Abstract
Antiproliferative and antibacterial activities of nine glutarimide derivatives (1–9) were reported. Cytotoxicity of compounds was tested toward three human cancer cell lines, HeLa, K562 and MDA-MB-453 by MTT assay. Compound 7 (2-benzyl-2-azaspiro[5.11]heptadecane-1,3,7-trione), containing 12-membered ketone ring, was found to be the most potent toward all tested cell lines (IC50 = 9–27 μM). Preliminary screening of antibacterial activity by a disk diffusion method showed that Gram-positive bacteria were more susceptible to the tested compounds than Gram-negative bacteria. Minimum inhibitory concentration (MIC) determined by a broth microdilution method confirmed that compounds 1, 2, 4, 6–8 and 9 inhibited the growth of all tested Gram-positive and some of the Gram-negative bacteria. The best antibacterial potential was achieved with compound 9 (ethyl 4-(1-benzyl-2,6-dioxopiperidin-3-yl)butanoate) against Bacillus cereus (MIC 0.625 mg/mL; 1.97 × 10−3 mol/L). Distinction between more and less active/inactive compounds was assessed from the pharmacophoric patterns obtained by molecular interaction fields.
Introduction
Both naturally occurring and synthetic cyclic imides, especially five- and six-membered systems, are an important group of bioactive molecules. They exhibit widespread pharmacological effects, including antitumorCitation1–4, anti-inflammatoryCitation5, immunomodulatory, antiangiogenic and anxiolyticCitation6–8.
Isolation and examination of pharmacologically active natural glutarimides started in 1960s. Initially, cycloheximideCitation9 and streptimidoneCitation10–12 were examined as antibiotics, but later it was found that they acted as very potent cytotoxic agentsCitation13,Citation14. The structurally related streptimidone derivative, 9-methylstreptimidone, exerts a significant inhibitory activity toward nuclear factor-κB (N-κB)Citation15. N-κB is involved in cancer and inflammations. Alkaloids (+)-sesbanimide A and (−)-sesbanimide B were isolated from the seeds of the leguminous plant Sesbania drummondiiCitation1,Citation16. Ethanol extracts of S. drummondii seeds showed significant inhibitory activity against P388 murine leukemia model in mice (in vivo)Citation17–19. Structurally related natural product lactimidomycin (LTM), 12-membered unsaturated macrolide antibiotic that comprise biosynthetically rare glutarimide side chain, produced by Streptomyces amphibiosporus R310-104 (ATCC 53964), display strong cytotoxicity against a number of human tumor cell lines in vitro, in vivo antitumor activity in mice model and potent antifungal activityCitation2.
In 1990s it was discovered that thalidomide, a well-known synthetic glutarimide derivative, has anti-inflammatory and antiangiogenic properties. It was approved as a drug for the treatment of certain cancers (newly diagnosed multiple myeloma) and for complication arised from leprosy. Later on, analogs of thalidomide with increased potency, 3-amino-thalidomid (pomalidomid, Pomalyst) and α-(3-aminophthalimido) glutarimide (lenalidomid, Revlimid) have been developedCitation20,Citation21. Lenalidomid has been used for the treatment of multiple myeloma, while pomalidomid has been recently approved by FDA for the treatment of relapsed and refractory multiple myelomaCitation22.
Some estrone derivatives with the d-ring replaced with the glutarimide moiety showed potent inhibition of steroid sulfatase, an enzyme involved in the pathway of the development of hormone-dependent breast tumorsCitation23; while aminoglutethimide, the non-steroidal aromatase inhibitor, is in use for the treatment of hormone-sensitive metastatic breast cancerCitation24,Citation25. In the past decade, antitumor activity in vitro of mitonafideCitation26, amonafideCitation27 and naphthalimideCitation28,Citation29 derivatives was intensively examined.
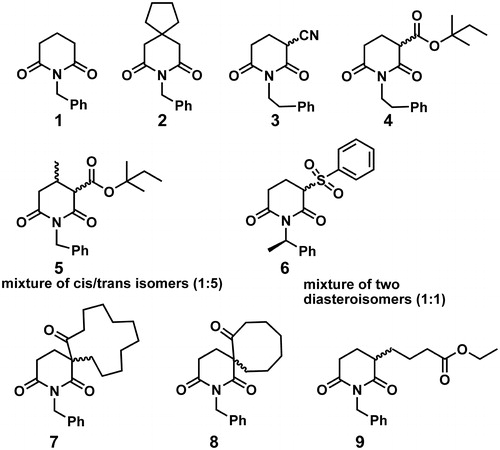
Upon detail analysis of bioactive compounds that comprise glutarimide moiety in their structure, as is briefly outlined in previous paragraphs, we concluded that data on antiproliferative and on antibacterial activity of compounds 1–9 () cannot be found in literature. Those compounds have been prepared in our group to demonstrate novel synthetic approach of glutarimide ring closure reactionCitation30. All compounds have a common N-substituted glutarimide moiety in their structure, and bear structurally diverse substituents in positions 3 and/or 4 of glutarimide ring. We have tested antiproliferative and antibacterial activity of compounds 1–9 in vitro, and hereby report on the results obtained.
Methods
Antiproliferative activity
Stock solutions of investigated compounds were prepared in a nutrient medium (RPMI-1640) supplemented with 3 mM l-glutamine, 100 μg/mL streptomycin, 100 IU/mL penicillin, 10% heat inactivated fetal bovine serum (FBS) and 25 mM Hepes, adjusted to pH 7.2 by bicarbonate solution. RPMI-1640, FBS, Hepes and l-glutamine were products of Sigma Chemical Co., St. Louis, MO. Human cervix adenocarcinoma HeLa, breast carcinoma MDA-MB-453 and normal lung fibroblast MRC-5 cells were cultured as monolayers in the nutrient medium, while human myelogenous leukemia K562 cells were maintained as suspension culture. The cells were grown at 37 °C in 5% CO2 and humidified air atmosphere.
HeLa (2000 cells per well), MDA-MB-453 (3000 c/w) and MRC-5 (5000 c/w) cells were seeded into 96-well microtiter plates and 20 h later, after the cell adherence, five different concentrations of investigated compounds were added to the wells. Final concentrations were in the range from 200 to 12.5 μM. Only nutrient medium was added to the cells in the control wells. Investigated compound was added to a suspension of leukemia K562 cells (5000 cells/ well) 2 h after cell seeding, in the same final concentrations applied to HeLa, MDA-MB-453 and MRC-5 cells. All experiments were done in triplicate. Nutrient medium with corresponding concentrations of compound, but void of cells, was used as blank.
