Abstract
3,6-Diazaphenothiazines were obtained in cyclization of 3-amino-3′-nitro-2,4′-dipyridinyl sulfide and the reaction of sodium 3-amino-2-pyridinethiolate with 4-chloro-3-nitropyridine followed by alkylation and heteroarylation. The thiazine ring formation ran via the Smiles rearrangement. The structure elucidation was based on 2D NMR and X-ray analysis of N-methylated product. 3,6-Diazaphenothiazines were investigated for antitumor activity using glioblastoma SNB-19, melanoma C-32 and breast cancer MCF-7 cells. 10H-3,6-diazaphenothiazine was 10 times more active (IC50 < 0.72 μg/mL) than cisplatin. Two diazaphenothiazines with the 2-pyrimidinyl and dimethylaminopropyl substituents were selectively active against MCF-7 and C-32 cells. The expressions of H3 (proliferation marker), TP53, CDKN1A (cell cycle regulators), BAX and BCL-2 (proapoptopic and antiapoptopic genes) were detected by RT-QPCR method. The expression analysis suggests the cell cycle arrest and the mitochondrial apoptosis pathway activation in MCF-7 and SNB-19 cells.
Introduction
Cancer has been one of the main causes of death worldwide for the last many years. The cancer therapy involves such a curative treatment as surgery, radiation, chemotherapy and biotherapyCitation1,Citation2. Although chemotherapy has been still improved in cancer therapy and the survival has been greatly increased, there is need to find new potent antitumor agents with better selectivity and minor or no side effects. Heterocyclic ring system plays an important role in the development of novel scaffold with improved pharmaceutical propertiesCitation3,Citation4.
Tricyclic phenothiazines (as 10-dialkylaminoalkyldibenzo-1,4-thiazines) are important class of fused heterocycles possessing important biological actions and interesting chemical properties. For many years, they have been recognized as drugs exhibiting neuroleptic, antihistaminic, antitussive and antiemetic activitiesCitation5. Classical phenothiazines are low toxic, inexpensive, easy to obtain, and some 10H-substituted compounds are even commercially available. The chemical modifications of the phenothiazine structures have been carried out mainly by introduction of new substituents at the thiazine nitrogen atom (using 10H-phenothiazines) and by the replacement of one or two benzene rings with the homoaromatic and heteroaromatic rings to form benzophenothiazines and azaphenothiazines. Such modifications are directed to alter a biological profile (its activity and potency). Many reports of last decade showed that both classical and modified phenothiazines exhibited very promising anticancer, antibacterial, antifungal, anti-inflammatory activities and reversal of multidrug resistance. This rich experimental material was summarized in review articles and chapters in monographsCitation6–14. Phenothiazines are also reported to exert a potential benefit in the treatment of Alzheimer’s, Creutzfeldt-Jakob’s and AIDS-associated diseasesCitation15–17.
The phenothiazine modification with the azine ring is most perspective as a new phenothiazine scaffold is formed (azaphenothiazine), which can be altered by introduction of new substituents at the thiazine nitrogen atom. The modification with the pyridine ring led to form pyridobenzothiazines (monoazaphenothiazines) and dipyridothiazines (diazaphenothiazines). One of pyridobenzothiazines is prothipendyl (1-azaphenothiazine,10-dimethylaminopropylpyrido[3,2-b][1,4]benzothiazine), a well-known drug possessing sedative and antiemetic propertiesCitation5. Recently, prothipendyl was found to exhibit antiviral activity against chikungunya virus (CHIKV), a mosquito-transmitting alphavirus causing CHIK feverCitation18,Citation19.
One of our strategies for phenothiazine modifications is based on the replacement of two benzene rings with the pyridine rings. We found new dipyridothiazines of the 1,6-, 1,8- and 2,7-diazaphenothiazine structures to exhibit promising anticancer activity against lung cancers HOP-62 and HOP-92, colon cancers COLO 205, HCT-116 and SW-948, renal cancers RXF393 and A498, and leukemia HL-60(TB) and L-1210Citation20,Citation21. 10H-2,7-diazaphenothiazine also shows immunosuppressant, inhibiting both humoral and cellular immune responses, and antioxidant propertiesCitation22–24.
In the literature, one can find the synthesis of two 7-substituted (Cl, OCH3) 10H-1-nitro-3,6-diazaphenothiazines in the reaction of 6-substituted 3-amino-2(1H)-pyridinethione with 3,5-dinitro-4-chloropyridine in methanolic potassium hydroxide. As the 1,4-thiazine ring formation can proceed directly as the Ullmann cyclization or indirectly through the Smiles rearrangement of the S–N type, the differentiation between possible structures, i.e. 7-substituted 10H-1-nitro-3,6-diazaphenothiazine and 10H-4-nitro-2,6-diazaphenothiazine, was based on the strong hydrogen bonding between the NH and NO2 groupsCitation25.
The aim of this paper is the synthesis of unknown 10H-3,6-diazaphenothiazine, transformation of this parent compound into 15 varied 10-substituted derivatives and determination of their anticancer activity against selected tumor cell lines. Since the synthesis can lead to one of the two 10H-diazaphenothiazines (2,6 or 3,6) and transformation into various N-substituted derivatives via alkylation and arylation of the thiazine nitrogen atom can be disturbed by the reaction of the pyridine nitrogen atom, the unquestionable elucidation of the product structure seems to be a crucial challenge.
