Abstract
Thirty new aryl-pyridazinone-substituted benzenesulphonylurea derivatives (I–XXX) were synthesized and evaluated for their anti-hyperglycaemic activity in glucose-fed hyperglycaemic normal rats. Twenty-three compounds (III–XI, XIV–XVII, XIX–XXIV, XXVI and XXVIII–XXX) showed more or comparable area under the curve (AUC) reduction percentage (ranging from 21.9% to 35.5%) as compared to the standard drug gliclazide (22.0%). On the basis of docking results, 18 compounds were screened for their in vitro ability to inhibit rat lens aldose reductase. Ten compounds (III–VI, XII, XVI–XVIII, XXI and XXVII) showed ARI activity with IC50 ranging from 34 to 242 μM. Out of these, two compounds IV and V showed best ARI activity which is comparable with that of quercetin. As a result, two compounds (IV and V) possessing significant dual action (anti-hyperglycaemic and aldose reductase inhibition) were identified and may be used as lead compounds for developing new drugs.
Introduction
Diabetes mellitus (DM) has become pandemic and according to reports, including a forecast by the World Health Organizations (WHO), there will be a sharp increase in the total number of cases world wide by 2030. In fact, India could be bracing itself for the dubious distinction of becoming the diabetes of capital of the worldCitation1. Alone, in India recently published report by ICMR-INDIAB national study stated that there are 62.4 million people with non-insulin-dependent diabetes mellitus (NIDDM) and 77 million people with prediabetes in India. NIDDM is the most common form of diabetes and has been found 90% of all diabetesCitation2.
NIDDM is a multifactorial metabolic disease characterized by abnormalities at multiple organ sites. These defects include insulin resistance and insulin deficiencyCitation3,Citation4. The former is primarily represented by decreased insulin-stimulated glucose uptake in skeletal muscle, augmented endogenous glucose production (predominately in the liver) and enhanced lipolytic activity in fatCitation5. The latter is an apparent progressive process with both functional defects in islet cell function and, eventually, apparent loss of β-cell massCitation6,Citation7. These defects are intimately linked with derangements in one system exacerbating those in the othersCitation8.
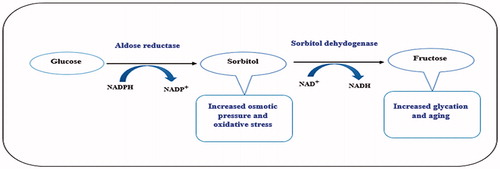
Patients with DM are at high risk of developing long-term complications including neuropathy, nephropathy, retinopathy and cataractCitation9,Citation10. Although strict glycaemic control is expected to prevent diabetic complications but perfect glycaemic control is not always possible. In hyperglycaemic conditions, aldose reductase catalyses an NADPH-dependent reduction of glucose to sorbitol, which in turn is oxidized to fructose by an NAD+-dependent sorbitol dehydrogenase (. Once sorbitol is accumulated inside the cells, it cannot diffuse easily across the cell membrane as a result osmotic pressure increases causing cellular damage. Moreover, the depletion of NADPH and NAD+ cofactors compromises body’s antioxidant defence system. In addition, high blood levels of fructose may account for increased glycation and accelerating ageingCitation11,Citation12 (. Thus, inhibition of AR is one of the important targets and its inhibition could be the key for the treatment of many diabetic complicationsCitation13. Aldose reductase inhibitors (ARIs) have been shown to reduce tissue sorbitol accumulation in diabetic animalsCitation14 and there are evidences that blockage of AR can have beneficial effect in diabetic complicationsCitation15,Citation16.
Various structurally different compounds have been identified as potent in vitro ARIs and some of them are in clinical trials to test their efficacy in the prevention and treatment of peripheral neuropathy in diabetesCitation10,Citation17. They can be categorized into various groups based on their chemical structures: acetic acid derivatives (e.g. epalrestat, Imirestat), cyclic imides (especially spirohydantoins, e.g. sorbinil, fidarestat) etc. Despite being structurally different, all ARIs possess the following two peculiar pharmacophoric elements: (i) an acidic moiety that is able to interact with the “anion-binding site” of the catalytic site and (ii) a lipophilic scaffold that can bind to the highly flexible specificity pocket of the catalytic siteCitation18. Our interest in studying the molecular determinants for binding to the aldose reductase inhibitor site led us to study the aldose reductase inhibitory activity of target compounds possessing benzenesulphonylurea as carboxylic acid surrogates. Besides this benzenesulphonamide derivatives have been already reported as ARICitation19,Citation20. The SO2 group of the benzenesulphonamide forms the hydrogen bond with the amino acid residues of the anionic binding site of the AR enzymeCitation21.
In curing, diabetes sulphonyl ureas have represented the backbone of oral therapy in NIDDM for more than 40 yearsCitation22. Recently, compounds containing pyridazinone nucleus have been reported as AR inhibitorsCitation23–26. In the view of above facts and as part of our research programme (to develop new lead compounds that not only act as antihyperglycaemic agents, but at the same time prevent diabetic complications caused by the aldose reductase or the associated pathway), we herein report the synthesis of new pyridazinone-substituted benzenesulphonyl urea derivatives (I–XXX) incorporating with known bioactive moieties highlighted in . We have introduced a diversity of aryl-substituted pyridazinones moiety at para position of benzenesulphonamide and other substitutions, such as isopropyl and 4-methylcyclohexyl groups, were made at N2 position of the uriedo group, keeping the core structure intact (.
Figure 2. Structures of clinically used antidiabetic drugs and pyridazinone derivatives reported as aldose reductase inhibitor. Rationally designed template for targeted compound (I–XXX).
The synthesized compounds (I–XXX) were evaluated for antihyperglycaemic effects in glucose-fed hyperglycaemic normal rats. In silico molecular docking studies of these compounds (I–XXX) were performed with respect to aldose reductase. Eighteen compounds displaying good docking results were evaluated for their ability to inhibit rat lens aldose reductase.
Experimental
Melting points were determined by open capillary tubes and are uncorrected. Purity of the compounds was checked on thin-layer chromatography (TLC) plates (silica gel G) that were visualized by exposing to iodine vapours. Infrared (IR) spectra were recorded (in KBr) on a BIO-RAD FTS-135 spectrophotometer and 1HNMR spectra were recorded on a Bruker Spectrospin DPX 300 MHz spectrometer using deuterated dimethyl sulphoxide (DMSO) as solvent and tetramethyl silane (TMS) as an internal standard. Chemical shifts are given in δ (ppm) scale and coupling constants (J values) are expressed in hertz (Hz). Elemental analysis was carried out on CHNS Elementar (Vario EL III). Mass spectra (MS) were scanned by using ESI Bruker Esquire 3000 (Billerica, MA). The m/z values of the more intense peaks are mentioned.
General procedure for synthesis of sulfonylureas (I-XXX)
A solution of appropriate pyridazinone (1 mmol) in dry acetone was refluxed over anhydrous K2CO3 (2 mmol) for 1–1.5 h. At this temperature, a solution of the appropriate isocyanate (1.2 mmol) in dry acetone 5 ml was added in a dropwise manner. It was refluxed for 24–72 h. Acetone was removed under reduced pressure. The solid residue thus obtained was suspended in water and acidified with dilute acetic acid. It was stirred for 30 min and filtered. The residue was washed with plenty of distilled water in order to make the residue free from potassium acetate. It was dried and crystallized from methanol. The purity of the compounds was checked on TLC plate (Silica gel G) in the solvent system toluene/ethyl acetate/formic acid (5:4:1, TEF).
1-{4-[3–(2,3-Dihydro-1H-inden-5-yl)-6-oxopyridazin-1-yl]benzenesulphonyl}-3-isopropylurea (I)
Yellow crystals, yield = 45%, m.p. 120–121 °C, Rf = 0.65 (TEF). IR νmax (KBr, in cm −1): 3335, 3037 and 2911 (NH-CO-NH), 1715 and 1525 (C=O) of urea, 1605 (C=N), 1320 and 1142 (SO2N). 1H NMR (300 MHz, DMSO-d6, δ): 0.95 (6H, d, J = 6 Hz, -CH-(CH3)2), 1.90–2.07 (2H, m, H-3″′), 2.91 (4H, d, J = 4.2 Hz, H-2″′, H-4″′), 3.53–3.66 (1H, m, -NH-CH-(CH3)2), 5.49 (1H, brs, -NH-(CH3)2), 7.14–8.15 (9H, m, H-5, H-5′, H-6′, H-2′, H-3″, H-5″, H-2″, H-6″, H-4), 8.31 (1H, s, SO2-NH-C=O). 13C NMR (75 MHz, DMSO-d6, δ, ppm): 43.20 (CH-NH), 125.71 (C-4 pyridazinone), 137.60 (C=O uriedo group), 144.22 (C-5 pyridazinone), 152.10 (C=O pyridazinone), 155.62 (C=N pyridazinone). ESI-MS (m/z): 452 [M+], 453 [M + 1], 451 [M-1], 450 [M-2]. CHNS Analysis for C23H24N4O4S Found (Calculated): C: 61.10 (61.08), H: 5.34 (5.39), N: 12.38 (12.30), O: 14.12(14.15), S: 7.07 (7.13).
3-Isopropyl-1–(4-{3-[4–(2-methylpropyl)phenyl]-6-oxopyridazin-1-yl} benzenesulphonyl) urea (II)
Yellow crystals, yield = 47%, m.p. 162–163 °C, Rf = 0.67 (TEF). IR νmax (KBr, in cm −1): 3360, 3025 and 2915 (NH-CO-NH), 1710 and 1510 (C=O) of urea, 1596 (C=N), 1325 and 1155 (SO2N). 1H NMR (300 MHz, DMSO-d6, δ): 0.87 (6H, d, J = 6.6 Hz, Ph-CH2-CH-(CH3)2), 0.98 (6H, d, J = 8.1 Hz, -NH-CH-(CH3)2), 1.88 (1H, m, Ph-CH2-CH-(CH3)2), 2.5 (2H, merged s, Ph-CH2-CH-(CH3)2), 3.58–3.67 (1H, m, -NH-CH-(CH3)2), 5.46 (1H, d, J = 4.5 Hz, -NH-CH-(CH3)2), 7.20 (1H, d, J = 10.2 Hz, H-5), 7.29 (2H, d, J = 6.9 Hz, H-3′, H-5′), 7.62 (1H, d, J = 9.8 Hz, -SO2-NH-C=O), 7.85 (2H, d, J = 7.2 Hz, H-2′, H-6′), 7.90 (2H, d, J = 7.8 Hz, H-3″, H-5″), 7.96 (2H, d, J = 8.4 Hz, H-2″, H-6″), 8.14 (1H, d, J = 9.9 Hz, H-4). 13C NMR (75 MHz, DMSO-d6, δ, ppm): 43.25 (CH-NH), 125.74 (C-4 pyridazinone), 137.62 (C=O uriedo group), 144.35 (C-5 pyridazinone), 151.20 (C=O pyridazinone), 156.44 (C=N pyridazinone). ESI-MS (m/z): 468 [M+], 469 [M + 1], 467 [M-1], 466 [M-2].CHNS Analysis for C24H28N4O4S Found (Calculated): C: 61.55 (61.56), H: 6.05 (6.08) N: 12.10 (12.15), O: 13.54 (13.59), S: 6.85 (6.90).