Cell survival was determined by MTT test according to the method of Mosmann and modified by Ohno and Abe, 72 h after addition of the compoundsCitation31,Citation32. Briefly, 20 µL of MTT solution (5 mg/mL in phosphate buffered saline) was added to each well. Samples were incubated for further 4 h at 37 °C in humidified atmosphere with 5% CO2. Then, 100 µL of 10% SDS was added to the wells. Absorbance was measured at 570 nm the next day. To achieve cell survival (S%), absorbance at 570 nm of a sample with cells grown in the presence of various concentrations of compounds tested was divided with absorbance of control sample (the absorbance of cells grown in nutrient medium only). Absorbance of blank was always subtracted from absorbance of a corresponding sample with cells. All experimentally obtained IC50 data were means of three measurements done in triplicate.
The antibacterial activity testing
The antibacterial activity of compounds 1, 2, 4–8 and 9 was determined against four Gram-positive (Staphylococcus aureus, Enterococcus faecalis, Bacillus cereus, Listeria monocytogenes) and seven Gram-negative bacterial species [Pseudomonas aeruginosa, Escherichia coli, Salmonella enteritidis, Proteus hauseri, Shigella sonnei, Yersinia enterocolitica, E. coli (O157:H7)]. Selected bacterial strains originated from ATCC (American Type Culture Collection, Rockville, MD). These microorganisms were chosen for the bioassay as the well-known food spoilage and pathogenic bacteria. Each species was maintained on Mueller–Hinton agar (MHA), which was also used to confirm the absence of contamination and the validity of the inocula. Before testing, each species was recovered by sub-culturing in Mueller–Hinton broth (MHB), aerobically, for 24 h, at 37 °C. Working concentrations of approximately 105–106 cfu/mL, used for antibacterial activity assays, were prepared by proper dilution of culture in microbiological medium. Compounds were dissolved in DMSO (2%) to prepare stock solutions at a concentration of 40 mg/mL, sterilized by filtration through a 0.22-μm membrane filter (Sartorius AG – Göttingen, Germany) according to Tepe et al. and further diluted in MHB to a working solutions. DMSO was chosen as a non-toxic solventCitation33.
Disk diffusion assay was performed using a slightly modified CLSICitation34. Each bacterial culture (approximately 105–106 cfu/mL) was added (0.1 mL) to Petri dishes (90 mm) containing MHA (20 mL). Three sterile blank paper disks (6 mm in diameter, Susceptibility Test Discs, SD 067-5CT, HiMedia, Mumbai, India) were placed on the surface of each agar plate and inoculated with 10 μL of the compound (20 mg/mL). After 2 h at 25 °C, the plates were incubated aerobically, for 24 h at 37 °C. After incubation period, inhibition zone (mm) was measured including the initial diameter of the disk. Tests were performed in triplicate and the results were analyzed for statistical significance. The plates with MHA were sterility controls. Negative controls were disks impregnated with DMSO. As positive controls disks (Sigma-Aldrich GmbH, Steinheim, Germany) with gentamicin (30 μg) and tetracycline (30 μg) were used.
Broth microdilution method was employed to determine minimum inhibitory concentrations (MICs)Citation34,Citation35. Concentrations of compound ranged from 10.0 to 0.048 mg/mL. Test bacterial culture (50 μL) in a MHB was added to the wells of a sterile 96-well microtiter plate (Sarstedt, Numbrecht, Germany) already containing 50 μL of twofold serially diluted compound in MHB. The final volume in each well was 100 μL. The microplates were prepared in triplicate and incubated aerobically, for 24 h at 37 °C. Wells with MHB was used as a sterility control, while negative controls were wells with tested compound in 50 μL of MHB, but void of bacteria. Positive controls were wells with a bacterial suspension in 50 μL of MHB and wells with a bacterial suspension in a MHB with DMSO, in amounts corresponding to the highest quantity present in the broth microdilution assay (to prove that DMSO had no inhibition effect on the bacterial growth). A microplate shaker (Lab Companion, VM-96B, Seoul, South Korea) was used for mixing the content of each well at 900 rpm for 1 min prior to incubation in the cultivation conditions described above. To indicate cellular respiration 2,3,5-triphenyltetrazolium chloride (TTC) (Aldrich Chemical Company Inc., Sigma-Aldrich, St. Louis, MO) was added to the culture medium. The final concentration of TTC after inoculation was 0.05%. Viable microorganisms enzymatically reduced white TTC to a pink TPF (1,3,5-triphenylformazan). The MIC was defined as the lowest sample concentration that prevented this change and exhibited complete inhibition of bacterial growth.
All measurements were done in triplicate and data were expressed as mean ± standard deviation. The experimental data were subjected to an one-way analysis of variance (ANOVA) and Fisher's LSD was calculated to detect significant difference (p ≤ 0.05) between the mean values.
Molecular modeling
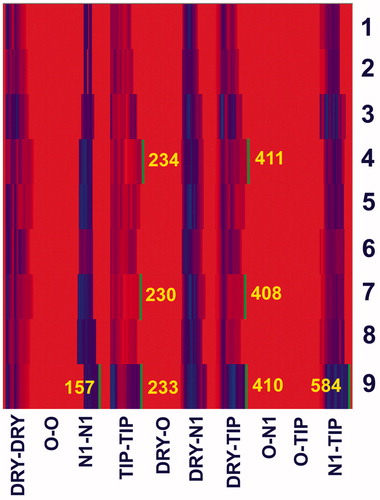
Initial 3D structures of compounds 1–9 were generated from SMILES notation in CORINA assuming R stereochemistry for all stereogenic centers, except for C2 of the phenethyl moiety of compound 6Citation36,Citation37. The S stereochemistry was ascribed to this stereogenic center, on the ground of experimental dataCitation30. Initial structures were imported in VegaZZCitation38. Up to 20 conformations, representing local energy minima, were obtained by conformational search on the molecular mechanics level (MMFF94s force field), using Boltzmann jump algorithm in AMMPCitation39,Citation40. Each conformation of each compound was minimized by the semi-empirical molecular orbital PM6 method, using implicit solvation in water (COSMO) to root mean square gradient of 0.01; by MOPAC2012Citation41,Citation42. Conformation of each compound that had the lowest heat of formation (implying the most stable one) was chosen for further modeling. 3D-dependent whole-molecular properties of compounds, surface area, polar surface area, apolar surface area, volume and virtual log P, were calculated in VegaZZ, using 1.4 Å probeCitation43. Molecular interaction fields (MIF) around molecules were calculated by a GRID method, as applied in Pentacle program, using grid resolution of 0.4 ÅCitation44. Hydrogen-bond donor (N1), hydrogen-bond acceptor (O), hydrophobic (DRY) and shape (TIP) probes were used. AMANDA algorithm were used for the extraction of hot spots (nodes) from the obtained MIFs (discretization); the distances and relative position of the nodes were described by maximum auto- and cross-correlation (MACC2) (encoding). For more exhaustive description of applied methodology see original referenceCitation45. Auto- and cross-correlograms, obtained by the Pentacle program are depicted in matrix-like representation, named as heatmap. Values of variables are color-coded from red (low value) to blue (high value). For color code in heatmap, depicted in , see on-line version of the article. Correlograms encode molecular descriptors grounded on two-point pharmacophoric pattern, which represent the local minima of two probes (nodes) around molecule, including distance between those nodes. All blocks of correlograms (DRY-DRY, O-O, N1-N1, TIP-TIP, DRY-O, DRY-N1, DRY-TIP, O-N1, O-TIP, N1-TIP) were considered during analysis.