Methods
Chemistry
Melting points were determined in open capillary tubes on a Boetius melting point apparatus (Stuart Equipment, Stone, UK) and are uncorrected. The 1H NMR, COSY, ROESY, HSQC, HMBC spectra were recorded on a Bruker Fourier 300 i AscendTM 600 spectrometer at 300 and 600 MHz (Bruker, Rheinstetten, Germany) in deuteriochloroform and dimethylsulfoxide-d6 with tetramethylsilane as the internal standard. The 13C NMR spectrum was recorded at 75 MHz. Electron impact mass spectra (EI MS) and chemical ionization mass spectra (CI MS) were run on a Finnigan MAT 95 spectrometer (Thermo Finnigan MAT, Bremen, Germany) at 70 eV. The thin layer chromatography was performed on silica gel 60 F254 (Merck 1.05735) with CHCl3-EtOH (5:1 and 10:1 v/v) and on aluminum oxide 60 F254 neutral (type E) (Merck 1.05581) with CHCl3-EtOH (10:1 v/v) as eluents.
Synthesis of sodium 3-amino-2-pyridinothiolate (1)
To a solution of 3,3′-dinitro-2,2′-dipyridinyl disulfide (310 mg, 1 mmol) in dry ethanol (30 mL), 2 tablets of NaBH4 (378 mg, 10 mmols) were added carefully and the mixture was refluxed for 2 h. After cooling, the solvent was evaporated in vacuo. The dry residue was recrystallized from ethanol yielding 230 mg (71%) of brown crystals of sodium 3-amino-2-pyridinethiolate (1) (230 mg, 71%), m.p. > 260 °C. After acidification of the aqueous solution of salt 1 with 10% solution of HCl, 3-aminopyridine-2(1H)-thione was obtained, m.p. 131–132 oC (Citation26; m.p. 131–132 oC).
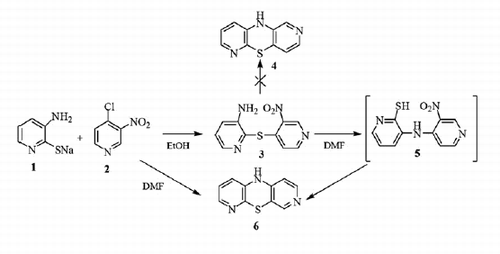
Synthesis of 3-amino-3′-nitro-2,4′-dipyridinyl sulfide (3)
To a solution of sodium 3-amino-2-pyridinethiolate (1) (148 mg, 1 mmol) in dry ethanol (10 mL), 2-chloro-3-nitropyridine (2) (158 mg, 1 mmol) was added. The mixture was stirred at room temperature for 3 h and next the resulting brown crystals were filtered off, washed with ethanol and air dried to give 3-amino-3′-nitro-2,4′-dipyridinyl sulfide (3). The dry product was recrystallized from ethanol yielding 218 mg (88%), m.p. 149–150 °C.
1H NMR (CDCl3) δ: 4.38 (broad s, 2H, NH2), 6.72(d, J = 5.7 Hz, 1H), 7.18 (dd, J =7.7 Hz, J = 1.3 Hz, 1H), 7.29 (dd, J = 7.7 Hz, J = 4.8 Hz, 1H), 8.18 (dd, J = 7.8 Hz, J = 1.3 Hz, 1H), 8.41 (d, J = 5.4 Hz, 1H), 9.39 (s, 1H). EI MS m/z: 248 (M, 27), 202 (M + 1-NO2 100). Anal. Calcd for: C10H8N4O2S, C 48.38, H 3.25, N 22.57. Found: C 48.49, H 3.32, N 22.47.
Synthesis of 10H-3,6-diazaphenothiazine (6)
From sodium 3-amino-2-pyridinethiolate (1) and 4-chloro-3-nitropyridine (2)
To a solution of sodium 3-amino-2-pyridinethiolate (1) (148 mg, 1 mmol) in dry DMF (10 mL), 4-chloro-3-nitropyridine (2) (158 mg, 1 mmol) was added. The reaction mixture was stirred at room temperature for 1 h and next was refluxed for 3 h. After cooling, the reaction mixture was evaporated in vacuo. The dry residue was dissolved in CHCl3 and purified by column chromatography (aluminum oxide, CHCl3) to give: 10H-3,6-diazaphenothiazine (6) (140 mg, 69%); m.p. 149–150 °C.
1H NMR (DMSOd6) δ: 6.48 (d, J = 5.4 Hz, 1H, H1), 6.83 (dd, J = 7.8 Hz, J = 1.2 Hz, 1H, H9), 6.95 (dd, J = 7.8 Hz, J = 4.8 Hz, 1H, H8), 7.79 (dd, J = 7.8 Hz, J = 1.2 Hz, 1H, H7), 7.86 (s, 1H, H4), 7.95 (d, J = 5.4 Hz, 1H, H2), 9.14 (broad s, 1H, NH). 13C NMR (DMSOd6) δ: 109.22 (C1), 114.05 (C4a), 121.08 (C9), 123.33 (C8), 136.30 (C9a), 140.52 (C7), 143.28 (C5a), 146.28 (C4), 146.46 (C2), 149.52 (C10a). EI MS m/z: 201 (M, 100). Anal Calcd for: C10H7N3S, C 59.68, H 3.51, N 20.88. Found: C 59.61, H 3.59, N 20.78.
