertain, the expression pattern suggests certain roles in the early brain development or differentiation of neuroprogenitor cells.
Figure 9. Distribution of Car11 mRNA in murine tissues. The relative expression pattern of Car11 mRNAs in 20 different mouse tissues analyzed using reverse transcription-quantitative PCR (RT-qPCR). The overall pattern of Car11 mRNA expression was similar to the expression of Car8 mRNA ( and ). The expression of mRNA and can be seen in most of the tissues. The Car11 mRNA was predominantly expressed in frontal cortex followed by significantly high level of expression in parietal cortex, cerebellum and midbrain. Low amount of Car11 mRNA signal was also seen in colon, kidney, ovary lung and eye (). Adopted from Aspatwar et al.Citation12
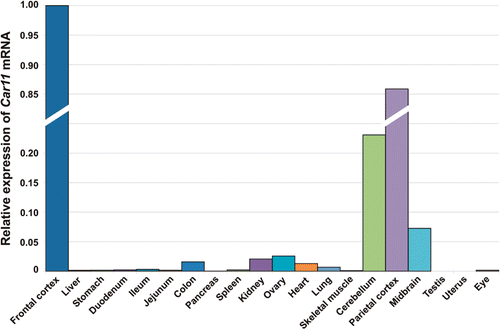
Figure 10. Immunohistochemical staining of carbonic anhydrase-related protein XI (CARP XI) protein in mouse tissues. Panel I. (A) Moderate expression of CARP XI in cerebellar Purkinje cells. (B) There was faint signal for CARP XI in cerebrum. Panel II. Weak immunoreactions for CARP XI in the crypts of Lieberkuhn (arrows in D), gastric glands (arrows in C), and renal tubule cells (F). Extremely weak signals can also be observed in the (A) liver, (E) colon and (I) testis. The (B) pancreas, (G) lung, and (H) skeletal muscle were all negative. Original magnifications are at ×20. Adopted from Aspatwar et al.Citation12
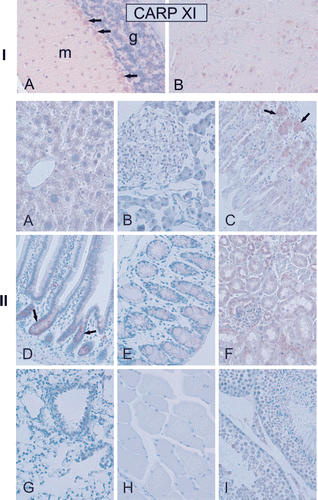
Figure 11. Expression pattern of CA11 gene in healthy human tissues analyzed by microarray method. The expression profile figure is adapted from BioGPS (http://biogps.gnf.org, accessed July 2012). The figures shows expression of CA11 gene in different human tissues samples and significant level of expression can only be seen in central nervous systemCitation20.
Very recently, we have studied the expression pattern of CA10B mRNA in zebrafish and found it fairly similar to the distribution in mouse tissues. There is no ortholog of CA11 in known fish genomes, but instead there are two descendants of CA10, namely CA10A and CA10BCitation12. CARP XB would seem to have been adapted to similar functions as CARP XI in mammals.
Bioinformatic analyses of CARP XI
In the previous bioinformatic analysis we identified 19 full-length CARP XI sequences only in tetrapod vertebrates with an amino acid identity of 70–100% among these organisms. Similar to CARP VIII and CARP X, the conservation of sequences was higher compared to the active CA isozymesCitation12. The study suggests that the CA11 genes have emerged from a gene duplication which has taken place either after the separation of the fish and tetrapod lineages or before this separation, with a subsequent loss of CA11 in the fish lineageCitation12.
CARP XI in pathological conditions
The expression of CARP XI was investigated in gastrointestinal stromal tumor (GIST) by using immunohistochemical analysis which showed clear cytoplasmic signals in 20 (91%) of 22 GIST tissue specimens. The positive signals for CARP XI were more intense in the periphery than in the central part of GISTs. The expression patterns of CARP XI were also studied in the cultured GIST cell line GIST-T1 by RT-PCR, Southern blot and immunocytochemistry. Ectopic expression of CARP XI in GIST-T1 cells induced cell proliferation and invasion in vitro. These findings indicate that CARP XI plays a role in the development of GISTCitation11.