Chemistry
Chemicals and solvents were purchased from Merck (Darmstadt, Germany), Sigma-Aldrich and Fluka (Basel, Switzerland). All reagents were of analytically pure. All solvents were dried by standard methods and distilled before use. The sodium hydride was used as a 60% dispersion in mineral oil. 18-Crown-6 ether was prepared according to a literature procedureCitation46. Reactions were monitored on silica gel precoated TLC plates, HF254 (Merck). The dry-flash chromatography on silica gel (12–16 μ, ICN Pharmaceuticals, Costa Mesa, CA) was used to purify the reaction products. Anhydrous reactions were carried out in oven-dried glassware in extra pure argon atmosphere. 1H and 13C NMR spectra were recorded on a Bruker Avance 500 (Bruker BioSpin GmbH, Karlsruhe, Germany), or Varian (Palo Alto, CA) Gemini 2000 instruments on 500/125 or 200/50 MHz, in CDCl3 with TMS as an internal reference. ESI-MS spectra were recorded on Agilent Technologies (Santa Clara, CA) 6210-1210 TOF-LC-ESI-HR/MS instrument in positive mode. Integrity and purity of all compounds used for biological tests are routinely checked by NMR and ESI/HR-MS.
General procedure for the synthesis of compounds 1 and 2
1-Benzyl-piperidine-2,6-dione (1, C12H13NO2). Toluene (20 mL) and sodium hydride (2.9 g, 72 mmol) were placed in a two-necked flask equipped with reflux condenser and dropping funnel. A solution of 4-benzylcarbamoyl-butyric acid methyl ester 1 c (5.0 g, 18 mmol) in toluene (∼30 ml) was added and mixture was refluxed for 3 h. After cooling at room temperature the mixture was filtered and the filtrate was evaporated under reduced pressure to give 65 % of compound 1 (C12H13NO2). 1H NMR (200 MHz, CDCl3): δ = 7.36–7.24 (m, 5H, Ar–H), 4.94 (s, 2H; CH2), 2.65 (t, J = 6.5 Hz, 4H, CH2), 1.98–1.88 (m, 2H, CH2) ppm; 13C NMR(50 MHz, CDCl3): δ = 172.5 (CCOimide), [137.3, 128.8, 128.4, 127.4 (CAr)], 42.6 (CCH2), 32.9 (CCH2), 17.0 (CCH) ppm; HR-MS (ESI, m/z): calcd. for C12H13NO2 [M + NH4]+: 221.1284; found: 221.1278.
8-Benzyl-8-aza-spiro[4.5]decane-7,9-dione (2, C16H19NO2). Prepared from methyl ester 2e. Yield 75%; 1H NMR (200 MHz, CDCl3): δ = 7.37–7.24 (m, 5H, Ar–H), 4.94 (s, 2H, CH2), 2.60 (s, 4H, CH2), 1.72–1.65 (m, 4H, CH2), 1.50–1.44 (m, 4H, CH2) ppm; 13C NMR (50 MHz, CDCl3): δ = 172.1(CCOimide), [137.2, 128.5, 128.3, 127.3 (CAr)], 44.7 (CCH2), 42.6 (CCH2), 39.4 (C), 37.4 (CCH2), 24.1 (CCH2) ppm; HR-MS (ESI, m/z): calcd. for C16H19NO2 [M + H]+: 258.1489; found: 258.1480.
General procedure described in our previous paper was used for preparation of compounds 3–9
2,6-Dioxo-1-phenethylpiperidine-3-carbonitrile (3, C14H14N2O2). Yield 65%; 1H NMR (200 MHz, CDCl3): δ = 7.34–7.19 (m, 5H, Ar–H), 4.05 (splitted t, J = 7.0 Hz; J = 2.1 Hz, 2H, CH2), 3.71 (dd, J = 9.3 Hz, J = 5.3 Hz, 1H, CH), 2.95–2.81 (m, 3H, CH2), 2.72–2.56 (m, 1H, CH2), 2.36–2.17 (m, 2H, CH2) ppm; 13C NMR (50 MHz, CDCl3,): δ = 169.6 (CCOimide), [137.7, 129.0, 128.5, 126.7 (CAr)], 115.1 (CCN), 41.6 (CCH2), 35.7 (CCH2), 33.6 (CCH), 30.7 (CCH2), 21.5 (CCH2) ppm; HR-MS (ESI, m/z): calcd. for C14H14N2O2 [M + NH4]+: 260.1393, found: 260.1389.Citation30
Tert-pentyl-2,6-dioxo-1-phenethylpiperidine-3-carboxylate (4, C19H25NO4). Yield 70%; 1H NMR (200 MHz, CDCl3) δ = 7.30–7.21 (m, 5H, Ar–H), 4.04–3.96 (m, 2H, CH2), 3.57–3.51 (m, 1H, CH), 2.86–2.78 (m, 2H, CH2), 2.67 (dt, J = 11.6 Hz, J = 5.8 Hz, 2H, CH2), 2.25–2.07 (m, 2H, CH2), 1.80 (q, J = 7.5 Hz, 2H, CH2), 1.46 (s, 6H, CH3), 0.90 (t, J = 7.5 Hz, 3H, CH3) ppm; 13C NMR (50 MHz, CDCl3): δ = 171.3 (CCOimide), 168.8 (CCOimide), 167.7 (CCOester), [138.4, 128.9, 126.4 (CAr)], 85.6 (C), 50.0 (CCH), 41.1 (CCH2), 33.8 (CCH2), 33.3 (CCH2), 30.8 (CCH2), 30.2 (CCH2), 25.3 (CCH3), 20.7 (CCH2), 8.1 (CCH3) ppm; HR-MS (ESI, m/z): calcd. for C19H25NO4 [M + H]+: 332.1856, found: 332.1841.