1-{4-[3–(3,4-Difluorophenyl)-6-oxopyridazin-1-yl]benzenesulphonyl}-3-isopropylurea (III)
Light brown crystals, yield = 51% m.p. 142–143 °C, Rf = 0.60 (TEF). IR νmax (KBr, in cm− 1): 3345, 3033 and 2940 (NH-CO-NH), 1698 and 1515 (C=O) of urea, 1613 (C=N), 1331 and 1151 (SO2N). 1H NMR (300 MHz, DMSO-d6, δ): 0.93 (6H, d, J = 6.5 Hz, -CH-(CH3)2), 3.75–3.84 (1H, m, -NH-CH-(CH3)2), 6.60 (1H, d, J = 5.4 Hz, -NH-CH-(CH3)2), 7.46 (1H, d, J = 7.2 Hz, H-5), 7.78 (1H, dd, J = 7.8 Hz, H-5′), 8.04 (1H, d, J = 6 Hz, H-2′), 8.17 (2H, d, J = 6.3 Hz, H-3″, H-5″), 8.22–8.28 (3H, m, H-6′, H-2″, H-6″), 8.40 (1H, d, J = 7.5 Hz, H-4), 10.75 (1H, s, SO2-NH-C=O). 13C NMR (75 MHz, DMSO-d6, δ, ppm): 43.22 (CH-NH), 125.67 (C-4 pyridazinone), 138.60 (C=O uriedo group), 145.31 (C-5 pyridazinone), 151.17 (C=O pyridazinone), 155.54 (C=N pyridazinone). ESI-MS (m/z): 448 [M+], 449 [M + 1], 447 [M-1], 446 [M-2]. CHNS Analysis for C20H18F2N4O4S Found (Calculated): C: 53.58 (53.59), H: 4.08 (4.09), F: 8.48 (8.49), N: 12.48 (12.49), O: 14.28 (14.29), S: 7.13 (7.14).
1-{4-[3–(4-Fluorophenyl)-6-oxopyridazin-1-yl] benzenesulphonyl}-3-isopropylurea (IV)
Yellow crystals, yield = 41%, m.p. 102–103 °C, Rf = 0.60 (TEF). IR νmax (KBr, in cm−1): 3327, 3050 and 2970 (NH-CO-NH), 1705 and 1503 (C=O) of urea, 1597 (C=N), 1339 and 1157 (SO2N). 1H NMR (300 MHz, DMSO-d6, δ): 0.99 (6H, d, J = 6.3 Hz, -NH-CH-(CH3)2), 3.61–3.69 (1H, m, -NH-CH-(CH3)2), 4.10 (1H, d, J = 5.7 Hz, -NH-CH-(CH3)2), 7.06 (1H, d, J = 9.6 Hz, H-5), 7.12 (2H, d, J = 9 Hz, H-2′, H-6′), 7.74–7.96 (6H, m, H-3′, H-5′, H-3″, H-5″, H-2″, H-6″), 8.01 (1H, d, J = 8.7 Hz, H-4). 13C NMR (75 MHz, DMSO-d6, δ, ppm): 42.91 (CH-NH), 125.40 (C-4 pyridazinone), 137.20 (C=O uriedo group), 146.10 (C-5 pyridazinone), 152.10 (C=O pyridazinone), 155.31 (C=N pyridazinone). ESI-MS (m/z): 430 [M+], 431 [M + 1], 429 [M-1], 428 [M-2].CHNS Analysis for C20H19FN4O4S Found (Calculated): C: 55.75 (55.76), H: 4.43 (4.44), F: 4.39 (4.4), N: 13.10 (13.12), O: 14.87 (14.88), S: 7.46 (7.47).
3-Isopropyl-1-{4-[6-oxo-3–(5, 6, 7, 8-tetrahydronaphthalen-2-yl) pyridazin-1-yl] benzenesulphonyl} urea (V)
White crystals, yield = 45%, m.p. 200–201 °C, Rf = 0.63 (TEF). IR νmax (KBr, in cm−1): 3356, 3071 and 2995 (NH-CO-NH), 1720 and 1518 (C=O) of urea, 1610 (C=N), 1345 and 1175 (SO2N). 1H NMR (300 MHz, DMSO-d6, δ): 1.01 (6H, d, J = 6.3 Hz, -NH-CH-(CH3)2), 1.76 (4H, brs, H-3″′, H-4″′), 2.77 (4H, brs, H-2″′, H-5″′), 3.59–3.61 (1H, m, -NH-CH-(CH3)2), 5.50 (1H, d, J = 6.3 Hz, -NH-CH-(CH3)2), 6.27 (1H, d, J = 15.6 Hz, SO2-NH-C=O), 6.97–7.28 (2H, m, H-5, H-5′), 7.64–7.75 (2H, m, H-2′, H-6′), 7.90 (2H, d, J = 8.1 Hz, H-3″, H-5″), 8.01 (2H, d, J = 6.9 Hz, H-2″, H-6″), 8.15 (1H, d, J = 9.6 Hz, H-4). 13C NMR (75 MHz, DMSO-d6, δ, ppm): 43.65 (CH-NH), 125.22 (C-4 pyridazinone), 138.52 (C=O uriedo group), 145.10 (C-5 pyridazinone), 151.37 (C=O pyridazinone), 155.72 (C=N pyridazinone). ESI-MS (m/z): 466 [M+], 467 [M + 1], 465 [M-1], 464 [M-2].CHNS Analysis for C24H26N4O4S Found (Calculated): C: 61.75 (61.74), H: 5.62 (5.60), N: 12.10 (12.11), O: 13.70 (13.72), S: 6.91(6.92).
1-{4-[3–(4-Benzylphenyl)-6-oxopyridazin-1-yl] benzenesulphonyl}-3-isopropylurea (VI)
Light yellow crystals, yield = 58%, m.p. 179–180 °C, Rf =0.61 (TEF). IR νmax (KBr, in cm −1): 3340, 3049 and 2998 (NH-CO-NH), 1705 and 1522 (C=O) of urea, 1596 (C=N), 1355 and 1153 (SO2N). 1H NMR (300 MHz, DMSO-d6, δ): 1.02 (6H, d, J = 8.4 Hz, -NH-CH-(CH3)2), 3.56–3.64 (1H, m, -NH-CH-(CH3)2), 4.01 (2H, s, Ph-CH2-Ph), 6.39 (1H, s, -NH-CH-(CH3)2), 7.18–7.32 (6H, m, H-2″′, H-3″′, H-4″′, H-5″′, H-6″′, H-5), 7.37 (2H, d, J = 6.3 Hz, H-3′, H-5′), 7.87 (2H, d, J = 6 Hz, H-2′, H-6′), 7.95 (2H, d, J = 6.3 Hz, H-3″, H-5″), 8.03 (2H, d, J = 6.6 Hz, H-2″, H-6″), 8.15 (1H, d, J = 7.2 Hz, H-4), 10.53 (1H, s, SO2-NH-C=O). 13C NMR (75 MHz, DMSO-d6, δ, ppm):43.72 (CH-NH), 125.82 (C-4 pyridazinone), 138.82 (C=O uriedo group), 145.70 (C-5 pyridazinone), 151.31 (C=O pyridazinone), 156.10 (C=N pyridazinone). ESI-MS (m/z): 502 [M+], 503 [M + 1], 501 [M-1], 500 [M-2]. CHNS Analysis for C27H26N4O4S Found (Calculated): C: 63.72 (63.73), H: 4.2 (4.1), N: 12.02 (12.03), O: 14.05 (14.06), S: 6.07 (6.08).
3-Isopropyl-1-{4-[6-oxo-3–(4- propylphenyl) pyridazin-1-yl] benzenesulphonyl} urea (VII)
Yellow crystals, yield = 47%, m.p. 185–186 °C, Rf = 0.60 (TEF). IR νmax (KBr, in cm− 1): 3340, 3071 and 2975 (NH-CO-NH), 1700 and 1510 (C=O) of urea, 1589 (C=N), 1337 and 1121 (SO2N). 1H NMR (300 MHz, DMSO-d6, δ): 0.90 (3H, t, Ph-CH2CH2CH3), 1.001 (6H, d, J = 6.3 Hz, -NH-CH-(CH3)2), 1.57–1.65 (2H, m, Ph-CH2CH2CH3), 2.61 (2H, t, Ph-CH2CH2CH3), 3.60–3.66 (1H, m, -NH-CH-(CH3)2), 5.48 (1H, d, J = 7.8 Hz, -NH-CH-(CH3)2), 6.35 (1H, brs, SO2-NH-C=O), 7.22 (1H, d, J = 9.6 Hz, H-5), 7.32 (2H, d, J = 8.1 Hz, H-3′, H-5′), 7.86 (2H, d, J = 7.8 Hz, H-2′, H-6′), 7.94 (2H, d, J = 8.7 Hz, H-3″, H-5″), 8.02 (2H, d, J = 8.4 Hz, H-2″, H-6″), 8.16 (1H, d, J = 9.6 Hz, H-4). 13C NMR (75 MHz, DMSO-d6, δ, ppm): 42.27 (CH-NH), 124.36 (C-4 pyridazinone), 137.85 (C=O uriedo group), 146.19 (C-5 pyridazinone), 150.89 (C=O pyridazinone), 157.12 (C=N pyridazinone). ESI-MS (m/z):454 [M+], 455 [M + 1], 453 [M-1], 452 [M-2]. CHNS Analysis for C23H26N4O4S Found (Calculated): C: 60.52 (60.53), H: 5.78 (5.77), N: 12.43 (12.45), O: 14.18 (14.17), S: 7.15 (7.16).