In cyclization of 3-amino-3′-nitro-2,4′-dipyridinyl sulfide (3)
The brown solution of 3-amino-3′-nitro-2,4′-dipyridinyl sulfide (3) (124 mg, 0.5 mmol) in dry DMF (5 mL) was refluxed for 3 h. After cooling, the reaction mixture was evaporated in vacuo. The dry residue was dissolved in CHCl3 and purified by column chromatography (aluminum oxide, CHCl3) to give 10H-3,6-diazaphenothiazine (6) (90 mg, 90%).
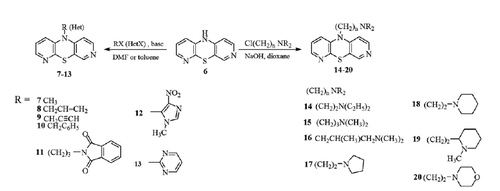
Synthesis of 10-substituted 3,6-diazaphenothiazines (7–10,12,13)
To a solution of 10H-3,6-diazaphenothiazine (6) (100 mg, 0.5 mmol) in dry DMF (5 mL), NaH (24 mg, 1 mmol, 60% NaH in mineral oil was washed out with hexane) was added. The reaction mixture was stirred at room temperature for 1 h and then alkyl or heteroaryl halide (methyl iodide, allyl bromide, benzyl chloride, 5-chloro-1-methyl-4-nitroimidazole, 2-chloropyrimidine, 1.5 mmol) was added and the stirring was continued for 24 h. The mixture was poured into water (15 mL), extracted with CHCl3 (3 × 10 mL) and dried using anhydrous Na2SO4. The obtained product was purified by column chromatography (aluminum oxide, CHCl3) to give:
10-Methyl-3,6-diazaphenothiazine (7) (87 mg, 79%); m.p. 158–159 °C: 1H NMR (CDCl3) δ: 3.30 (s, 3H, CH3), 6.60 (d, J = 5.4 Hz, 1H, H1), 6.98 (dd, J =7.8 Hz, J = 1.2 Hz, 1H, H9), 7.07 (dd, J = 7.8 Hz, J = 4.8 Hz, 1H, H8), 8.06 (dd, J = 4.8 Hz, J = 1.2 Hz, 1H, H7), 8.15 (s, 1H, H4), 8.27 (d, J = 5.4 Hz, 1H, H2). 13C NMR (CDCl3) δ: 34.36 (NCH3), 108.52 (C1), 118.61 (C4a), 120.37 (C9), 122.14 (C8), 139.05 (C9a), 143.52 (C7), 145.79 (C5a), 146.76 (C4), 149.15 (C2), 150.36 (C10a). EI MS m/z: 215 (M, 100), 200 (M-CH3, 58). Anal Calcd for: C11H9N3S C 61.37, H 4.21, N 19.52. Found: C 61.26, H 4.27, N 19.41.
10-Allyl-3,6-diazaphenothiazine (8) (92 mg, 78%); an oil: 1H NMR (CDCl3) δ: 4.37 (m, 2H, N-CH2), 5.21 (m, 1H, =CH), 5.39 (m, 1H, =CH), 5.97 (m, 1H, CH), 6.59 (d, J = 5.4 Hz, 1H, H1), 6.97 (m, 2H, H9, H8), 8.02 (dd, 1H, J = 4.8 Hz, J = 1.2 Hz, H7), 8.06 (s, 1H, H4), 8.17 (d, J = 5.4 Hz, 1H, H2). EI MS m/z: 241 (M, 45), 39 (C3H3, 100). Anal Calcd for: C13H11N3S C 64.70, H 4.59, N 17.41. Found: 64.72, H 4.54, N 17.29.
10-Benzyl-3,6-diazaphenothiazine (10) (85 mg, 58%); an oil: 1H NMR (CDCl3) δ: 5.05 (s, 2H, CH2), 6.49 (d, J = 5.4 Hz, 1H, H1), 6.83 (dd, J =7.8 Hz, J = 1.2 Hz, 1H, H9), 7.07 (dd, J = 7.8 Hz, J = 4.8 Hz, 1H, H8), 7.39 (m, 5H, C6H5), 8.08 (m, 3H, H7, H4, H2). EI MS m/z: 291 (M, 20), 200 (M-CH2C6H5, 60), 91 (C6H5CH2, 100). Anal Calcd for: C17H13N3S C 70.08, H 4.50, N 14.42. Found: C 70.10, H 4.59, N 14.31.