Tert-pentyl-1-benzyl-4-methyl-2,6-dioxopiperidine-3-carboxylate (5, C19H25NO4). Yield 46.6%; 1H NMR (200 MHz, CDCl3): δ = 7.38–7.24 (m, 5H, Ar–H), 5.04–4.88 (m, 2H, CH2), 3.59 (d, J = 4.9 Hz, 0.13 H, CH), 3.21 (d, J = 9.0 Hz, 0.59 H, CH), 2.83 (dd, J = 16.5, J = 3.9 Hz, 1 H, CH), 2.74–2.68 (m, 0.41 H, CH2), 2.60–2.46 (m, 1H, CH2), 2.43–2.29 (m, 1H, CH2), 1.76 (dt, J = 9.4 Hz, J = 4.8 Hz, 2H, CH2), 1.44 (s, 5H, CH3), 1.36 (d, J = 3.5 Hz, 1H, CH3), 1.14–1.07 (m, 3H, CH3), 0.91–0.75 (m, 3H, CH3) ppm; 13C NMR (50 MHz, CDCl3): δ = 170.9 (CCOimide), 169.1(CCOimide), 167.4 (CCOester), [136.9, 128.7, 128.4, 127.5 (CAr)], 85.4 (C), 57.9 (CCH), 43.0 (CCH2), 38.6 (CCH2), 33.4 (CCH2), 27.7 (CCH), 25.3 (CCH3), 25.2 (CCH2), 19.1 (CCH3), 8.1 (CCH3) ppm; HR-MS (ESI, m/z): calcd. for C19H25NO4 [M + NH4]+: 349.2122, found: 349.2128.
1-((R)-1-Phenylethyl)-3-(phenylsulfonyl)piperidine-2,6-dione (6, C19H19NO4S). Yield 68%; 1H NMR (200 MHz, CDCl3): δ = 7.90–7.26 (m, 10H, Ar–H), 6.19–6.00 (m, 1H, CH), 4.13–3.942 (m, 1H, CH), 3.44–3.14 (m, 1H, CH2), 2.92–2.71 (m, 2H, CH2), 2.45–2.25 (m, 1H, CH2), 1.77–1.71 (m, 3H, CH3) ppm; 13C NMR (50 MHz, CDCl3): δ = 171.1 (CCOimide), 170.8 (CCOimide), [139.6, 134.5, 134.4, 129.2, 129.0, 128.1, 128.0, 127.7, 127.0, 126.8 (CAr)], 66.2 (CCH), 50.3 (CCH), 50.1 (CCH), 29.8 (CCH2), 29.4 (CCH2), 17.6 (CCH3), 17.4, (CCH3), 15.8 (CCH2) ppm; HR-MS (ESI, m/z): calcd. for C19H19NO4S [M + H]+: 358.1099, found: 358.1108.
2-Benzyl-2-azaspiro[5.11]heptadecane-1,3,7-trione (7, C23H31NO3). Yield 42.1%; 1H NMR (500 MHz, CDCl3): δ = 7.34–7.23 (m, 5H, Ar–H), 4.95 (ABq, 2H, CH2, J = 14.6 Hz), 3.23–3.17 (ddd, 1H, CH2, J = 7.6 Hz, J = 5.3 Hz, J = 1.0 Hz), 2.68–2.56 (m, 2H, CH2), 2.59–2.53 (m, 2H, CH2), 2.00–1.95 (m, 1H, CH2), 1.60–1.47 (m, 4H, CH2), 1.33–1.19 (m, 15H, CH2), 0.96–0.89 (m, 1H, CH2) ppm; 13C NMR (125 MHz, CDCl3): δ = 204.8 (CCOketo), 172.7 (CCOimide), 171.9 (CCOimide), [136.9, 128.8, 128.4, 127.5 (CAr)], 60.0 (CCH), 43.4 (CCH2), 34.7 (CCH2), 33.9 (CCH2), 29.9 (CCH2), 26.3 (CCH2), 26.2 (CCH2), 23.3 (CCH2), 23.1 (CCH2), 22.0 (CCH2), 21.7 (CCH2), 18.8 (CCH2) ppm; HR-MS (ESI, m/z): calcd. for C23H31NO3 [M + H]+: 370.2377, found: 370.2371.
2-Benzyl-2-azaspiro[5.7]tridecane-1,3,7-trione (8, C19H23NO3). Yield 53%; 1H NMR (200 MHz, CDCl3): δ = 7.28–7.23 (m, 5H, Ar–H), 4.91 (s, 2H, CH2), 3.19–2.40 (m, 6H, CH2), 2.25 (ddd, J = 12.3 Hz, J = 6.2 Hz, J = 3.4 Hz, 1H, CH2), 1.88–1.53 (m, 9H, CH2) ppm; 13C NMR (50 MHz, CDCl3): δ = 213.3 (CCOketo), 172.3 (CCOimide), 171.6 (CCOimide), [137.0, 128.4, 128.2, 127.3 (CAr)], 57.9 (C), 43.1 (CCH2), 38.3 (CCH2), 31.4 (CCH2), 30.0 (CCH2), 29.5 (CCH2), 25.7 (CCH2), 23.9 (CCH2), 23.8 (CCH2), 22.5 (CCH2) ppm; HR-MS (ESI, m/z): calcd. for C19H23NO3 [M + H]+: 314.1751, found: 314.1738.
Ethyl 4-(1-benzyl-2,6-dioxopiperidin-3-yl)butanoate (9, C18H23NO4). Yield 42%; 1H NMR (200 MHz, CDCl3): δ = 7.37–7.22 (m, 5H, Ar–H), 4.94 (s, 2H, CH2), 4.13 (q, J = 7.1 Hz, 2H, CH2), 2.82 (dt, J = 17.6 Hz, J = 4.9 Hz, 1H, CH2), 2.64 (dd, J = 11.1 Hz, J = 5.3 Hz, 1H, CH2), 2.54–2.42 (m, 1H, CH), 2.34 (t, J = 7.2 Hz, 2H, CH2), 2.12–1.90 (m, 2H, CH2), 1.82–1.56 (m, 4H, CH2), 1.25 (t, J = 7.1 Hz, 3H, CH3) ppm; 13C NMR (50 MHz, CDCl3): δ = 174.5 (CCOimide), 173.2 (CCOester), 172.2 (CCOimide), [137.3, 128.5, 128.3, 127.3 (CAr)], 60.3 (CCH2), 42.9 (CCH2), 41.9 (CCH2), 33.9 (CCH2), 31.9 (CCH2), 29.6 (CCH2), 22.2 (CCH2), 22.0 (CCH2), 14.1 (CCH3) ppm; HR-MS (ESI, m/z): calcd. for C18H23NO4 [M + H]+: 318.1700, found: 318.1691.