1-{4-[3–(4-Ethoxyphenyl)-6-oxopyridazin-1-yl] benzenesulphonyl}-3-isopropylurea (VIII)
Yellow crystals, yield = 50%, m.p. 148–149 °C, Rf = 0.62 (TEF). IR νmax (KBr, in cm− 1): 3317, 3090 and 2981 (NH-CO-NH), 1718 and 1520 (C=O) of urea, 1585 (C=N), 1331 and 1142 (SO2N). 1H NMR (300 MHz, DMSO-d6, δ): 1.007 (6H, d, J = 6.3 Hz, -NH-CH-(CH3)2), 1.34 (3H, t, Ph-O-CH2-CH3), 3.61–3.67 (1H, m, -NH-CH-(CH3)2), 4.09 (2H, q, Ph-O-CH2-CH3), 5.40 (1H, brs, -NH-CH-(CH3)2), 7.03 (2H, d, J = 8.7 Hz, H-3′, H-5′), 7.15 (1H, d, J = 11.1 Hz, H-5), 7.85–7.97 (6H, m, H-2″, H-6″, H-3″, H-5″, H-2′, H-6′), 8.02 (1H, d, J = 8.7 Hz, H-4), 8.27 (1H, s, SO2-NH-C=O). 13C NMR (75 MHz, DMSO-d6, δ, ppm): 42.22 (CH-NH), 126.11 (C-4 pyridazinone), 136.87 (C=O uriedo group), 146.13 (C-5 pyridazinone), 153.15 (C=O pyridazinone), 157.12 (C=N pyridazinone). ESI-MS (m/z): 456 [M+], 457 [M + 1], 455 [M-1], 454 [M-2]. CHNS Analysis for C22H24N4O5S Found (Calculated): C: 58.68 (58.67), H: 4.9 (5.01), N: 12.1 (12.09), O: 17.32 (17.31), S: 7.02 (7.01).
3-Isopropyl-1–(4-{6-oxo-3-[4–(2-phenylethyl) phenyl]pyridazin-1-yl}benzenesulphonyl) urea (IX)
White crystals, yield = 51%, m.p. 190–191 °C, Rf = 0.62 (TEF). IR νmax (KBr, in cm−1): 3321, 3080 and 2983 (NH-CO-NH), 1729 and 1522 (C=O) of urea, 1582 (C=N), 1325 and 1144 (SO2N). 1H NMR (300 MHz, DMSO-d6, δ): 0.98 (6H, d, J = 6.6 Hz, -NH-CH-(CH3)2), 2.92 (4H, s, Ph-CH2-CH2-Ph), 3.50–3.59 (1H, m, -NH-CH-(CH3)2), 7.16–7.26 (6H, m, H-5, H-2 ″′, H-3 ″′, H-4 ″′, H-5 ″′, H-6 ″′), 7.34 (2H, d, J = 7.2 Hz, H-3′, H-5′), 7.72–8.13 (7H, m, H-4, H-2 ″, H-6 ″, H-3 ″, H-5 ″, H-2′, H-6′), 8.30 (1H, s, SO2-NH-C=O). 13C NMR (75 MHz, DMSO-d6, δ, ppm): 45.10 (CH-NH), 125.55 (C-4 pyridazinone), 137.67 (C=O uriedo group), 144.29 (C-5 pyridazinone), 152.19 (C=O pyridazinone), 155.81 (C=N pyridazinone).). ESI-MS (m/z): 516 [M+], 517 [M + 1], 515 [M-1], 514 [M-2]. CHNS Analysis for C28H28N4O4S Found (Calculated): C: 65.09 (65.1), H: 5.46 (5.44), N: 10.83 (10.84), O: 12.45 (12.46), S: 6.20 (6.22).
3-Isopropyl-1-{4-[6-oxo-3–(4-phenoxyphenyl) pyridazin-1-yl]benzenesulphonyl}urea (X)
White crystals, yield = 53%, m.p. 127–128 °C, Rf = 0.61 (TEF). IR νmax (KBr, in cm −1): 3336, 3061 and 2981 (NH-CO-NH), 1732 and 1530 (C=O) of urea, 1591 (C=N), 1345 and 1165 (SO2N). 1H NMR (300 MHz, DMSO-d6, δ): 0.97 (6H, d, J = 6.9 Hz, -NH-CH-(CH3)2), 3.03–3.64 (1H, m, -NH-CH-(CH3)2), 5.48 (1H, brs, -NH-CH-(CH3)2), 5.83 (1H, brs, SO2-NH-C=O), 7.07–8.16 (15H, m, H-2′, H-6′, H-3′, H-5′, H-3″, H-5 ″, H-2 ″, H-6 ″, H-2 ″′, H-3 ″′, H-4 ″′, H-5 ″′, H-6 ″′, H-4, H-5), 8.30 (1H, s, SO2-NH-C=O). 13C NMR (75 MHz, DMSO-d6, δ, ppm): 42.72 (CH-NH), 125.81 (C-4 pyridazinone), 137.74 (C=O uriedo group), 144.67 (C-5 pyridazinone), 152.21 (C=O pyridazinone), 155.81 (C=N pyridazinone). ESI-MS (m/z): 504 [M+], 505 [M + 1], 503 [M-1], 502 [M-2]. CHNS Analysis for C26H24N4O5S Found (Calculated): C: 61.9 (61.89), H: 4.78 (4.79), N: 11.11 (11.12), O: 15.85 (15.84), S: 6.36 (6.37).
1-{4-[3–(4-Cyclohexylphenyl)-6-oxopyridazin-1-yl] benzenesulphonyl}-3-isopropylurea (XI)
Yellow crystals, yield = 60%, m.p. 188–189 °C, Rf = 0.68 (TEF). IR νmax (KBr, in cm− 1): 3329, 3071 and 2977 (NH-CO-NH), 1740 and 1540 (C=O) of urea, 1605 (C=N), 1350 and 1168 (SO2N). 1H NMR (300 MHz, DMSO-d6, δ): 1.06 (6H, d, J = 6.6 Hz, -NH-CH-(CH3)2), 1.22–1.97 (11H, m, cyclohexyl ring), 3.62–3.73 (1H, m, -NH-CH-(CH3)2), 5.56 (1H, d, J = 7.5 Hz, -NH-CH-(CH3)2), 6.28 (1H, brs, SO2-NH-C=O), 7.27 (1H, d, J = 9.9 Hz, H-5), 7.41 (2H, d, J = 8.1 Hz, H-3′, H-5′), 7.90–8.01 (4H, m, H-3″, H-5 ″, H-2′, H-6′), 8.04 (2H, d, J = 8.7 Hz, H-2 ″, H-6 ″), 8.21 (1H, d, J = 9.6 Hz, H-4). 13C NMR (75 MHz, DMSO-d6, δ, ppm): 45.31 (CH-NH), 124.93 (C-4 pyridazinone), 138.20 (C=O uriedo group), 145.32 (C-5 pyridazinone), 153.10 (C=O pyridazinone), 154.92 (C=N pyridazinone). ESI-MS (m/z): 494 [M+], 495 [M + 1], 493 [M-1], 492 [M-2]. CHNS Analysis for C26H30N4O4S Found (Calculated): C: 63.17 (63.19), H: 6.25 (6.24), N: 11.3 (11.29), O: 12.87 (12.86), S: 6.45 (6.44).
1-{4-[3–(4-Bromophenyl)-6-oxopyridazin-1-yl] benzenesulphonyl}-3-isopropylurea (XII)
Dark yellow crystals, yield = 43%, m.p. 168–169 °C, Rf = 0.70 (TEF). IR νmax (KBr, in cm− 1): 3341, 3102 and 2988 (NH-CO-NH), 1743 and 1538 (C=O) of urea, 1610 (C=N), 1341 and 1149 (SO2N); 1H NMR (300 MHz, DMSO-d6, δ): 1.16 (6H, d, J = 6.3 Hz, -NH-CH-(CH3)2), 4.12–4.14 (1H, m, -NH-CH-(CH3)2), 6.006 (1H, brs, -NH-CH-(CH3)2), 7.20–8.16 (11H, m, H-2′, H-3′, H-5′, H-6′, H-2″, H-3″, H-5″, H-6″, H-4, H-5, SO2-NH C=O), 13C NMR (75 MHz, DMSO-d6, δ, ppm): 44.18 (CH-NH), 125.62 (C-4 pyridazinone), 136.71 (C=O uriedo group), 145.15 (C-5 pyridazinone), 152.24 (C=O pyridazinone) 154.31 (C=N pyridazinone). ESI-MS (m/z): 492 [M + 2], 491 [M + 1], 490 [M+], 489 [M-1]. CHNS Analysis for C20H19BrN4O4S Found (Calculated): C: 49.21 (49.19), H: 3.4 (3.38), Br: 16.17 (16.18), N: 11.45(11.44), O: 13.3(13.29), S: 6.54(6.55).
1-{4-[3–(4-Iodophenyl)-6-oxopyridazin-1-yl] benzenesulfonyl}-3-isopropylurea (XIII)
Yellow crystals, yield = 41%, m.p. 171–172 °C, Rf = 0.66 (TEF). IR νmax (KBr, in cm −1): 3307, 3175 and 2991 (NH-CO-NH), 1750 and 1541 (C=O) of urea, 1613 (C=N), 1345 and 1153 (SO2N). 1H NMR (300 MHz, DMSO-d6, δ): 1.04 (6H, d, J = 7.2 Hz, -NH-CH-(CH3)2), 3.64–3.72 (1H, m, -NH-CH-(CH3)2), 5.56 (1H, d, J = 7.5 Hz, -NH-CH-(CH3)2), 6.01 (1H, s, SO2-NH-C=O), 7.27 (1H, d, J = 9.3 Hz, H-5), 7.59 (2H, d, J = 19.8 Hz, H-2′, H-6′), 7.81 (2H, d, J = 7.8 Hz, H-3′, H-5′), 7.91–8.01 (4H, m, H-2″, H-3″, H-5″, H-6″), 8.22 (1H, d, J = 5.4 Hz, H-4). 13C NMR (75 MHz, DMSO-d6, δ, ppm): 45.11 (CH-NH), 126.31 (C-4 pyridazinone), 135.13 (C=O uriedo group), 146.22 (C-5 pyridazinone), 153.10 (C=O pyridazinone), 156.10 (C=N pyridazinone). ESI-MS (m/z): 538 [M+], 539 [M + 1], 537 [M-1], 536 [M-2]. CHNS Analysis for C20H19IN4O4S Found (Calculated): C: 44.73 (44.71), H: 3.45 (3.47), I: 23.75 (23.74), N: 10.43 (10.44), O: 11.78 (11.77), S: 5.88 (5.87).