10-(1'-Methyl-4′-nitro-5′-imidazolyl)-3,6-diazaphenothiazine (12) (100 mg, 69%); m.p. 82–83 °C: 1H NMR (DMSO-d6) δ: 3.65 (s, 3H, CH3), 6.03 (d, J = 5.4 Hz, 1H, H1), 6.85 (m, 2H, H9, H8), 7.49 (s, 1H), 7.86 (dd, 1H, J = 4.8 Hz, J = 1.2 Hz, 1H, H7), 8.13 (m, 2H, H2, H4). EI MS m/z: 326 (M, 45), 42 (C2H4N, 100). Anal Calcd for: C14H10N6SO2 C 51.53, H 3.09, N 25.75. Found: C 51.29, H 3.13, N 25.61.
10-(2′-Pyrimidinyl)-3,6-diazaphenothiazine (13) (119 mg, 85%); m.p. 199–200 °C: 1H NMR (DMSOd6) δ: 6.09 (d, J = 5.4 Hz, 1H, H1), 6.97 (m, 2H, H9, H8), 7.43 (t, J =4.5 Hz, 1H, H5′), 7.79 (dd, J = 4.8 Hz, J = 1.2 Hz, 1H, H7), 8.08 (s, 1H, H4), 8.34 (d, J = 5.4 Hz, 1H, H2) 8.84 (d, J = 4.5 Hz, 2H, H4′,H6′). EI MS m/z: 279 (M, 100), 200 (M-C4H3N2, 25). Anal Calcd for: C14H9N5S C 60.20, H 3.25, N 25.07. Found: C 60.31, H 3.29, N 24.95.
Synthesis of 10-propargyl-3,6-diazaphenothiazines (9)
To a suspension of 10H-3,6-diazaphenothiazine (6) (100 mg, 0.5 mmol) in dry DMF (10 mL), potassium tert-butoxide (80 mg, 0.72 mmol) was added. The mixture was stirred at room temperature for 1 h. Then the 80% solution of propargyl bromide (80 mg, 0.64 mmol) in dry toluene was added dropwise. The solution stirred at room temperature for 24 h and poured into water (20 mL), extracted with methylene chloride (20 mL), dried with anhydrous Na2SO4, evaporated to the brown oil. The residue was purified by column chromatography (silica gel, CHCl3) to yield 70 mg (61%) of 10-propargyl-3,6-diazaphenothiazine (9), m.p. 139–141 °C.
1H NMR (CDCl3): δ 2.31 (t, J = 2.4 Hz, 1H), 4.43 (d, J = 2.4 Hz, 2H), 6.95 (d, J = 5.7 Hz, 1H, H1), 7.09 (dd, J = 7.8 Hz, J = 4.8 Hz, 1H, H8), 7.35 (dd, J =7.8 Hz, J = 1.2 Hz, 1H, H9), 8.11 (dd, J = 4.8 Hz, J = 1.2 Hz, 1H, H7), 8.20 (s, 1H, H4), 8.32 (d, J = 5.7 Hz, 1H, H2). CI MS: 240 (M + 1, 100). Anal Calcd for: C13H9N3S C 65.25, H 3.79, N 17.56. Found: C 65.22, H 3.71, N 17.46.
Synthesis of 10-phthalimidopropyl-3,6-diazaphenothiazines (11)
To a stirred solution of 10H-3,6-diazaphenothiazine (6) (100 mg, 0.5 mmol) in dry toluene (20 mL), NaH (120 mg, 5 mmol, washed out with hexane) was added. The mixture was stirred at room temperature for 30 min, then refluxed for 1 h and a solution of N-(3-bromopropyl)phthalimide (405 mg, 1.5 mmol) in toluene (10 mL) was added. The mixture was refluxed for 48 h. After cooling, the resulted solid was filtered off, toluene was evaporated in vacuo and the residue was purified by column chromatography (aluminum oxide, CHCl3) to give 10-(3′-phthalimidopropyl)-3,6-diazaphenothiazine (11) (117 mg, 74%), m.p. 45–47 °C.
1H NMR (DMSOd6) δ: 2.11 (m, 2H, CH2), 3.58 (t, J = 6.1 Hz, 2H, NCH2), 3.77 (t, J = 6.0 Hz, 2H, NCH2), 5.92 (d, J = 7.2 Hz, 1H, H1), 6.42 (d, J = 1.5 Hz, 1H, H4), 6.68 (dd, J = 7.8 Hz, J = 4.7 Hz, 1H, H8), 6.81 (dd, J = 7.8 Hz, J = 1.5 Hz, 1H, H9), 6.85 (dd, J = 7.2 Hz, J = 1.5 Hz, 1H, H2), 7.69 (dd, J = 4,7 Hz, J = 1.5 Hz, 1H, H7), 7.71 (m, 2Hphthalimide), 7.86 (m, 2Hphthalimide). CI MS m/z: 389 (M + H, 100), 201 (M + 1-(CH2)3N(CO)2C6H4, 10). Anal Calcd for: C21H16N4O2S: C 64.93, H 4.15, N 14.42. Found: C 64.82, H 4.22, N 14.21.