8-Oxaspiro[4.5]decane-7,9-dione (2c) was prepared according to a modified literature procedureCitation47.
7,9-Dioxo-8-azaspiro[4.5]decane-6,10-dicarbonitrile (2a) and 2,2′-cyclopentane-1,1-diyldi acetic acid (2b) were prepared by a known literature procedureCitation48. Methyl 5-chloro-5-oxopentanoate (1b) and methyl [1-(2-chloro-2-oxoethyl)cyclopentyl] acetate (2d) were prepared by a modification of a known literature procedureCitation49.
Methyl 5-(benzylamino)-5-oxopentanoate (1c) and methyl 2-(1-(2-(benzylamino)-2-oxoethyl) cyclopentyl) acetate (2e) were prepared according to a modified literature procedureCitation50.
Methyl 2-methylbutan-2-yl propanedioate (3b) was prepared by a literature procedureCitation51. Methyl 2-(phenylsulfonyl)acetate (3c) was prepared by a literature procedureCitation52.
Acrylamides (4a, 4c, 4d) and (E)-N-benzylbut-2-enamide (4b) were prepared according to a modified literature procedureCitation50.
β-Ketoesters (5a–c) were prepared by a modification of a literature procedureCitation53.
Results and discussion
Chemistry
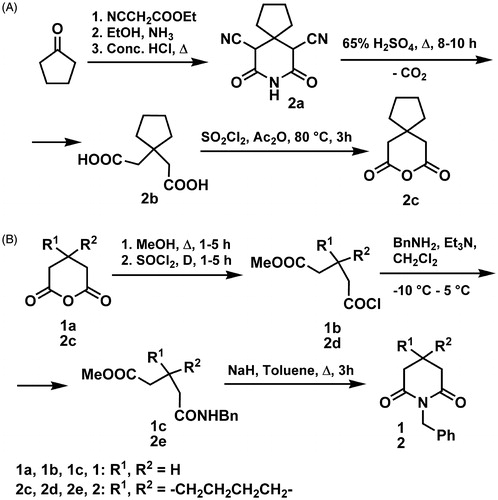
Glutarimide derivatives, depicted in , were synthesized according to previously reported procedures of ourCitation30 or other research groups. Compounds 1 and 2 were prepared according to a modified literature procedure. Cyclization of amido-esters, derived from corresponding glutaric acid anhydrides, in presence of the base (NaH), was carried out by reflux in toluene (∼3 h), yielding glutarimides 1 and 2 (Scheme 1). Amido-esters 1c and 2e were prepared from 1a and 2c, respectively, by a modification of standard methodsCitation47,Citation49,Citation50.
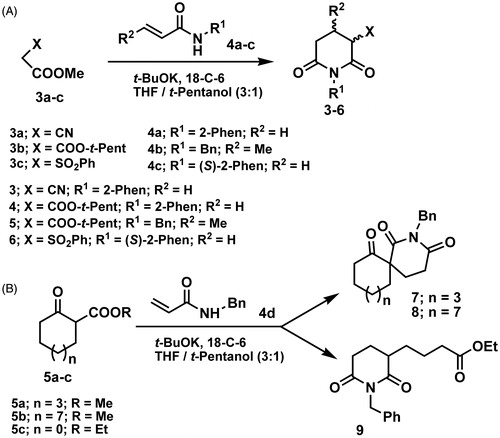
Compounds 3–9 were synthesized by tandem process described in our previous paperCitation30. The process involved a base-catalyzed Michael addition of active methylene compounds to secondary acrylamides or crotonamides, followed by intramolecular N-acylation of the carboxamido group. Synthesis of derivatives 3–6 was performed by reacting methyl l,2-cyanoacetate (3a), methyl t-pentyl malonate (3b) and methyl 2-(phenylsulfonyl)acetate (3c) with N-substituted acryl- and crotonamides (4a–c), under the reaction conditions (Scheme 2A). Yields of products were 42–72%. In the reaction of β-keto esters, comprising 5, 8 and 12 member rings, 5a–c, with N-benzyl acrylamide, imides 9, 8 and 7, respectively, were obtained (Scheme 2B).
Antiproliferative activity
The cytotoxicity of compounds 1–9 was tested toward selected human cancer cell lines: cervix adenocarcinoma HeLa, human myelogenous leukemia K562 and human breast carcinoma MDA-MB-453 cells. Cell survival was determined by MTT test, after 72 h of exposure to compoundsCitation31,Citation32. IC50 values () are shown in molar concentrations, as the mean ± SD, determined from three independent measurements.
Table 1. Concentrations of compounds 1–9 that induced 50% decrease in cell survival (IC50).
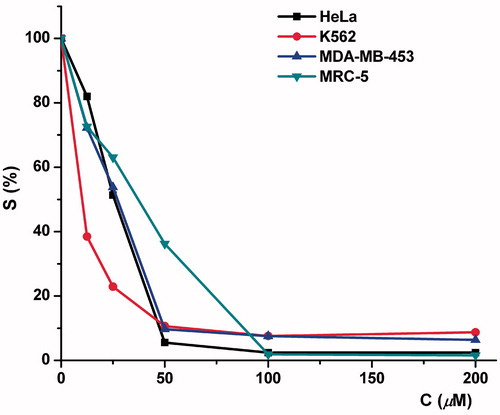
IC50 was defined as the concentration of the compound inhibiting cell survival by 50%, compared with a vehicle-treated control cells. The most potent compound, 7, was also tested toward MRC-5, normal lung fibroblast cells. Compounds 2, 4, 5–8 and 9 exerted dose-dependent cytotoxicity toward malignant cells, while compound 7 appeared as the most active. Compound 7 exerted humble selectivity, with IC50 toward normal cells ∼37 μM. The decrease in survival of target cells induced by compound 7 is shown in .