1-{4-[3–(4-Hexylphenyl)-6-oxopyridazin-1-yl] benzenesulphonyl}-3-isopropylurea (XIV)
Yellow crystals, yield = 42%, m.p. 174–175 °C, Rf = 0.63 (TEF). IR νmax (KBr, in cm−1): 3319, 3091 and 2981 (NH-CO-NH), 1738 and 1529 (C=O) of urea, 1602 (C=N), 1336 and 1149 (SO2N). 1H NMR (300 MHz, DMSO-d6, δ): 0.81 (3H, t, -Ph-(CH2)5-CH3), 1.07 (6H, d, J = 6.3 Hz, -NH-CH-(CH3)2), 1.26 (6H, s, -Ph-CH2-CH2-(CH2)3-CH3), 1.55 (2H, q, -Ph-CH2-CH2-(CH2)3-CH3), 2.57 (2H, t, -Ph-CH2-(CH2)4-CH3), 3.71–3.82 (1H, m, -NH-CH-(CH3)2), 4.27 (1H, d, J = 6.3 Hz, -NH-CH-(CH3)2), 6.19 (1H, d, J = 9.6 Hz, SO2-NH-C=O), 7.05 (1H, d, J = 9.6 Hz, H-5), 7.20 (2H, d, J = 7.8 Hz, H-3′, H-5′) 7.63–7.69 (3H, m, H-2′, H-6′, H-4′), 7.89 (2H, d, J = 8.4 Hz, H-3″, H-5″), 8.008 (2H, d, J = 8.1 Hz, H-2″, H-6″). 13C NMR (75 MHz, DMSO-d6, δ, ppm): 42.78 (CH-NH), 125.42 (C-4 pyridazinone), 135.81 (C=O uriedo group), 144.85 (C-5 pyridazinone), 153.14 (C=O pyridazinone), 155.41 (C=N pyridazinone). ESI-MS (m/z): 496 [M+], 497 [M + 1], 495 [M-1], 494 [M-2]. CHNS Analysis for C26H32N4O4S Found (Calculated): C: 62.76 (62.77), H: 6.52 (6.53), N: 11.31 (11.3), O: 12.92 (12.91), S: 6.51 (6.52).
3-Isopropyl-1-{4-[3–(4-isopropylphenyl)-6-oxopyridazin-1-yl]benzenesulphonyl}urea (XV)
Light yellow crystals, yield = 50%, m.p. 157–158 °C, Rf = 0.65 (TEF). IR νmax (KBr, in cm− 1): 3362, 3055 and 2975 (NH-CO-NH), 1725 and 1521 (C=O) of urea, 1597 (C=N), 1340 and 1151 (SO2N). 1H NMR (300 MHz, DMSO-d6, δ): 1.01 (12H, d, J = 6.3 Hz, -Ph-CH-(CH3)2, -NH-CH-(CH3)2), 2.96 (6H, d, J = 6.9 Hz, Ph-CH-(CH3)2), 2.97 (1H, h, Ph-CH-(CH3)2), 3.61–3.68 (1H, m, -NH-CH-(CH3)2), 5.43 (1H, s, -NH-CH-(CH3)2), 7.17 (1H, d, J = 7.8 Hz, H-5), 7.37 (2H, d, J = 7.8 Hz, H-3′, H-5′), 7.75 (2H, d, J = 8.1 Hz, H-2′, H-6′), 7.84–7.98 (4H, m, H-2″, H-6″, H-3″, H-5″), 8.10 (1H, d, J = 7.2 Hz, H-4), 8.20 (1H, brs, SO2-NH-C=O). 13C NMR (75 MHz, DMSO-d6, δ, ppm): 42.95 (CH-NH), 125.54 (C-4 pyridazinone), 136.72 (C=O uriedo group), 145.45 (C-5 pyridazinone), 152.23 (C=O pyridazinone), 156.12 (C=N pyridazinone). ESI-MS (m/z): 454 [M+], 455 [M + 1], 453 [M-1], 452 [M-2]. CHNS Analysis for C23H26N4O4S Found (Calculated): C: 60.6 (60.59), H: 5.7 (5.71), N: 12.39 (12.41), O: 14.25 (14.26), S: 7.08 (7.1).
3-{4-[3–(2,3-Dihydro-1H-inden-5-yl)-6-oxopyridazin-1-yl]benzenesulphonyl}-1–(4-methylcyclohexyl) urea (XVI)
White crystals, yield = 65%, m.p. 225–226 °C, Rf = 0.68 (TEF). IR νmax (KBr, in cm − 1): 3331, 3028 and 2907 (NH-CO-NH), 1718 and 1531 (C=O) of urea, 1609 (C=N), 1329 and 1139 (SO2N). 1H NMR (300 MHz, DMSO-d6, δ): 0.90 (3H, d, J = 6.3 Hz, -cyclohexyl-CH3), 1.006–1.92 (9H, m, cyclohexyl ring protons), 2.08–2.13 (2H, m, H-3″′), 2.97 (4H, d, J = 5.1 Hz, H-2″′, H-4″′), 3.20 (1H, m, proton at C-1 of cyclohexyl), 5.50 (1H, brs, -NH-cyclohexyl ring), 7.23 (1H, d, J = 9.9 Hz, H-5), 7.40 (1H, d, J = 7.8 Hz, H-5′), 7.74 (1H, d, J = 8.1 Hz, H-6′), 7.83 (1H, s, H-2′), 7.95 (2H, d, J = 8.7 Hz, H-3″, H-5″), 8.02 (2H, d, J = 8.4 Hz, H-2″, H-6″), 8.17 (1H, d, J = 9.6 Hz, H-4). 13C NMR (75 MHz, DMSO-d6, δ, ppm): 48.75 (CH-NH), 125.51 (C-4 pyridazinone), 145.42 (C-5 pyridazinone), 152.17 (C=O pyridazinone), 155.32 (C=N pyridazinone), 157.60 (C=O uriedo group). ESI-MS (m/z): 506 [M+], 507 [M + 1], 505 [M-1], 503 [M-2]. CHNS Analysis for C27H30N4O4S Found (Calculated): C: 65.91 (65.89), H: 5.85 (5.84), N: 11.9 (11.91), O: 10.88 (10.89), S: 5.54 (5.55).
1–(4-Methylcyclohexyl)-3–(4-{3-[4–(2-methylpropyl) phenyl]-6-oxopyridazin-1-yl} benzenesulphonyl) urea (XVII)
White crystal, yield = 57%, m.p. 251–252 °C, Rf = 0.63 (TEF). IR νmax (KBr, in cm− 1): 3357, 3021 and 2913 (NH-CO-NH), 1713 and 1508 (C=O) of urea, 1597 (C=N), 1331 and 1142 (SO2N). 1H NMR (300 MHz, DMSO-d6, δ): 0.89–0.98 (9H, m, cyclohexyl-CH3, Ph-CH2-CH-(CH3)2), 1.05–1.97 (9H, m, cyclohexyl ring), 3.26 (1H, m, proton at C-1 of cyclohexyl), 5.50 (1H, d, J = 5.4 Hz, -NH-cyclohexyl ring), 7.25 (1H, d, J = 9.9 Hz, H-5), 7.35 (2H, d, J = 7.5 Hz, H-3′, H-5′), 7.90 (2H, d, J = 6.9 Hz, H-2′, H-6′), 7.96 (2H, d, J = 7.8 Hz, H-3″, H-5″), 8.02 (2H, d, J = 8.4 Hz, H-2″, H-6″), 8.022 (1H, d, J = 9.6 Hz, H-4), 8.34 (1H, s, SO2-NH-C=O). 13C NMR (75 MHz, DMSO-d6, δ, ppm): 50.13 (CH-NH), 126.74 (C-4 pyridazinone), 145.35 (C-5 pyridazinone), 152.20 (C=O pyridazinone), 155.44 (C=N pyridazinone), 158.93 (C=O uriedo group). ESI-MS (m/z): 522 [M+], 523 [M + 1], 521 [M-1], 520 [M-2]. CHNS Analysis for C28H34N4O4S Found (Calculated): C: 64.43 (64.41), H: 6.5 (6.49), N: 10.82 (10.81), O: 12.15 (12.17), S: 6.10 (6.12).
3-{4-[3–(3,4-Difluorophenyl)-6-oxopyridazin-1-yl] benzenesulphonyl}-1–(4-methylcyclohexyl) urea (XVIII)
Light yellow crystals, yield = 61%, m.p. 212–213 °C, Rf = 0.62 (TEF). IR νmax (KBr, in cm− 1): 3341, 3027 and 2935 (NH-CO-NH), 1690 and 1510 (C=O) of urea, 1597 (C=N), 1329 and 1147 (SO2N). 1H NMR (300 MHz, DMSO-d6, δ): 0.90 (3H, d, J = 6.3 Hz, -cyclohexyl-CH3), 1.008–1.89 (9H, m, cyclohexyl ring protons), 3.31 (1H, m, proton at C-1 of cyclohexyl), 5.54 (1H, d, J = 7.5 Hz, -NH-cyclohexyl ring),): 7.26–8.28 (10H, m, H-3′, H-5′, H-6′, H-4, H-5, H-2″, H-6″, H-3″, H-5″, SO2-NHC=O). 13C NMR (75 MHz, DMSO-d6, δ, ppm): 49.10 (CH-NH), 124.66 (C-4 pyridazinone), 144.73 (C-5 pyridazinone), 153.11 (C=O pyridazinone), 156.41 (C=N pyridazinone), 156.44 (C=O uriedo group).). ESI-MS (m/z): 502 [M+], 503 [M + 1], 501 [M-1], 500 [M-2]. CHNS Analysis for C20H18F2N4O4S Found (Calculated): C: 57.33 (57.35), H: 4.91 (4.89), F: 7.71 (7.73), N: 11.22 (11.21), O: 12.52 (12.54), S: 6.35 (6.37).