Synthesis of 10-substituted 3,6-diazaphenothiazines (14–20)
To a solution of 10H-3,6-diazaphenothiazine (6) (100 mg, 0.5 mmol) in dry dioxane (10 mL), NaOH (200 mg, 5 mmol) was added. The mixture was refluxed for 2 h and hydrochloride of dialkylaminoalkyl chloride (2-diethylaminoethyl, 3-dimethylaminopropyl, 3-dimethylamino-2-methylpropyl) or hydrochloride of cycloaminoethyl chloride [1–(2-chloroethyl)pyrrolidine, 2–(2-chloroethyl)-1-methylpiperidine, 1–(2-chloroethyl)piperidine, 1–(2-chloroethyl)mor-pholine 1.5 mmol] was added. The reaction mixture was refluxed for 24 h. After cooling, dioxane was evaporated in vacuo and residue was dissolved in CHCl3 (10 mL). The solution was washed with water (10 mL), dried with anhydrous Na2SO4 and evaporated in vacuo. The obtained product was purified by column chromatography (aluminum oxide, CH2Cl2) to give:
10-(2′-Diethylaminoethyl)-3,6-diazaphenothiazine (14) (102 mg, 71%); an oil: 1H NMR: δ 1.10 (t, J = 7.2 Hz, 6H, 2CH3), 2.65 (q, J = 7.2 Hz, 4H, 2CH2), 2.82 (t, J = 7.2 Hz, 2H, CH2), 3.94 (t, J = 7.2 Hz, 2H, CH2), 6.73 (d, J = 5.4 Hz, 1H, H1), 7.03 (m, 2H, H9, H8), 8.02 (dd, 1H, J = 4.8 Hz, J = 1.2 Hz, H7), 8.10 (s, 1H, H4), 8.24 (d, J = 5.4 Hz, 1H, H2). CI MS m/z: 301 (M + 1, 100), Anal Calcd for: C16H20N4S C 63.97; H 6.71; N 18.65. Found: C 63.92; H 6.78; N 18.50.
10-(3′-Dimethylaminopropyl)-3,6-diazaphenothiazine (15) (100 mg, 70%); an oil: 1H NMR: δ 1.83 (m, 2H, CH2), 2.21 (s, 6H, 2CH3), 2.37 (t, J = 7.5 Hz, 2H, NCH2), 3.79 (t, J = 7.5 Hz, 2H, NCH2), 6.64 (d, J = 5.4 Hz, 1H, H1), 7.01 (m, 2H, H9, H8), 7.97 (dd, J = 4.8 Hz, J = 1.2 Hz, 1H, H7), 8.06 (s, 1H, H4), 8.17 (d, J = 5.4 Hz, 1H, H2).CI MS m/z: 287 (M + 1, 100), 202 (M + 1-C3H6NC2H6, 15). Anal Calcd for C15H18N4S C 62.91; H 6.33; N 19.56. Found: C 62.99; H 6.27; N 19.45.
10-(3′-Dimethylamino-2′-methylpropyl)-3,6-diazaphenothiazine (16) (120 mg, 82%); an oil: 1H NMR: δ 1.02 (d, J = 6.5 Hz, 3H, CH3), 2.29 (m, 9H, 2CH3, CH2, CH), 4.05 (m, 2H, CH2), 6.79 (d, J = 5.4 Hz, 1H, H1), 7.05 (m, 2H, H9, H8), 8.08 (dd, J = 4.8 Hz, J = 1.2 Hz, 1H, H7), 8.10 (s, 1H, H4), 8.26 (d, J = 5.4 Hz, 1H, H2). CI MS m/z: 301 (M + 1, 100). Anal Calcd for: C16H20N4S C 63.97; H 6.71; N 18.65. Found: C 63.87; H 6.79; N 18.52.
10-(2′-Pyrrolidinylethyl)-3,6-diazaphenothiazine (17) (110 mg, 74%); an oil: 1H NMR (CDCl3) δ: 1.85 (m, 4H, 2CH2), 2.65 (m, 4H, 2CH2), 2.86 (t, J = 7.5 Hz, 2H, CH2), 3.93 (t, J = 7.5 Hz, 2H, NCH2), 6.71 (d, J = 5.4 Hz, 1H, H1), 7.04 (m, 2H, H9, H8), 8.02 (dd, 1H, J = 4.8 Hz, J = 1.2 Hz, H7), 8.10 (s, 1H, H4), 8.23 (d, J = 5.4 Hz, 1H, H2). CI MS m/z: 299 (M + 1, 100). Anal Calcd for: C16H18N4S C 64.40; H 6.08; N 18.78. Found: C 64.25; H 6.09; N 18.65.
10-(2′-Piperydinylethyl)-3,6-diazaphenothiazine (18) (120 mg, 72%); an oil: 1H NMR (CDCl3) δ: 1.48 (m, 2H, CH2), 2.48 (m, 4H, 2CH2) 2.55 (m, 4H, 2CH2), 2.68 (t, J = 6.8 Hz, 2H, CH2), 3.87 (t, J = 6.8 Hz, 2H, NCH2), 6.72 (d, J = 5.4 Hz, 1H, H1), 7.03 (m, 2H, H9, H8), 8.01 (dd, J = 4.8 Hz, J = 1.2 Hz, 1H, H7), 8.08 (s, 1H, H4), 8.20 (d, J = 5.4 Hz, 1H, H2). CI MS m/z: 313 (M + 1, 100). Anal Calcd for: C17H20N4S: C 65.35; H 6.45; N 17.93. Found: C 65.22; H 6.49; N 17.80.