Antibacterial activity
Antibacterial activity of compounds 1, 2, 4–8 and 9 was examined against selected foodborne pathogenic bacteria, Gram-positive species S. aureus (ATCC 25923), E. faecalis (ATCC 29212), B. cereus (ATCC 10876), L. monocytogenes (ATCC 19115) and Gram-negative species P. aeruginosa (ATCC 27853), E. coli (ATCC 25922), S. enteritidis (ATCC 13076), P. hauseri (ATCC 13315), S. sonnei (ATCC 29930), Y. enterocolitica (ATCC 27729), E. coli O157:H7 (ATCC 12900). A preliminary screening, done by a disc diffusion method, indicated the ability of bacteria to produce visible growth in the presence of compounds 1, 2, 4–8 and 9. In most cases, Gram-negative bacteria were more resistant to the tested compounds than Gram-positive bacteria (). The diameters of the inhibition zones, determined by a disk diffusion method, ranged from 6.4 to 14.6 mm, in the presence of 200 μg of compounds tested.
Table 2. Antibacterial activity of compounds 1, 2, 4, 6–8 and 9, determined by the disk diffusion method.
The Gram-positive bacteria B. cereus were the most sensitive to compounds 4 and 9 (14.6 and 14.2 mm, respectively). Such activity was comparable to the effect of commercial antibiotic gentamicin (14.8 mm). Also, there was no statistically significant difference between the susceptibility of B. cereus on commercial antibiotic tetracycline (12.4 mm) and compounds 2, 7 and 8 (12.1, 12.9 and 12.4 mm, respectively). Among Gram-negative bacteria, S. sonnei was the most sensitive to compounds 2, 4 and 6 (11.5, 10.4 and 10.2 mm, respectively), comparable to antibiotic tetracycline (11.4 mm). Other compounds did not show significant antimicrobial activity against tested bacteria in the applied concentration. Compound 5 did not show any antimicrobial effect on tested Gram-positive and Gram-negative bacteria.
Due to the fact that the disk diffusion method is not entirely reliable in determining the antimicrobial properties, the broth microdilution method, as a rapid and quantitative method for determining of MIC, based on the color change caused by the enzymatic activity of viable microorganisms, was applied (). Compounds 1, 2, 4, 6–8 and 9 inhibited the growth of all tested Gram-positive and some of the Gram-negative bacteria. Achieved MICs were in the range of 0.625–10.0 mg/mL (1.97 × 10−3– 4.92 × 10−2 mol/L).
Table 3. Antibacterial activity of compounds 1, 2, 4, 6–8 and 9 expressed as MIC (mg/mL), determined by the broth microdilution method.
The highest antibacterial potential was reached with compound 9 against B. cereus (MIC was 0.625 mg/mL = 1.97 × 10−3 mol/L). MICs against Gram-negative bacteria exceeded 10.0 mg/mL, for the majority of compounds, including compound 5, proved to be inactive in concentrations up to 10.0 mg/mL against all bacterial strains tested. Exceptions were compound 4, whose antibacterial activity ranged from 5.0 to 10.0 mg/mL against all tested Gram-negative bacteria and compounds 2, 6 and 7, with MICs of 10.0 mg/mL (3.89 × 10−2, 2.80 × 10−2 and 2.71 × 10−2 mol/L, respectively) against S. sonnei.
Structure–activity relationship
Although exerting humble potency, derivatives 9 and 4 appeared most potent against bacteria, while derivative 7 was the one with fair potency toward human tumor cells. In order to rationalize structural features associated with the most potent compounds we calculated molecular descriptors derived from the 3D structures of compounds. For modeling studies structures of all compounds (1–9) were prepared as described in our previous paperCitation54, see section “Methods”. Due to low number of compounds tested against bacteria, or toward human tumor cells, and limited potencies, we did not build novel quantitative structure–activity models, but envisaged to use molecular properties and to use previously built model to draw conclusions on structural characteristics needed for significant potency. To predict potency of compound 7 toward K562 cells, we projected structures of compounds 2, 7 and 8 (all spirocyclic compounds in our dataset) on model previously derived for antiproliferative activity data of glutarimide derivatives toward K562 cells, collected from National Cancer Institute (NCI) repositoryCitation54. Unfortunately, our prediction failed. Compounds 2 and 7 were predicted as more potent comparing to compound 8 (data not shown). There are several possible reasons for the poor prediction. First, in the model of antiproliferative activity of NCI glutarimides toward K562 cells, potency data span range from 4 to 8.6 (p(GI50) values). Approximating equality between p(IC50) and p(GI50) values, which for sure is not the most stringent criteria; compounds 2 and 7, with p(IC50) of 3.98 and 3.82, respectively, are out of the range of potencies that NCI model covers. Next, antiproliferative data of compounds tested by NCI were obtained by Sulforhodamine B assay after 48 h of exposure of cells to compounds, while we used MTT assay and exposed cells to compounds during 72 h. The most potent compounds in NCI set comprise unsubstituted glutarimide nitrogen, while all compounds in our set bear bulky substituent in this position. Finally, our model of antiproliferative activity of NCI glutarimides was built on the pharmacophoric similarity patterns (including spatial positions of HBA, HBD, hydrophobic and shape features), but did not include whole-molecule properties, as surfaces and volumes. Next, we compared whole-molecular properties of compounds 1–9 (surface area, polar surface area, apolar surface area, volume, virtual log P; Supplemental Information, Table S1) derived from the 3D structures of compounds. We concluded that derivative 7, most potent toward human tumor cells tested, had the largest molecular volume. This observation is in accordance with the fact that compounds having largest molecular volumes are among most potent in NCI glutarimide set (Figure 4a in referenceCitation54). To obtain more data on structural features associated with most potent compounds in our set, we calculated MIF, with hydrogen-bond donor (N1), hydrogen-bond acceptor (O), hydrophobic (DRY) and shape (TIP) probes around compounds 1–9 in program Pentacle, and visually inspected interaction patterns, obtained by two-point pharmacophoric features offered by AMANDA algorithmCitation45,Citation55. Blocks of correlograms were depicted as a heatmap (). Features that separate the most potent compounds from the rest appeared straightforwardly.