3-{4-[3–(4-Fluorophenyl)-6-oxopyridazin-1-yl] benzenesulphonyl}-1–(4 methylcyclohexyl) urea (XIX)
Yellow crystals, yield = 50%, m.p. 188–189 °C, Rf = 0.64 (TEF). IR νmax (KBr, in cm− 1): 3332, 3059 and 2981 (NH-CO-NH), 1715 and 1510 (C=O) of urea, 1603 (C=N), 1344 and 1162 (SO2N). 1H NMR (300 MHz, DMSO-d6, δ): 091 (3H, d, J = 6.6 Hz, -cyclohexyl- CH3), 0.95–2.15 (9H, m, cyclohexyl ring), 3.22 (1H, m, proton at C-1 of cyclohexyl), 7.30 (1H, d, J = 9.6 Hz, H-5), 7.42 (2H, d, J = 8.4 Hz, H-2′, H-6′), 7.97–8.07 (6H, m, H-3′, H-5′, H-2″, H-6″, H-3″, H-5″), 8.25 (1H, d, J = 9.9 Hz, H-4). 13C NMR (75 MHz, DMSO-d6, δ, ppm): 48.66 (CH-NH), 125.56 (C-4 pyridazinone), 145.63 (C-5 pyridazinone), 154.22 (C=O pyridazinone), 155.74 (C=O uriedo group), 156.32 (C=N pyridazinone). ESI (m/z): 484 [M+], 485 [M + 1], 483 [M-1], 482 [M-2]. CHNS Analysis for C24H25FN4O4S Found (Calculated): C: 59.47 (59.45), H: 5.28 (5.30), F: 3.88 (3.90), N: 11.62 (11.63), O: 13.21 (13.20), S: 6.58 (6.57).
1–(4-Methylcyclohexyl)-3-{4-[6-oxo-3–(5,6,7,8-tetrahydronaphthalen-2-yl) pyridazin-1-yl] benzenesulphonyl} urea (XX)
White crystals, yield = 45%, m.p. 240–241 °C, Rf = 0.67 (TEF). IR νmax (KBr, in cm − 1): 3362, 3078 and 3003 (NH-CO-NH), 1729 and 1525 (C=O) of urea, 1621 (C=N), 1350 and 1188 (SO2N). 1H NMR (300 MHz, DMSO-d6, δ): 0.89 (3H, d, J = 6.3 Hz, cyclohexyl-CH3), 1.006–2.11 (9H, m, cyclohexyl ring), 1.79 (4H, s, H-3″′, H-4″′), 2.98 (4H, s, H-2″′, H-5″′), 3.26 (1H, m, proton at C-1of cyclohexyl), 5.57 (1H, brs, -NH-cyclohexyl ring), 7.24 (2H, m, H-5, H-5′), 7.67–7.74 (2H, m, H-2′, H-6′), 7.79–7.99 (4H, m, H-2″, H-6″, H-3″, H-5″), 8.17–8.34 (2H, m, H-4, SO2-NH-C=O), 13C NMR (75 MHz, DMSO-d6, δ, ppm): 47.75 (CH-NH), 125.66 (C-4 pyridazinone), 145.77 (C-5 pyridazinone), 153.66 (C=O pyridazinone), 155.81 (C=O uriedo group), 156.32 (C=N pyridazinone). ESI-MS (m/z):520 [M+], 521 [M + 1], 519 [M-1], 518 [M-2]. CHNS Analysis for C28H32N4O4S Found (Calculated): C: 64.38 (64.40), H: 6.22 (6.20), N: 10.55 (10.56), O: 12.67 (12.68), S: 6.25 (6.24).
3-{4-[3–(4-Benzylphenyl)-6-oxopyridazin-1-yl] benzenesulphonyl}-1–(4-methylcyclohexyl) urea (XXI)
White crystals, yield = 68%, m.p. 239–240 °C, Rf = 0.7 (TEF). IR νmax (KBr, in cm− 1): 3347, 3053 and 3007 (NH-CO-NH), 1710 and 1529 (C=O) of urea, 1608 (C=N), 1361 and 1170 (SO2N). 1H NMR (300 MHz, DMSO-d6, δ): 0.84 (3H, d, J = 6.3 Hz, cyclohexyl-CH3), 0.94–1.90 (9H, m, cyclohexyl ring), 3.51 (1H, m, proton at C-1 of cyclohexyl), 3.99 (2H, s, Ph-CH2-Ph),): 5.48 (1H, d, J = 7.8 Hz, -NH-cyclohexyl ring), 5.71 (1H, brs, SO2-NH-C=O), 7.16–7.38 (8H, m, H-5, H-2″′, H-3″′, H-4″′, H-5″′, H-6″′, H-3′, H-5′), 7.63–8.09 (6H, m, H-2″, H-6″, H-3″, H-5″, H-2′, H-6′), 8.12 (1H, d, J = 7.2 Hz, H-4). 13C NMR (75 MHz, DMSO-d6, δ, ppm): 47.80 (CH-NH), 125.44 (C-4 pyridazinone), 145.22 (C-5 pyridazinone), 155.33 (C=O uriedo group), 154.21 (C=O pyridazinone), 155.11 (C=N pyridazinone). ESI-MS (m/z): 556 [M+], 557 [M + 1], 555 [M-1], 554 [M-2]. CHNS Analysis for C31H32N4O4S Found (Calculated): C: 66.68 (66.69), H: 5.99 (5.97), N: 10.25 (10.26), O: 11.32 (11.31), S: 5.80 (5.79).
1–(4-Methylcyclohexyl)-3-{4-[6-oxo-3–(4-propylphenyl)pyridazin-1-yl] benzenesulphonyl} urea (XXII)
Light yellow crystals, yield = 57%, m.p. 246–247 °C, Rf = 0.66 (TEF). IR νmax (KBr, in cm− 1): 3335, 3063 and 2968 (NH-CO-NH), 1696 and 1507 (C=O) of urea, 1581 (C=N), 1332 and 1125 (SO2N). 1H NMR (300 MHz, DMSO-d6, δ): 0.825 (3H, d, J = 5.4 Hz, cyclohexyl-CH3), 1.03–1.90 (9H, m, cyclohexyl ring), 3.20 (1H, m, proton at C-1 of cyclohexyl): 5.44 (1H, d, J = 8.1 Hz, -NH-cyclohexyl), 5.70 (1H, brs, SO2-NH-C=O), 7.16–8.17 (10H, m, H-5, H-3′, H-5′, H-2′, H-6′, H-3″, H-5″, H-2″, H-6″, H-4). 13C NMR (75 MHz, DMSO-d6, δ, ppm): 50.10 (CH-NH), 125.44 (C-4 pyridazinone), 145.22 (C-5 pyridazinone), 154.18 (C=O pyridazinone), 155.12 (C=N pyridazinone), 155.33 (C=O uriedo group). ESI-MS (m/z): 508 [M+], 509 [M + 1], 507 [M-1], 506 [M-2]. CHNS Analysis for C27H32N4O4S Found (Calculated): C: 63.36 (63.37), H: 6.74 (6.73), N: 10.97 (10.96), O: 12.58 (12.59), S: 6.35 (6.37).
3-{4-[3–(4-Ethoxyphenyl)-6-oxopyridazin-1-yl] benzenesulphonyl}-1–(4-methylcyclohexyl) urea (XXIII)
Yellow crystals, yield = 49%, m.p. 241–242 °C, Rf = 0.71 (TEF). IR νmax (KBr, in cm −1): 3320, 3092 and 2986 (NH-CO-NH), 1721 and 1530 (C=O) of urea, 1590 (C=N), 1333 and 1146 (SO2N). 1H NMR (300 MHz, DMSO-d6, δ): 0.96 (3H, d, J = 8.7 Hz, cyclohexyl-CH3), 1.002–1.24 (5H, m, cyclohexyl ring), 1.30 (3H, t, Ph-O-CH2-CH3), 1.37–1.73 (4H, m, cyclohexyl ring), 3.18 (1H, m, proton at C-1 of cyclohexyl), 4.09 (2H, q, Ph-O-CH2-CH3), 6.38 (1H, d, J = 5.1 Hz, -NH-cyclohexyl), 7.04 (2H, d, J = 6.6 Hz, H-3′, H-5′), 7.21 (1H, d, J = 7.2 Hz, H-5), 7.90 (2H, d, J = 6.6 Hz, H-2′, H-6′), 7.96 (2H, d, J = 6.6 Hz, H-3″, H-5″), 8.03 (2H, d, J = 6.6 Hz, H-2″, H-6″), 8.16 (1H, d, J = 7.5 Hz, H-4), 10.51 (1H, s, SO2-NH-C=O). 13C NMR (75 MHz, DMSO-d6, δ, ppm): 46.91 (CH-NH), 125.33 (C-4 pyridazinone), 145.80 (C-5 pyridazinone), 153.42 (C=O pyridazinone), 155.35 (C=N pyridazinone), 156.81 (C=O uriedo group). ESI-MS (m/z):510 [M+], 511 [M + 1], 509 [M-1], 508 [M-2]. CHNS Analysis for C26H30N4O5S Found (Calculated): C: 61.35 (61.33), H: 5.80 (5.81), N: 10.86 (10.87), O: 15.75 (15.77), S: 6.25 (6.27).
1–(4-Methylcyclohexyl)-3–(4-{6-oxo-3-[4–(2-phenylethyl)phenyl]pyridazin-1-yl} benzenesulphonyl) urea (XXIV)
White crystals, yield = 63%, m.p. 287–288 °C, Rf = 0.61 (TEF). IR νmax (KBr, in cm− 1): 3323, 3084 and 2987 (NH-CO-NH), 1733 and 1528 (C=O) of urea, 1589 (C=N), 1335 and 1151 (SO2N). 1H NMR (300 MHz, DMSO-d6, δ): 0.84 (3H, d, J = 6 Hz, cyclohexyl-CH3), 0.89–1.24 (9H, m, cyclohexyl ring), 2.92 (4H, s, Ph-CH2-CH2-Ph), 3.12 (1H, m, proton at C-1 of cyclohexyl), 7.15–8.16 (17H, m, H-2′, H-3′, H-5′, H-6′, H-2″, H-3″, H-5″, H-6″, H-2″′, H-3″′, H-4″′, H-5″′, H-6″′, H-4, H-5, SO2-NH-C=O, -NH-cyclohexyl ring), 0.84 (3H, d, J = 6 Hz, cyclohexyl-CH3). 13C NMR (75 MHz, DMSO-d6, δ, ppm): 44.88 (CH-NH), 125.66 (C-4 pyridazinone), 145.17 (C-5 pyridazinone), 153.17 (C=O pyridazinone), 155.22 (C=N pyridazinone), 156.55 (C=O uriedo group). ESI-MS (m/z): 570 [M+], 571 [M + 1], 569 [M-1], 568 [M-2]. CHNS Analysis for C32H34N4O4S Found (Calculated): C: 67.41 (67.43), H: 5.81 (5.80), N: 9.81 (9.82), O: 11.30 (11.29), S: 5.70 (5.71).