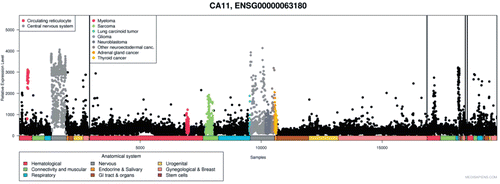
Additionally, expression pattern of human CA11 gene from nearly 20,000 microarrays is available in MediSapiens. The expression pattern of CA11 gene has been studied in normal, neoplastic tissues and other pathological conditions, and the study showed upregulation of CA11 mRNA in several cancers and pathological conditions ()Citation21.
Figure 12. Expression of CA11 gene in normal and pathological conditions in human: Microarray data of the mRNA expression levels of human genes in normal and pathological tissues from MediSapiens. (http://www.medisapiens.com, accessed February 2012)Citation21.
In summary, the studies done in the past and recent information available from public databases and the information generated in our lab on CARP VIII, CARP X and CARP XI have given better insights into evolution of CARPs, their expression patterns and possible roles. The preliminary knockdown studies in zebrafish have already produced some valuable information on their roles during embryonic development. Nevertheless, further studies are needed to understand the exact function of CARPs, especially in the brain and CNS. In addition, screening of large numbers of patients with unknown neurological disorders for variations in CARP X and CARP XI may reveal involvement of these proteins in neurological disorders. Future research on these inactive CA isoforms may provide us with the information on how these proteins work, possibly by coordination with other proteins through protein-protein interactions, and might help us to design novel drugs to treat the diseases caused by variations in these proteins.
Declaration of interest
This work was supported by the competitive Research Funding of the Tampere University Hospital (Grant 9N054) and the grants from Academy of Finland, and Sigrid Juselius Foundation for the original work done in our laboratory.
References
- Hewett-Emmett D, Tashian RE. Functional diversity, conservation, and convergence in the evolution of the alpha-, beta-, and gamma-carbonic anhydrase gene families. Mol Phylogenet Evol 1996;5:50–77.
- Kato K. Sequence of a novel carbonic anhydrase-related polypeptide and its exclusive presence in Purkinje cells. FEBS Lett 1990;271:137–140.
- Hirota J, Ando H, Hamada K, Mikoshiba K. Carbonic anhydrase-related protein is a novel binding protein for inositol 1,4,5-trisphosphate receptor type 1. Biochem J 2003;372:435–441.
- Lakkis MM, Bergenhem NC, O’Shea KS, Tashian RE. Expression of the acatalytic carbonic anhydrase VIII gene, Car8, during mouse embryonic development. Histochem J 1997;29:135–141.
- Taniuchi K, Nishimori I, Takeuchi T, Fujikawa-Adachi K, Ohtsuki Y, Onishi S. Developmental expression of carbonic anhydrase-related proteins VIII, X, and XI in the human brain. Neuroscience 2002a;112:93–99.
- Taniuchi K, Nishimori I, Takeuchi T, Ohtsuki Y, Onishi S. cDNA cloning and developmental expression of murine carbonic anhydrase-related proteins VIII, X, and XI. Brain Res Mol Brain Res 2002b;109:207–215.
- Kaya N, Aldhalaan H, Al-Younes B, Colak D, Shuaib T, Al-Mohaileb F et al. Phenotypical spectrum of cerebellar ataxia associated with a novel mutation in the CA8 gene, encoding carbonic anhydrase (CA) VIII. Am J Med Genet B Neuropsychiatr Genet 2011;156B:826–834.
- Türkmen S, Guo G, Garshasbi M, Hoffmann K, Alshalah AJ, Mischung C et al. CA8 mutations cause a novel syndrome characterized by ataxia and mild mental retardation with predisposition to quadrupedal gait. PLoS Genet 2009;5:e1000487.
- Picaud SS, Muniz JR, Kramm A, Pilka ES, Kochan G, Oppermann U et al. Crystal structure of human carbonic anhydrase-related protein VIII reveals the basis for catalytic silencing. Proteins 2009;76:507–511.
- Kato K. A Collection of cDNA Clones with Specific Expression Patterns in Mouse Brain. Eur J Neurosci 1990;2:704–711.
- Morimoto K, Nishimori I, Takeuchi T, Kohsaki T, Okamoto N, Taguchi T et al. Overexpression of carbonic anhydrase-related protein XI promotes proliferation and invasion of gastrointestinal stromal tumors. Virchows Arch 2005;447:66–73.
- Aspatwar A, Tolvanen ME, Parkkila S. Phylogeny and expression of carbonic anhydrase-related proteins. BMC Mol Biol 2010;11:25.