10-(1'-Methyl-2′-piperydinylethyl)-3,6-diazaphenothiazine (19) (0.119 g, 74%); an oil: 1H NMR (CDCl3) δ: 1.25–2.20 (m, 12H), 2.26 (s, 3H, NCH3), 2.87 (m, 1H, CH), 3.85 (m, 2H, NCH2), 6.63 (d, J = 5.4 Hz, 1H, H1), 7.01 (m, 2H, H9, H8), 8.02 (dd, J = 4.8 Hz, J = 1.2 Hz, 1H, H7), 8.11 (s, 1H, H4), 8.22 (d, J = 5.4 Hz, 1H, H2). CI MS m/z 327 (M + H, 100). Anal Calcd for: C18H22N4S C 66.22; H 6.79; N 17.16. Found: C 66.15; H 6.79; N 17.11.
10-(2′-Morpholinylethyl)-3,6-diazaphenothiazine (20) (0.112 g, 69%); an oil: 1H NMR (CDCl3) δ: 1.67 (m, 4H, 2CH2), 2.59 (m, 4H, 2CH2), 2.82 (t, J = 6.6 Hz, 2H, CH2), 4.22 (t, J = 6.6 Hz, 2H, NCH2) 6.73 (d, J = 5.4 Hz, 1H, H1), 7.01 (m, 2H, H9, H8), 8.04 (dd, J = 4.8 Hz, J = 1.2 Hz, 1H, H7), 8.10 (s, 1H, H4), 8.25 (d, J = 5.4 Hz, 1H, H2). CI MS m/z: 315 (M + 1, 40), 202 (M + 1-C2H4NOC4H8, 15), 114 (C2H4NC5H10, 100). Anal Calcd for: C16H18N4OS: C 61.12; H 5.77; N 17.82. Found: C 61.19; H 5.57; N 17.71.
Crystal data
C11H9N3S, M = 215.27, orange needle, 0.48 × 0.22 × 0.15 mm3, orthorhombic, space group P212121, V = 941.6(1) Å3, Z = 4, Dc = 1.519 g/cm3, F000 = 448, KappaApexII, Mo-Kα radiation, λ = 0.71073 Å, T = 100(2) K, 2θmax = 55.0°, 4688 reflections collected, 2123 unique (Rint = 0.030). The structure was solved and refined using the programs SHELXS-97Citation27 and SHELXL-2013Citation28, respectively. Final GooF = 1.11, R = 0.056, wR = 0.113, R indices based on 1907 reflections with I > 2σ(I) (refinement on F2), 137 parameters, 0 restraints. Lp and absorption corrections applied, μ = 0.307 mm − 1. Absolute structure parameter = 0.2(1)Citation29.
Cytotoxic and antiproliferative effects in vitro
Cell culture
Compounds were evaluated for their anticancer activity using three cultured cell lines: SNB-19 (human glioblastoma, DSMZ – German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany), C32 (human amelanotic melanoma, ATCC – American Type Culture Collection, Manassas, VA) and MCF-7 (human breast cancer, ATCC, Manassas, VA). The cultured cells were kept at 37 °C and 5% CO2. The cells were seeded (1 × 104 cells/well/100 μL DMEM supplemented with 10% FCS and streptomycin and penicillin) using 96-well plates (Corning).
Cell proliferation and viability
In recent years, tetrazolium salts have been described to be used for the measurement of cell proliferation and viability. The tetrazolium salts are cleaved to formazan by cellular enzymes. An expansion in the number of viable cells results in an increase in the overall activity of mitochondrial dehydrogenases in the sample. This augmentation in enzyme activity leads to an increase in the amount of formazan dye formed, which directly correlates to the number of metabolically active cells in the culture. The formazan dye produced by metabolically active cells is quantified by a scanning ELISA reader by measuring the absorbance of the dye solution at appropriate wavelengths (λ = 420–480 nm with a reference wavelength λ = 600 nm).
WST-1 assay
The WST-1 assay (Roche Diagnostics, Mannheim, Germany) was used to evaluate the effect of compounds on the number of cells in cultures, which has the cytotoxic effect of the tested compounds and their influence on the proliferation of cells. After exposure to tested compounds (at concentrations between 0 and 100 μg/mL) for 72 h, cells were incubated with WST-1 (10 μL) for 1 h, and the absorbance of the samples against a background control was read at 450 nm using with a reference wavelength λ = 600 nm a microplate reader. Results are expressed as means of at least two independent experiments performed in triplicate.
The RT-QPCR method
Genes transcriptional activity (H3, TP53, CDKN1A, BCL-2, BAX) was evaluated by real time RT-QPCR method with OPTICON TM DNA Engine (MJ Research, Watertown, NY) and QuantTect® SYBR® Green RT-PCR Kit (Quiagen, Valencia, CA). Cells were exposed to compounds 6 and 13 at concentration of 0.5 μg/mL for 24 h. The RNA extraction was made by using Quick-RNA™ Kit MiniPrep (ZYMO RESEARCH, Irvine, CA). Total RNA integrity was analyzed in 1.2% agarose electrophoresis with added ethidium bromide compound. The quantity and purity of extracted total RNA were determined by using spectrophotometric analysis with HP845 (Hewlett Packard, Waldbronn, Germany) spectrophotometer. The statistical analysis was performed using the Statistica 8.0 software (StatSoft, Tulsa, OK). All values were expressed as means ± SE.