1–(4-Methylcyclohexyl)-3-{4-[6-oxo-3–(4-phenoxyphenyl)pyridazin-1-yl]benzenesulphonyl}urea (XXV)
White crystals, yield = 51%, m.p. 236–237 °C, Rf = 0.72 (TEF). IR νmax (KBr, in cm− 1): 3340, 3062 and 2989 (NH-CO-NH), 1738 and 1545 (C=O) of urea, 1597 (C=N), 1358 and 1167 (SO2N). 1H NMR (300 MHz, DMSO-d6, δ): 0.84 (1H, d, J = 6.3 Hz, cyclohexyl-CH3), 1.03–1.90 (9H, m, cyclohexyl ring), 3.27 (1H, m, proton at C-1 of cyclohexyl), 5.44 (1H, brs, -NH-cyclohexyl ring), 5.50 (1H, brs, SO2-NH-C=O), 7.07–7.45 (10H, m, H-2′, H-3′, H-5′, H-6′, H-2″′, H-3″′, H-4″′, H-5″′, H-6′″, H-5), 7.88 (2H, d, J = 8.1 Hz, H-3″, H-5″), 7.95 (2H, d, J = 8.4 Hz, H-2″, H-6″), 8.11 (1H, d, J = 9.3 Hz, H-4). 13C NMR (75 MHz, DMSO-d6, δ, ppm): 46.20 (CH-NH), 126.12 (C-4 pyridazinone), 146.40 (C-5 pyridazinone), 155.11 (C=O pyridazinone), 155.16 (C=N pyridazinone), 155.52 (C=O uriedo group). ESI-MS (m/z): 558 [M+], 559 [M + 1], 557 [M-1], 556 [M-2]. CHNS Analysis for C30H30N4O5S Found (Calculated): C: 64.55 (64.56), H: 5.26 (5.28), N: 10.15 (10.13), O: 14.37 (14.36), S: 5.75 (5.74).
3-{4-[3–(4-Cyclohexylphenyl)-6-oxopyridazin-1-yl] benzenesulphonyl}-1–(4-methylcyclohexyl)urea (XXVI)
Yellow crystals, yield = 60%, m.p. 280–281 °C, Rf = 0.73 (TEF). IR νmax (KBr, in cm− 1): 3322, 3065 and 2971 (NH-CO-NH), 1736 and 1528 (C=O) of urea, 1601 (C=N), 1347 and 1161 (SO2N). 1H NMR (300 MHz, DMSO-d6, δ): 0.90 (3H, d, J = 6.3 Hz, cyclohexyl-CH3), 0.96–2.08 (20H, m, cyclohexyl rings), 3.23 (1H, m, proton at C-1 of cyclohexyl),): 5.52 (1H, d, J = 8.1 Hz, -NH-cyclohexyl ring), 6.13 (1H, brs, SO2-NH-C=O), 7.25 (1H, d, J = 9.6 Hz, H-5), 7.40 (2H, d, J = 8.4 Hz, H-3′, H-5′), 7.90 (2H, d, J = 7.8 Hz, H-3″, H-5″), 8.01 (2H, d, J = 8.1 Hz, H-2′, H-6′), 8.19 (1H, d, J = 9.6 Hz, H-4). 13C NMR (75 MHz, DMSO-d6, δ, ppm): 47.11 (CH-NH), 125.32 (C-4 pyridazinone), 145.43 (C-5 pyridazinone), 156.12 (C=O uriedo group), 156.20 (C=N pyridazinone), 156.71 (C=O pyridazinone). ESI-MS (m/z): 548 [M+], 549 [M + 1], 547 [M-1], 546 [M-2]. CHNS Analysis for C30H36N4O4S Found (Calculated): C: 65.81 (65.83), H: 6.53 (6.54), N: 10.30 (10.29), O: 11.59 (11.6), S: 5.80 (5.81).
3-{4-[3–(4-Bromophenyl)-6-oxopyridazin-1-yl] benzenesulphonyl}-1–(4-methylcyclohexyl)urea (XXVII)
Light yellow crystals, yield = 55%, m.p. 231–232 °C, Rf = 0.62 (TEF). IR νmax (KBr, in cm−1): 3338, 3100 and 2982 (NH-CO-NH), 1740 and 1533 (C=O) of urea, 1608 (C=N), 1339 and 1142 (SO2N). 1H NMR (300 MHz, DMSO-d6, δ): 1.17 (3H, d, J = 8.1 Hz, cyclohexyl-CH3), 1.22–1.92 (9H, m, cyclohexyl ring), 3.21 (1H, m, proton at C-1 of cyclohexyl), 6.50 (1H, brs, -NH-cyclohexyl ring), 7.45 (1H, d, J = 7.5 Hz, H-5), 7.91 (2H, d, J = 6.3 Hz, H-2′, H-6′), 8.12 (2H, d, J = 6.3 Hz, H-3′, H-5′), 8.15 (2H, d, J = 6.6 Hz, H-3″, H-5″), 8.23 (2H, d, J = 6.3 Hz, H-2″, H-6″), 8.39 (2H, d, J = 6.3 Hz, H-2″, H-6″), 8.39 (1H, d, J = 7.2 Hz, H-4), 10.72 (1H, s, SO2-NH-C=O). 13C NMR (75 MHz, DMSO-d6, δ, ppm): 49.10 (CH-NH), 125.70 (C-4 pyridazinone), 145.77 (C-5 pyridazinone), 154.35 (C=O pyridazinone), 155.66 (C=N pyridazinone), 156.35 (C=O uriedo group). ESI-MS (m/z):546 [M + 2], 544 [M+], 545 [M + 1]. CHNS Analysis for C24H25BrN4O4S Found (Calculated): C: 52.88 (52.86), H:4.7 (4.71), Br: 14.61 (14.60), N: 10.28 (10.29), O: 11.71 (11.73), S: 5.87 (5.88).
3-{4-[3–(4-Iodophenyl)-6-oxopyridazin-1-yl] benzenesulphonyl}-1–(4-methylcyclohexyl) urea (XXVIII)
Yellow crystals, yield = 47%, m.p. 253–2545 °C, Rf = 0.63 (TEF). IR νmax (KBr, in cm−1): 3312, 3182 and 2997 (NH-CO-NH), 1752 and 1545 (C=O) of urea, 1617 (C=N), 1354 and 1166 (SO2N). 1H NMR (300 MHz, DMSO-d6, δ): 0.90 (3H, d, J = 6.3 Hz, cyclohexyl-CH3), 0.96–1.91 (9H, m, cyclohexyl ring), 3.2 (1H, m, proton at C-1 of cyclohexyl), 5.52 (1H, d, J = 7.8 Hz, -NH-cyclohexyl ring), 6.03 (1H, brs, SO2-NH-C=O), 7.28 (1H, d, J = 6.9 Hz, H-5), 7.62 (2H, d, J = 9.6 Hz, H-2′, H-6′), 7.80 (2H, d, J = 8.4 Hz, H-3′, H-5′), 7.87–8.05 (4H, m, H-2″, H-3″, H-5″, H-6″), 8.23 (1H, d, J = 5.4 Hz, H-4). 13C NMR (75 MHz, DMSO-d6, δ, ppm): 50.13 (CH-NH), 125.66 (C-4 pyridazinone), 146.55 (C-5 pyridazinone), 154.17 (C=O pyridazinone), 155.45 (C=O uriedo group), 156.22 (C=N pyridazinone). ESI-MS (m/z): 592 [M+], 593 [M + 1], 591 [M-1], 590 [M-2]. CHNS Analysis for C24H25IN4O4S Found (Calculated): C: 48.65 (48.63), H: 4.27 (4.26), I: 21.57 (21.55), N: 9.43 (9.41), O: 10.76 (10.75), S: 5.39 (5.41).
3-{4-[3–(4-Hexylphenyl)-6-oxopyridazin-1-yl] benzenesulphonyl}-1–(4-methylcyclohexyl) urea (XIXX)
White crystals, yield = 53%, m.p. 281–282 °C, Rf = 0.65 (TEF). IR νmax (KBr, in cm −1): 3325, 3101 and 2989 (NH-CO-NH), 1744 and 1538 (C=O) of urea, 1609 (C=N), 1341 and 1150 (SO2N), 1H NMR (300 MHz, DMSO-d6, δ): 0.80 (3H, t, -Ph-(CH2)5-CH3), 0.90 (3H, d, J = 6.3 Hz, cyclohexyl ring -CH3), 0.96–1.33 (9H, m, cyclohexyl ring), 1.33 (6H, s, -Ph-CH2-CH2-(CH2)3-CH3), 1.74 (2H, q, -Ph-CH2-CH2-(CH2)3-CH3), 2.68 (2H, t, -Ph-CH2-(CH2)4-CH3), 3.24 (1H, m, proton at C-1 of cyclohexyl), 5.51 (1H, d, J = 7.8 Hz, -NH-cyclohexyl ring), 6.50 (1H, brs, SO2-NH-C=O), 7.25 (1H, d, J = 9.6 Hz, H-5), 7.37 (2H, d, J = 8.4 Hz, H-3′, H-5′), 7.88–8.05 (6H, m, H-2′, H-6′, H-3″, H-5″, H-2″, H-6″), 8.18 (1H, d, J = 9.6 Hz, H-4). 13C NMR (75 MHz, DMSO-d6, δ, ppm): 48.35 (CH-NH), 125.64 (C-4 pyridazinone), 146.22 (C-5 pyridazinone), 155.25 (C=O uriedo group), 155.30 (C=N pyridazinone), 156.80 (C=O pyridazinone). ESI-MS (m/z): 550 [M+], 551 [M + 1], 549 [M-1], 548 [M-2]. CHNS Analysis for C30H38N4O4S Found (Calculated): C: 65.73 (65.75), H: 6.97 (6.96), N: 10.13 (10.11), O: 11.51 (11.50), S: 5.69 (5.71).