- Tashian RE, Hewett-Emmett D, Carter ND, Bergenhem NCH, eds. Carbonic anhydrase (CA)-related proteins (CA-RPs), and trans-membrane proteins with CA or CA RP membrane proteins with CA or CA RP domains. Basel: New Horizons, Birkhauser, 2000.
- Akisawa Y, Nishimori I, Taniuchi K, Okamoto N, Takeuchi T, Sonobe H et al. Expression of carbonic anhydrase-related protein CA-RP VIII in non-small cell lung cancer. Virchows Arch 2003;442:66–70.
- Lu SH, Takeuchi T, Fujita J, Ishida T, Akisawa Y, Nishimori I et al. Effect of carbonic anhydrase-related protein VIII expression on lung adenocarcinoma cell growth. Lung Cancer 2004;44:273–280.
- Miyaji E, Nishimori I, Taniuchi K, Takeuchi T, Ohtsuki Y, Onishi S. Overexpression of carbonic anhydrase-related protein VIII in human colorectal cancer. J Pathol 2003;201:37–45.
- Nishikata M, Nishimori I, Taniuchi K, Takeuchi T, Minakuchi T, Kohsaki T et al. Carbonic anhydrase-related protein VIII promotes colon cancer cell growth. Mol Carcinog 2007;46:208–214.
- Jiao Y, Yan J, Zhao Y, Donahue LR, Beamer WG, Li X et al. Carbonic anhydrase-related protein VIII deficiency is associated with a distinctive lifelong gait disorder in waddles mice. Genetics 2005;171:1239–1246.
- Aspatwar A, Tolvanen ME, Ortutay C, Parkkila S. Carbonic anhydrase related protein VIII and its role in neurodegeneration and cancer. Curr Pharm Des 2010;16:3264–3276.
- Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol 2009;10:R130.
- Kilpinen S, Autio R, Ojala K, Iljin K, Bucher E, Sara H et al. Systematic bioinformatic analysis of expression levels of 17,330 human genes across 9,783 samples from 175 types of healthy and pathological tissues. Genome Biol 2008;9:R139.
- Yoon CH. Waddler, a new mutation, and its interaction with quivering. J Hered 1959;50:238–244.
- Okamoto N, Fujikawa-Adachi K, Nishimori I, Taniuchi K, Onishi S. cDNA sequence of human carbonic anhydrase-related protein, CA-RP X: mRNA expressions of CA-RP X and XI in human brain. Biochim Biophys Acta 2001;1518:311–316.
- Tashian RE. Genetics of the mammalian carbonic anhydrases. Adv Genet 1992;30:321–356.
- Kleiderlein JJ, Nisson PE, Jessee J, Li WB, Becker KG, Derby ML et al. CCG repeats in cDNAs from human brain. Hum Genet 1998;103:666–673.
- Ortutay C, Olatubosun A, Parkkila S, Vihinen M, Tolvanen, MEE. An evolutionary analysis of insect carbonic anhydrases. In: Berhardt LV, ed. Advances in Medicine and Biology. Volume 7, Hauppauge, NY: Nova Science Publishers Inc; 2010, 145–168.
- Mikoshiba K, Okano H, Miyawaki A, Furuichi T, Ikenaka K. Molecular genetic analyses of myelin deficiency and cerebellar ataxia. Prog Brain Res 1995;105:23–41.
- Bellingham J, Gregory-Evans K, Gregory-Evans CY. Sequence and tissue expression of a novel human carbonic anhydrase-related protein, CARP-2, mapping to chromosome 19q13.3. Biochem Biophys Res Commun 1998;253:364–367.
- Lovejoy DA, Hewett-Emmett D, Porter CA, Cepoi D, Sheffield A, Vale WW et al. Evolutionarily conserved, “acatalytic” carbonic anhydrase-related protein XI contains a sequence motif present in the neuropeptide sauvagine: the human CA-RP XI gene (CA11) is embedded between the secretor gene cluster and the DBP gene at 19q13.3. Genomics 1998;54:484–493.
- Fujikawa-Adachi K, Nishimori I, Taguchi T, Yuri K, Onishi S. cDNA sequence, mRNA expression, and chromosomal localization of human carbonic anhydrase-related protein, CA-RP XI. Biochim Biophys Acta 1999;1431:518–524.
- Bergenhem NC, Hallberg M, Wisén S. Molecular characterization of the human carbonic anhydrase-related protein (HCA-RP VIII). Biochim Biophys Acta 1998;1384:294–298.
- Supuran C, Scozzafava A, Conway, J, ed. Carbonic anhydrase: its inhibitors and activators. Washington, DC: CRC Press, 2004.