Results and discussion
Chemistry
Synthesis and 2D NMR analysis
The synthetic routes to phenothiazines proceed mainly through the Smiles rearrangement depending on the substrate structures and reaction conditions. Very often the rearrangement was observed under basic conditions (sodium or potassium hydroxide in methanol or ethanol), rarely under neutral or acidic media. In some cases, the rearrangement is hard to monitor for the rearranged and non-rearranged products that can have the same structure or can differ very subtly (the place of a substituent or a nitrogen atom in the case of the azaphenothiazinesCitation30–33.
We started the azaphenothiazine synthesis with the cyclization of 3-amino-3′-nitro-2,4′-dipyridinyl sulfide (3) (obtained from sodium 3-amino-2-pyridinethiolate (1) and 4-chloro-3-nitropyridine (2) in ethanol) in refluxing DMF solution to give 90% yield of the product. The same product was obtained (in 69%) directly from sodium 3-amino-2-pyridinethiolate (1) and 4-chloro-3-nitropyridine (2) in refluxing DMF (Scheme 1). Those reactions were monitored by TLC analysis as the product chromatogram, unlike to the substrate chromatograms, showed characteristic for azaphenothiazines color changing during irradiation with UV light from pale yellow (6–11) and yellow-celadon (14–20) to beige, and yellow (12,13) to orange-red. All azaphenothiazine chromatograms gave yellow color after sprayed with the mixture of sulfuric acid-water-ethanol (1:1:8).
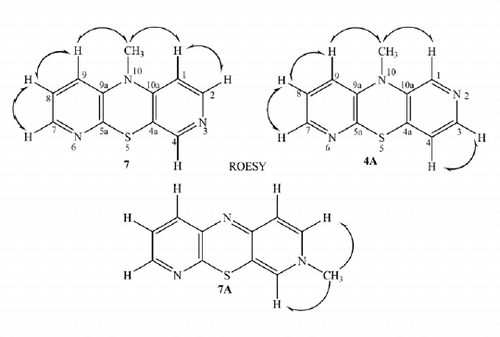
To distinguish the structure of the azaphenothiazine (4 or 6), the obtained compound was methylated with methyl iodide. The alkylation of azaphenothiazines was reported as the process occurring mainly at the thiazine nitrogen atom but one can find a few reports on alkylation at the azine nitrogen atom giving azaphenothiazinium salts and neutral N-alkylazaphenothiazinesCitation34–38. The reaction of the product with methyl iodide in dry DMF in the presence of sodium hydride led to the methylated product, which possessed only one methyl group (observed in the 1H NMR spectrum). The lack of the ammonium function could point at 10-methyl-3,6-diazaphenothiazine (7) (the Smiles product) or 10-methyl-2,6-diazaphenothiazine (4A) (the Ullmann product) or 3-methyl-3,6-diazaphenothiazine (7A) (an alternative alkylation product). To elucidate the product structure, we recorded 2D NMR (ROESY, COSY, HSQC and HMBC) spectra of the N-methyl product. The ROESY experiment with irradiation of the methyl protons at 3.30 ppm showed the proximity of the methyl group to the protons at 6.60 ppm (a doublet) and 6.98 ppm (a doublet of doublet), which excludes structures 4A and 7A (a singlet proton signal would be involved) and pointed at structure 7. The full proton signal assignment was achieved by study of other proton spatial proximity (ROESY) and 1H–1H connectivities in COSY spectrum. The signals at 6.60 and 6.98 ppm were assigned as H1 and H9 protons, respectively. The confirmation of the proton assignment came from the 13C NMR spectrum, which was solved by the use of HSQC and HMBC spectra indicating the 13C–1H relationship. The HSQC spectra showed which proton was bonded to the carbon atom (the C–H relationship through one bond, 1JC,H connectivity) and the HMBC spectra indicated the C–H relationship through three (predominantly), two and four (exceptionally) bonds (3JC,H, 2JC,H and 4JC,H connectivities). Selected spatial proton–proton proximity, proton–proton and proton–carbon connectivities for compound 7 are shown in Scheme 2. The all 1H–1H and 1H–13C connectivities are included in the “Supplementary Material” (Table 3, Scheme 4). The methylated product was identified as 10-methyl-3,6-diazaphenothiazine (10-methyldipyrido[2,3-b;4′,3′-e][1,4]thiazine) (7).
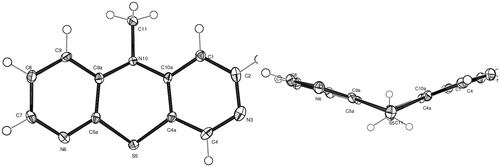
The X-ray structure analysis
As the indirect method of the structure elucidation based on the 1H NMR analysis is subtle and can lead to false conclusions, a single crystal X-ray diffraction study of the methyl derivative was carried out. The X-ray analysis fully confirmed the product structure concluded from the 1H NMR spectra as the 3,6-diazaphenothiazine system 7 and showed spatial arrangement in the molecule in a solid state (.