3-{4-[3–(4-Isopropylphenyl)-6-oxopyridazin-1-yl]benzenesulphonyl}-1–(4-methylcyclohexyl)urea (XXX)
White crystals, yield = 57%, m.p. 248–249 °C, Rf = 0.65 (TEF). IR νmax (KBr, in cm− 1): 3364, 3056 and 2978 (NH-CO-NH), 1729 and 1523(C=O) of urea, 1607 (C=N), 1343 and 1157 (SO2N). 1H NMR (300 MHz, DMSO-d6, δ): 0.79 (3H, d, J = 4.8 Hz, cyclohexyl ring -CH3), 0.86–1.17 (9H, m, cyclohexyl ring), 1.23 (6H, d, J = 6 Hz, Ph-CH-(CH3)2), 2.86 (1H, h, Ph-CH-(CH3)2), 3.19 (1H, m, proton at C-1 of cyclohexyl), 3.20–3.31 (1H, m, -NH-CH-(CH3)2), 4.47 (1H, d, J = 8.1 Hz, -NH-cyclohexyl ring), 7.08 (1H, d, J = 9.9 Hz, H-5), 7.27 (2H, d, J = 8.4 Hz, H-3′, H-5′), 7.52 (1H, s, SO2-NH-C=O), 7.75 (2H, d, J = 8.4 Hz, H-2′, H-6′), 7.86 (2H, d, J = 6.9 Hz, H-3″, H-5″), 7.93–8.04 (3H, m, H-2″, H-6″, H-4). 13C NMR (75 MHz, DMSO-d6, δ, ppm): 47.80 (CH-NH), 125.44 (C-4 pyridazinone), 145.22 (C-5 pyridazinone), 154.21 (C=O pyridazinone), 155.11 (C=N pyridazinone), 155.33 (C=O uriedo group). ESI-MS (m/z): 508 [M+], 509[M + 1], 507 [M-1], 506 [M-2].CHNS Analysis for C27H32N4O4S Found (Calculated): C: 63.60 (63.59), H: 6.38 (6.39), N: 11.10 (11.09), O: 12.62 (12.63), S: 6.35 (6.33).
Effect of synthesized compounds on oral glucose tolerance in normal rats
All the experiments were performed utilizing albino rats of Wistar strain (either sex) weighing 210 ± 10 g which were provided by the Animal House of Department of Pharmacology, State Company for Drug Industries and Medical Appliances, Samarra/Baghdad (1871/SDI) and were housed in the same location under standardized conditions. Animals were fed commercial chaw and had free access to water ad libitum. The experiments were performed in congruence with the instructions for the care and use of laboratory animals, ordained by the State Company for Drug Industries and Medical Appliances SDI. Animals delineated as fasted were bereaved of food for at least 16 h but permitted free access to water. Carboxymethyl cellulose CMC (1%w/v) in distilled water was used as vehicle for dosing in all the experiments. All treatments were given orally using gauge. Fasted rats were divided into groups of six animals each. Animals of group I were fed with the vehicle (CMC 1% w/v in distilled water) in a volume of 10 ml/kg, while animals of group II were given gliclazide 0.05 mM/kg suspended in the vehicle. Animals of the experimental group (I–XXX) were administered the suspension of the test compounds at a dosage of 0.05 mM/kg b.w. A glucose load (3 g/kg) was given to each animal exactly after 30-min post-administration of the respective treatments. Blood samples were collected from retro-orbital plexus just prior to and 60 min after the glucose loading and blood glucose levels were measured with an autoanalyser (Accu Check Active glucose kit)Citation27.
Docking
Docking studies were implemented utilizing Glide module of the Schrodinger-9 software on the aldose reductase (PDB id: 1EL3, 1USO, 2FZD and 2PDK). Receptor preparation was done using protein preparation wizard with defaults settings. The structures were sketched using maestro graphical user interface (GUI) and were energy minimized/cleaned up by Ligprep module of the same software using OPLS_2005 force field and proper protonation states were assigned with the ionizer subprogram at pH 7.2 ±0.2Citation28. A grid space of 20 Å around cocrystallized ligand was set for the docking calculations. Glide XP module was used for final docking studiesCitation29.
Preparation of rat lens aldose reductase
Crude AR was prepared from rat lens through reported methodCitation30. Institutional and national guidelines for the care and use of animals were followed and all experimental procedures involving animals were approved by the IAEC (institutional animal ethical committee) of the National Institute of Nutrition. Lenses were homogenized in nine volumes of 100 mM potassium phosphate buffer, pH 6.2. The homogenate was centrifuged at 15 000 × g for 30 min at 4 °C and the resulting supernatant was used as the source of ALR2.
Aldose reductase activity assay
Aldose reductase (AR) activity was assayed as described by SuryanarayanaCitation30. All the experiments were performed in triplicate. The assay mixture in 1 ml contained 50 mM potassium phosphate buffer, pH 6.2, 0.4 M lithium sulphate, 5 mM β-mercaptoethanol, 10 mM DL-glyceraldehyde, 0.1 mM NADPH and enzyme preparation. Appropriate blanks were employed for corrections. The assay mixture was incubated at 37 °C and the reaction was initiated by the addition of NADPH at 37 °C. The change in the absorbance at 340 nm due to NADPH oxidation was followed in a spectrophotometer.
Enzyme inhibition studies
For inhibition studies, a concentrated stock of pyridazinone-substituted benzenesulphonylurea derivatives (I–XXX) was prepared in DMSO. Aliquots drawn from a working solution were added to the enzyme assay mixture and incubated for 5 min before initiating the reaction by NADPH as described above. The per cent inhibition with test compound was calculated considering the enzyme activity in the absence of inhibitor as 100%. The concentration of each test sample giving 50% inhibition (IC50) was determined by non-linear regression analysis of log concentration of compound versus percentage inhibition.
Results and discussion
Synthesis of compounds
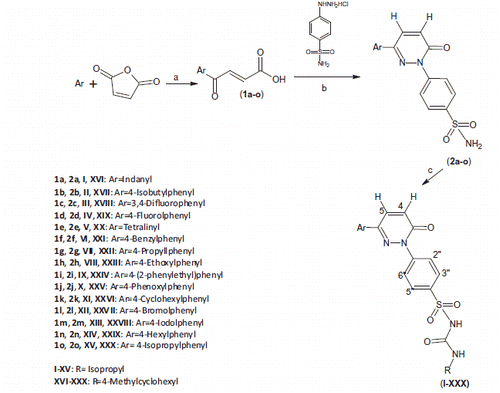
The synthetic route used to synthesize title compounds (I–XXX) is outlined in Scheme 1. The 4-aryl-pyridazinone-substituted benzenesulphonamides derivatives (2a–o) synthesized through reported methodCitation31 were converted to corresponding sulphonyl urea derivatives by refluxing with appropriate isocyanate in dry acetone containing K2CO3.
Scheme 1. Synthesis of pyridazinone-based benzenesulphonylurea derivatives. Reagents and conditions: (a) maleic anhydride, 1,1,2,2-tetrachloroethane, AlCl3; (b) Absolute alcohol, reflux 12–18 h; (c) Appropriate isocyanate, K2CO3, dry acetone, reflux 24–72 h.
The structure of synthesized pyridazinones-substituted benzenesulphonylurea derivatives (I–XXX) were determined on the basis of elemental analysis and various spectroscopic methods, such as IR, 1H NMR, 13C NMR and MS. Elemental analysis (C, H, N and S) data were within ±0.5% of the theoretical values. All the peaks corresponding to IR, 1H NMR and 13C NMR were observed at expected positions in respective spectra (for details see experimental section).
Oral glucose tolerance test in normal rats (OGGT)
In the current study, oral hypoglycaemic effects of 30 newly synthesized compounds (I–XXX) were assessed in glucose-fed hyperglycaemic normal rats at the dose of 0.05 mM/kg b.w. The clinically used sulphonyl urea drug gliclazide at the dose 0.05 mM/kg b.w. was used as positive control.
Quantitative glucose tolerance of each animal was calculated by area under curve (AUC) method by using prism software. Comparing AUC of experimental and control groups determined the percentage antihyperglycaemic activity. Statistical comparison was made by Dunnett’s test. Samples showing significant inhibition (p < 0.05) on postprandial hyperglycaemia were considered as active samples.
The results are summarized in . Twenty-three compounds (III–XI, XIV–XVII, XIX–XXIV, XXVI and XXVIII–XXX) out of 30 compounds showed more or comparable area under the curve (AUC) reduction percentage (ranging from 21.9% to 35.5%) as compared to the standard drug gliclazide (22.0%). The compound VIII is the most active compound and exhibited 35.5% AUC reduction.
Table 1. In vivo OGTT results for the compounds (I–XXX).
With regard to the SAR, it seems impossible to extract an obvious structure–activity relationship from the data shown in .
Docking
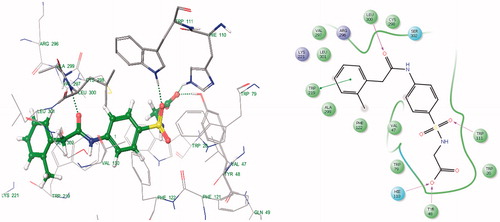
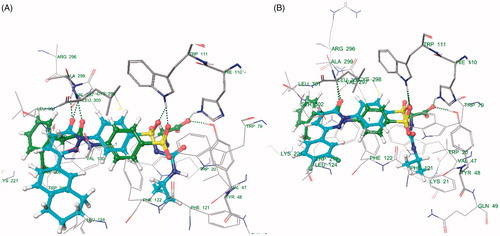
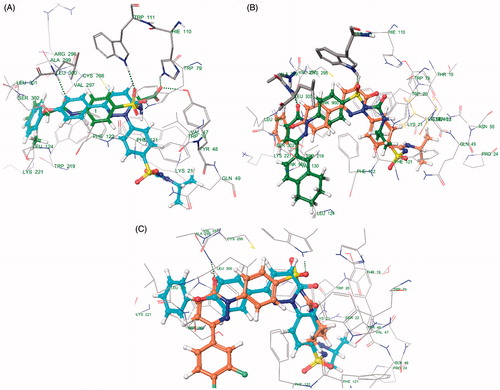
Docking studies of 30 synthesized compounds (I to XXX) were performed in order to get better comprehension of the aldose reductase inhibitory potency at molecular level and to shed light on the interactions in the active site of aldose reductase. Docking studies were performed using Glide module of the Schrodinger-9.4 software on the Aldose reductase (ALR) receptor (PDB id: 1EL3, 1USO, 2FZD, 2PDK). The most relevant poses were obtained with PDBID: 1EL3. However, the docking studies were performed with PDB ID: 1El3 because this structure is bound with IDD384, which is a sulphonamide derivative and is relevant with our study. The re-docking of reference ligand (IDD384) into active site of aldose reductase enzyme reveals that it occupies the same binding pocket with root mean square deviation (RMSD) of 0.61A which further validates the present docking protocol ().