We observed TIP-TIP and DRY-TIP blocks of variables which were broader for compounds 4, 7 and 9, comparing with the same block of variables for the rest of compounds. Along with this, the N1-TIP block of variables is significantly broader for compound 9, compared to other compounds. So, variables in those three blocks, with the largest distances between nodes, made distinction between compounds 4, 7 and 9 and the rest. Variables TIP-TIP-230, encoding nodes of shape probe on distance of ∼17.1 Å (Supplemental Information, Figure S19a), and DRY-TIP-408, encoding node of hydrophobic probe and the node of shape probe on distance of ∼17.4 Å (Supplemental Information, Figure S19b), clearly separated compound 7, most potent toward tumor cells tested, from the rest. Along the largest molecular volume, compound 7 comprises bulky cycloalkyl moiety distal from the benzyl ring bound to glutarimide N. We observed similar structural features for compounds 9 and 4, most active against Gram-positive bacteria. Variables TIP-TIP-233 (nodes of the shape probe on distance of ∼18.1 Å) and DRY-TIP-410 (node of the hydrophobic probe on distance of ∼18.1 Å from the node of shape probe) encoded compound 9. Variables TIP-TIP-234 (nodes of the shape probe on distance of ∼18.4 Å) and DRY-TIP-411 (node of the hydrophobic probe on distance of ∼18.4 Å from the node of shape probe) encode compound 4. Along with those two variables, comparable with variables associated with compound 7, that described overall shape and spatial position of benzyl moiety and branched alkyl chains attached to position 3 of glutarimide ring in compounds 9 and 4; we observed additional variables characteristic for compound 9. Variables N1-TIP-584 (HBD associated with keto group in glutarimide ring, and alkyl moiety of the ester, associated with the shape probe, on spatial distance of ∼17.1 Å) and N1-N1-157 (HBDs associated with keto group in glutarimide ring and with ester keto moiety) are typical for compound 9. We did not observe similar variables for compound 4, most probably because ester keto group, as a HBA, was hindered with branched alkyl moiety. All variables described for compounds 9 and 4 are depicted in Figures S20 and S21 (Supplemental Information). So, along with overall large molecular volumes, due to alkyl chains bound to position 3 of glutarimide moiety, compound 9, which exerted better antibacterial activity than the rest of compounds, has HBA moiety in this alkyl chain and this HBA is not hindered by the vicinal parts of molecule.
Conclusion
The cytotoxicity study of compounds 1–9 toward selected human cancer cell lines showed that compound 7 was the most potent, with IC50 values 8.98, 26.8 and 27.36 μM, toward K562, HeLa and MDA-MB-453 cells, respectively. This compound exerted modest selectivity toward normal control MRC-5 cells with IC50 ∼37 μM. Preliminary screening of antibacterial activity by a disk diffusion method showed that Gram-positive bacteria were more susceptible to tested compounds than Gram-negative bacteria. Compounds 4 and 9 expressed the highest inhibition on growth of Gram-positive bacteria B. cereus and it is comparable to antibiotic gentamicin. S. sonnei (Gram-negative bacteria) was the most sensitive to the compound 2 (comparable to antibiotic tetracycline). Using a quantitative method for determination of MIC the highest antibacterial potential was achieved with compound 9 against B. cereus. Compound 5 did not show any antimicrobial effect on tested Gram-positive and Gram-negative bacteria in any of applied tests. Whole-molecular descriptors derived from 3D structures of examined compounds and structure–activity model based on MIF, calculated by the GRID method, rationalized structural features associated with the most potent compounds. Compound with bulky hydrophobic moiety distal from glutarimide ring exerted best antiproliferative potency within studied set.
We found that compound 7 exerts the highest antiproliferative potency within studied set. This derivative will be a starting point for further structural modifications in order to obtain more potent compounds. As compound 7 exerts modest selectivity, the second goal of structural modifications will be improvement of selectivity of new derivatives.
Supplementary materials available online
IENZ_1070844_Supp.pdf
Download PDF (4.4 MB)Declaration of interest
The authors have no conflicts of interest. Authors acknowledge the Ministry of Education, Science and Technological Development of the Republic of Serbia for financial support (Grant Nos. 172032 and 175011).
References
- Matsuda F, Terashima S. Total synthesis of natural (+)-sesbanimide A and (−)-sesbanimide B. Tetrahedron 1988;44:4721–36
- Ju J, Rajski SR, Lim SK, et al. Lactimidomycin iso-migrastatin and related glutarimide-containing 12-membered macrolides are extremely potent inhibitors of cell migration. J Am Chem Soc 2009;131:1370–1
- Schneider-Poetsch T, Ju J, Eyler DE, et al. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Chem Biol 2010;6:209–17
- Sugawara K, Nishiyama Y, Toda S, et al. Lactimidomycin, a new glutarimide group antibiotic. Production, isolation, structure and biological activity. J Antibiot 1992;45:1433–41
- Machado AL, Lima LM, Araujo Jr JX, et al. Design, synthesis and antiinflammatory activity of novel phthalimide derivatives, structurally related to thalidomide. Bioorg Med Chem Lett 2005;15:1169–72
- Bartlett JB, Dredge K, Dalgleish AG. The evolution of thalidomide and its IMiD derivatives as anticancer agents. Nat Rev Cancer 2004;4:314–22
- Wu YH, Rayburn JW, Allen LE, et al. Psychosedative agents. 2. 8-(4-Substituted l-piperazinylalkyl)-8-azaspiro[4.5]decane-7,9-diones. J Med Chem 1972;15:477–9
- Barrdell LB, Fitton A. Tandospirone. CNS Drugs 1996;5:147–52
- Obrig TG, Culp WJ, Mckeehan WL, Hardesty B. The mechanism by which cycloheximide and related glutarimide antibiotics inhibit peptide synthesis on reticulocyte ribosomes. J Biol Chem 1971;246:174–81
- Kondo H, Oritani T, Kiyota H. Synthesis and antifungal activity of the four stereoisomers of streptimidone, a glutarimide antibiotic from Streptomyces rimosus forma paromomycinus. Eur J Org Chem 2000;2000:3459–62
- Frohardt RP, Dion HW, Jakubowski ZL, et al. Chemistry of streptimidone, a new antibiotic. J Am Chem Soc 1959;81:5500–6