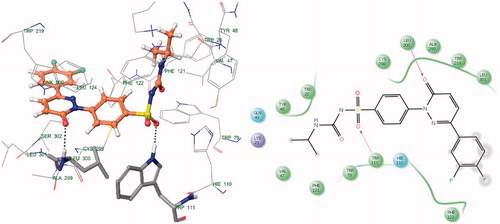
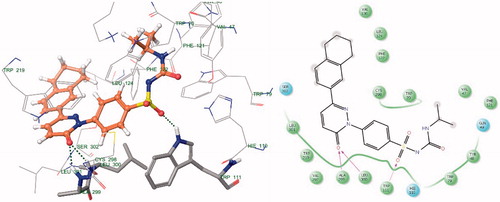
Docking results reveal that the compounds III and V bound tightly in the active site of aldose reductase (ALR2). These compounds (III and V) well occupied in the receptor cavity and forms hydrogen bonds and hydrophobic interactions ( and ). The sulfonamide group of III and V is anchored into the anion-binding site formed by Ala299, Leu300 and Trp111 and forms hydrogen-bonding with Trp111, which is key residue in binding and catalysisCitation32,Citation33. The 1,2,3,4-tetrahydronaphthalene ring core gets tightly trapped in the hydrophobic pocket formed by Phe122, Leu300, Leu301, Leu124 and Val130. Further docking analysis of compounds V and III with cocrystallized ligand (1DD384) by superimposition; it gives the same interaction pattern of binding with catalytic domain of ALR enzyme. The fact is also validated by where III and V were superimposed on cocrystal ligand. The binding pose of compound X () reveals why it is inactive. It does not fit well in the receptor binding site and does not show hydrogen bonding like III and V. This may be explained on the basis that the cyclohexyl attached to the sulphonamide moiety renders the configuration of molecule. Moreover, all other groups attached to the pyridazinone ring show different poses when compared to the III and V (). Therefore, it misses all the important interactions required for ALR inhibition. The fact is also validated by (), where X was superimposed on V and it is clear that compound V has more relaxed conformation that compound X.
Aldose reductase inhibition
Eighteen synthesized compounds were evaluated in vitro for their ability to inhibit activity of partially purified rat lens aldose reductase (AR). It has been shown that there is an approximately 85% sequence similarity between rat lens and human aldose reductase (ALR2), while the proposed active sites of both enzymes are identicalCitation32. The performed assay was based on a spectrometric measurement, which is proven to be a reliable methodCitation33, with DL-glyceraldehyde as the substrate and NADPH as the cofactor. Quercetin, a known ARI was used as a positive control. Results are presented in . Out of the tested 18 compounds, only ten compounds showed aldose reductase activity with IC50 ranging from 34 to 242 μM. Among these derivatives, compound IV (IC50 = 45 μM) and V (IC50 = 34 μM) were found comparable with the known ARI quercetin (IC50 = 41 μM). As regards to the structural activity relationship, no definite SAR could be extracted from .
Table 2. Aldose reductase (AR) inhibitory data.
Conclusion
In the present study, 30 new pyridazinone-substituted benzenesulphonylurea derivatives (I–XXX) were synthesized. The synthesized compounds are well supported by the spectroscopic data and elemental analysis. These compounds were screened for their antihyperglycaemic activity using in vivo OGTT assay (albino rats of Wistar strain). Eighteen compounds displaying good docking results were screened for their in vitro ability to inhibit rat lens aldose reductase. As a result, two compounds (IV and V) were identified possessing significant dual action (antihyperglycaemic and aldose reductase inhibition) and may be used as lead compounds for developing new drugs.
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
References
- Whiting DR, Guariguata L, Weil C, Shaw J. Global estimates of the prevalence of diabetes for 2011 and 2030. Diabetes Res Clin Pract 2011;94:311–21
- Anjana RM, Pradeepa R, Deepa M, et al. ICMR-INDIAB collaborative study group: prevalence of diabetes and prediabetes (impaired fasting glucose and/or impaired glucose tolerance) in urban and rural India: phase I results of the Indian Council of Medical Research-India diabetes (ICMR-INDIAB) study. Diabetologia 2011;54:3022–7
- Ferrannini E. Insulin resistance versus insulin deficiency in non-insulin-dependent diabetes mellitus: problems and prospects. Endocr Rev 1998;19:477–90
- Lillioja S, Mott DM, Spraul M, et al. Insulin resistance and insulin secretory dysfunction as precursors of non-insulin-dependent diabetes mellitus: prospective studies of Pima Indians. Engl J Med 1993;329:1988–92
- Sulman GI. Cellular mechanisms of insulin resistance. J Clin Invest 2000;106:171–6
- Kahn SE. The relative contributions of insulin resistance and beta cell dysfunction to the pathophysiology of type 2 diabetes. Diabetologia 2003;46:3–19
- LeRoith D. β-Cell dysfunction and insulin resistance in type 2 diabetes: role of metabolic and genetic abnormalities. Am J Med 2002;113:3–11
- Robertson RP, Harmon J, Tran PO. Beta-cell glucose toxicity, lipotoxicity, and chronic oxidative stress in type 2 diabetes. Diabetes 2004;53:5119–24
- Oates PJ. Polyol pathway and diabetic peripheral neuropathy. Int Rev Neurobiol 2002;50:325–92
- Alexiou P, Pegklidou K, Chatzopoulou M, et al. Aldose reductase enzyme and its implication to major health problems of the 21(st) century. Curr Med Chem 2009;16:734–52
- Jez JM, Bennett MJ, Schlegel BP, et al. Comparative anatomy of the aldo–keto reductase superfamily. Biochem J 1997;326:625–36
- Vander Jagt DL, Kolb NS, Vander Jagt TJ, et al. Substrate specificity of human aldose reductase: identification of 4-hydroxynonenal as an endogenous substrate. Biochim Biophys Acta 1995;1249:117–26
- Kumar PA, Reddy GB. Focus on molecules: aldose reductase. Exp Eye Res 2007;85:739–40
- Peterson MJ, Sarges R, Aldinger CE, McDonald DP. CP-45,634: a novel aldose reductase inhibitor that inhibits polyol pathway activity in diabetic and galactosemic rats. Metab Clin Exp 1979;28:456–61
- Kador FP, Kinoshita JH, Sharpless NE. Aldose reductase inhibitors: a potential new class of agents for the pharmacological control of certain diabetic complications. J Med Chem 1985;28:841–9
- Carvalho VF, Barreto EO, Serra MF, et al. Aldose reductase inhibitor zopolrestat restores allergic hyporesponsiveness in alloxan-diabetic rats. Eur J Pharmacol 2006;549:173–8
- (a) Settimo FD, Primofiore G, Motta CL, et al. Naphtho[1,2-d] isothiazole acetic acid derivatives as a novel class of selective aldose reductase inhibitors. J Med Chem 2005;48:6897–907 (b) El-Kabbani O, Carbone V, Darmanin C, et al. Structure of aldehyde reductase holoenzyme in complex with the potent aldose reductase inhibitor fidarestat: implications for inhibitor binding and selectivity. J Med Chem 2005;48:5536–42 (c) Schemmel KE, Padiyara RS, D’Souza JJ. Aldose reductase inhibitors in the treatment of diabetic peripheral neuropathy: a review. J Diabetes Complicat 2010;24:354–60
- Ramunno A, Cosconati S, Sartini S, et al. Progresses in the pursuit of aldose reductase inhibitors: the structure-based lead optimization step. Eur J Med Chem 2012;51:216–26
- Donkora IO, Abdel-Ghanyb YS, Kadorc PF, et al. Synthesis and biological activities of aldose reductase inhibitors bearing acyl benzenesulfonamides as carboxylic acid surrogates. Eur J Med Chem 1998;33:15–22
- Alexiou P, Demopoulos VJ. Novel inhibitors of rat lens aldose reductase: N-[[(substituted amino)phenyl]sulfonyl]glycines. J Med Chem 2010;53:7756–66
- C. Zhu Aldose reductase inhibitors as potential therapeutic drugs of diabetic complications (Chapter 2). In: Oluwafemi O. Oguntibeju, ed. Diabetes mellitus – insights and perspectives. Croatia – European Union: INTECH; 2013:17–46
- Kecskemeti V, Bagi Z, Posa I, et al. New trends in the development of oral antidiabetic drugs. Curr Med Chem 2002;9:53–71
- Costantino L, Rastelli G, Vescovini K, et al. Synthesis, activity, and molecular modeling of a new series of tricyclic pyridazinones as selective aldose reductase inhibitors. J Med Chem 1996;39:4396–405
- Costantino L, Rastelli G, Cignarella G, Barlocco D. Synthesis and aldose reductase inhibitory activity of a new series of benz[h]cinnolinone derivatives. Farmaco 2000;55:544–52
- Courdert P, Duroux E, Bastide P, Couquelet J. Synthesis and evaluation of the aldose reductase inhibitory activity of new diaryl pyridazine-3-ones. J Pharm Belg 1991;46:375–80
- Mylari BL, Armento SJ, Beebe DA, et al. A novel series of non-carboxylic acid, non-hydantoin inhibitors of aldose reductase with potent oral activity in diabetic rat models: 6-(5-chloro-3-methylbenzofuran-2-sulfonyl)-2H-pyridazin-3-one and congeners. J Med Chem 2005;48:6326–39
- Srivastava BK, Joharapurkar A, Raval S, et al. Diaryl dihydropyrazole-3-carboxamides with significant in vivo antiobesity activity related to CB1 receptor antagonism: synthesis, biological evaluation, and molecular modeling in the homology model. J Med Chem 2007;50:5951–66
- LigPrep, Version 2.3, New York: Schrödinger, LLC; 2009
- Glide, Version 5.5, New York: Schrödinger, LLC; 2009
- Suryanarayana P, Kumar PA, Saraswat M, et al. Inhibition of aldose reductase by tannoid principles of Emblica officinalis: implications for the prevention of sugar cataract. Mol Vis 2004;10:148–54
- Yaseen R, Ekinci D, Senturk M, et al. Pyridazinone substituted benzenesulfonamides as potent carbonic anhydrase inhibitors. Bioorg Med Chem Lett. [Epub ahead of print]. doi:10.1016/j.bmcl.2015.12.016
- Bohren KM, Grimshaw CE, Lai CJ, et al. Tyrosine-48 is the proton donor and histidine-110 directs substrate stereochemical selectivity in the reduction reaction of human aldose reductase: enzyme kinetics and crystal structure of the Y48H mutant enzyme. Biochemistry 1994;33:2021–32
- Grimshaw CE, Bohren KM, Lai CJ, Gabbay KH. Human aldose reductase: pK of tyrosine 48 reveals the preferred ionization state for catalysis and inhibition. Biochemistry 1995;34:14374–84