- Kim BS, Moon SS, Hwang BK. Isolation, antifungal activity, and structure elucidation of the glutarimide antibiotic, streptimidone, produced by Micromonospora coerulea. J Agric Food Chem 1999;47:3372–80
- Ha DKK, Lau WH. Effect of recombinant human tumor necrosis factor on human nasopharyngeal carcinoma cell line in vitro. Cancer Lett 1988;41:217–24
- Andres MI, Sanz P, Garfia A, et al. Induction of cell differentiation and other in vitro effects of cycloheximide on neuroblastoma cells. In Vitro Mol Toxicol 1977;10:319–28
- Ishikawa Y, Tachibana M, Matsui C, et al. Synthesis and biological evaluation on novel analogs of 9-methylstreptimidone, an inhibitor of NF-κB. Bioorg Med Chem Lett 2009;19:1726–8
- Powell RG, Smith Jr CR, Weisleder D. Sesbanimide A and related tumor inhibitors from Sesbania drummondii: structure and chemistry. Phytochemistry 1984;23:2789–96
- Powell RG, Smith CR, Weisleder D, et al. Sesbanimide, a potent antitumor substance from Sesbania drummondii seed. J Am Chem Soc 1983;105:3739–41
- Powel RG, Smith Jr CR. An investigation of the antitumor activity of Sesbania drummondii. J Nat Prod 1981;44:86–90
- Powel RG, Smith Jr CR, Madrigal RV. Antitumor activity of Sesbania vesicaria, S. punicea, and S. drummondii seed extracts. Planta Med 1976;30:1–8
- Armoiry X, Aulagner G, Facon T. Lenalidomide in the treatment of multiple myeloma: a review. J Clin Pharm Ther 2008;33:219–26
- Lentzsch S, Rogers MS, LeBlanc R, et al. S-3-Amino-phthalimido-glutarimide inhibits angiogenesis and growth of B-cell neoplasias in mice. Cancer Res 2002;62:2300–5
- McCarthy PL, Owzar K, Hofmeister CC, et al. Lenalidomide after stem-cell transplantation for multiple myeloma. N Engl J Med 2012;366:1770–81
- Fischer DS, Woo LW, Mahon MF, et al. D-ring modified estrone derivatives as novel potent inhibitors of steroid sulfatase. Bioorg Med Chem 2003;11:1685–700
- Brueggemeier RW, Hackett JC, Diaz-Cruz ES. Aromatase inhibitors in the treatment of breast cancer. Endocr Rev 2005;26:331–45
- Santen RJ, Brodie H, Simpson ER, et al. History of aromatase: saga of an important biological mediator and therapeutic target. Endocr Rev 2009;30:343–75
- Antonini I, Volpini R, Dal Ben D, et al. Design, synthesis, and biological evaluation of new mitonafide derivatives as potential antitumor drugs. Bioorg Med Chem 2008;16:8440–6
- Norton JT, Witschi MA, Luong L, et al. Synthesis and anticancer activities of 6-amino amonafide derivatives. Anti-Cancer Drugs 2008;19:23–36
- Wu A, Xu Y, Qian X, et al. Novel naphthalimide derivatives as potential apoptosis-inducing agents: design, synthesis and biological evaluation. Eur J Med Chem 2009;44:4674–80
- Machado KE, Navakoski de Oliveira K, Santos-Bubniak L, et al. Evaluation of apoptotic effect of cyclic imide derivatives on murine B16F10 melanoma cells. Bioorg Med Chem 2011;19:6285–91
- Popović-Đorđević JB, Ivanović MD, Kiricojević VD. A novel tandem process leading to functionalized glutarimides. Tetrahedron Lett 2005;46:2611–14
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 1983;65:55–63
- Ohno M, Abe T. Rapid colorimetric assay for the quantification of leukemia inhibitory factor (LIF) and interleukin-6 (IL-6). J Immunol Methods 1991;145:199–203
- Tepe B, Daferera D, Sokmen A, et al. Antimicrobial and antioxidant activities of the essential oil and various extracts of Salvia tomentosa Miller (Lamiaceae). Food Chem 2005;90:333–40
- Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing: 15th informational supplement 2005. CLSI Document M100-S15. Pennsylvania: CLSI
- Klančnik A, Piskernik S, Jeršek B, Smole Možina S. Evaluation of diffusion and dilution methods to determine the antibacterial activity of plant extracts. J Microbiol Methods 2010;81:121–6
- Sadowski J, Gasteiger J. From atoms and bonds to three-dimensional atomic coordinates: automatic model builders. Chem Rev 1993;93:2567–81
- Sadowski J, Gasteiger J, Klebe G. Comparison of automatic three-dimensional model builders using 639 X-ray structures. J Chem Inform Model 1994;34:1000–8
- Pedretti A, Villa L,Vistoli G. VEGA – an open platform to develop chemo-bio-informatics applications, using plug-in architecture and script programming. J Comput-Aided Mol Des 2004;18:167–73
- Halgren TA. MMFF VI. MMFF94s option for energy minimization studies. J Comput Chem 1999;20:720–9
- Harrison RW. Stiffness and energy conservation in molecular dynamics: an improved integrator. J Comput Chem 1993;14:1112–22
- Stewart JJP. Optimization of parameters for semiempirical methods V: modification of NDDO approximations and application to 70 elements. J Mol Model 2007;13:1173–213
- Stewart JJP. MOPAC: a semiempirical molecular orbital program. J Comput-Aided Mol Des 1990;4:1–103
- Gaillard P, Carrupt PA, Testa B, Boudon A. Molecular lipophilicity potential, a tool in 3D QSAR: method and applications. J Comput-Aided Mol Des 1994;8:83–96
- Goodford PJ. A Computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J Med Chem 1985;28:849–57
- Durán Á, Martínez GC, Pastor M. Development and validation of AMANDA, a new algorithm for selecting highly relevant regions in molecular interaction fields. J Chem Inform Model 2008;48:1813–23 . Pentacle 1.0.6. http://www.moldiscovery.com/software/pentacle/
- Gokel GW, Cram DJ, Liotta CL, et al. 18-Crown-6 (1,4,7,10,13,16-hexaoxacycloöctadecane). Org Syn 1988;VI:301–2
- Cason J. β-Methylglutaric anhydride. Org Syn 1963;IV:630–1
- Farmer HH, Rebjohn N. β-Ethyl-β-methylglutaric acid. Org Syn 1963;IV:441–2
- Cason J. β-Carbomethoxypropionyl chloride. Org Syn 1955;III:169–70
- Vogel AI. Vogel’s text book of practical organic chemistry. 5th ed. Harlow: Longman Group, UK Limited; 1989
- Strube RE. Ethyl tert-butyl malonate. Org Syn 1963;IV:417–18
- Paquet LA, ed. Encyclopedia of reagents for organic synthesis. Vol. 7. Pennsylvania State University, John-Wiley; 1995. Available from: http://eu.wiley.com/WileyCDA/WileyTitle/productCd-0470017546.html [last accessed 21 Jul 2015]
- Krapcho AP, Diamanti J, Cayen C, Bingham R. 2-Carbethoxycyclooctanone. Org Syn 1973;V:198–9
- Popović-Djordjević JB, Došen-Mićović LjI, Juranić IO, Drakulić BJ. Antiproliferative activity of NCI-DTP glutarimide derivatives: an alignment independent 3D QSAR study. J Serbian Chem Soc 2010;75:1167–79
- Cruciani G, ed. Methods and principles in medicinal chemistry, molecular interaction fields: applications in drug discovery and ADME prediction. Weinheim: Wiley-VCH; 2006