The tricyclic ring system is not planar but folded along the S–N axis with the butterfly angle of 134.7(2)° between two pyridine ring planes. The central thiazine ring is in boat conformation with the angle between two halves (SCCS) of 139.3(2)°. The methyl group is located in equatorial position with the S5···N10–C11 angle of 170.5(2)°. The important bond angles of the central ring C9a–N10–C10a and C4a–S5–C5a are 120.11(18)° and 98.1(2)°, respectively. The sum of three C–N10–C angles is 353.1° indicating the pyramidal configuration of the bonds around the central nitrogen atom. Whereas the N–C bond lengths in the pyridine rings are in the range of 1.328–1.354 Å, the N–C bond lengths in the thiazine ring are significantly longer 1.409–1.410 Å. The N–C bond to the methyl group was the longest one (1.460 Å) being a result of the sp3 hybridization of the carbon atom. The only known X-ray structures of dipyridothiazines, deposited in the Cambridge Structural DatabaseCitation39, concern 10-substituted 1,8- and 2,7-diazaphenothiazines, and their methyl iodides. Whereas 1,8- and 2,7-diazaphenothiazines are folded, the methyl iodide salts are folded or planar depending on the character of the nitrogen atom and number of alkylated atomsCitation32,Citation33,Citation40,Citation41.
In solid state, the molecules are arranged into layer-type stacks along the c-axis. The arrangement within a layer involves four weak C–H…N hydrogen bonding to neighboring molecules (Supplementary Material).
The transformation into 10-substituted derivatives
The parent product 6 was transformed into other derivatives possessing the allyl (8), propargyl (9), benzyl (10), phthalimidopropyl (11), imidazolyl (12), pyrimidinyl (13) and dialkylaminoalkyl (with cyclic and non-cyclic amine groups) (14–20) substituents in the reactions with appropriate halides in neutral solvents (DMF, toluene, dioxane) in the presence of base (NaH, NaOH, t-BuOK) (Scheme 3).
Anticancer activity
The anticancer activity of 3,6-diazaphenothiazines (6–20) was investigated in vitro using cultured glioblastoma SNB-19, melanoma C-32 and breast cancer MCF-7 cell lines and cisplatin as a reference drug. Normal human fibroblast (HFF-1) cell line was used as a control. To compare the influence of the nitrogen atoms in the azaphenothiazine system on the anticancer activity, the classical monoazaphenothiazine drug, prothipendyl, was also tested. The tested 3,6-diazaphenothiazines exhibited different activities against the cell lines. Two derivatives exhibited very strong activity with IC50 < 1 μg/mL ().
Table 1. The anticancer activity of 3,6-diazaphenothiazines.
The parent compound (10H-phenothiazine) (6) exerted very strong action against all tumor lines with IC50 = 0.46–0.72 μg/mL), being over 10 times more active than a reference drug – cisplatin. Similar strong and selective action was found for 10-pyrimidinyl derivative (13) against breast cancer MCF-7 cell line. 10-Dimethylaminopropyl derivative (15) exhibited as good activity against melanoma C-32 cell line as cisplatin and moderate activity against MCF-7 cell line. Other 3,6-diazaphenothiazines exhibited weaker activity with IC50 > 28 μg/mL. Prothipendyl turned out to be less active than 3, 6, and 8 derivatives of 3,6-diazaphenothiazine against investigated cell lines, respectively. All 3,6-diazaphenothiazines were found to be non-toxic (IC50 > 50 μg/mL) or almost non-toxic (IC50 38.7–43.9 μg/mL) against normal human fibroblast (HFF-1) cell line in comparison with toxic cisplatin (IC50 = 8.2 μg/mL).
Apoptosis assay
It has been known that the growth, division and eventual death of the cells in the body are processes that are controlled by hundreds of genes working togetherCitation42. The most active compounds 6 and 13 (with the hydrogen and pyrimidinyl substituents) were selected for efforts to understand the mechanism of anticancer action. To determine one of the antiproliferative mechanisms, the gene transcriptional activities of proliferation marker (H3) cell cycle regulator (TP53 and CDKNIA) and intracellular apoptosis pathway (BACL-2 and BAX) were analyzed with the use of RT-QPCR method. The results of analysis of H3, TP53, CDKN1A, BCL-2, BAX genes in MCF-7, SNB-19 and C-32 cells after 24 h of treatment are collected in .
Table 2. The influence of compounds 6 and 13 on the expression of genes encoding: H3, TP53, CDKN1A, BCL-2, BAX in: breast cancer cell line MCF-7, glioblastoma SNB-19 and melanoma C-32.
The gene encoding the histone H3 is considered as an indicator proliferation, which plays an important role in regulation of the expression of the genetic information encoded in DNACitation43. For both compounds, the number of transcripts significantly decreased, what could suggest the alteration in chromatin conformation.
The importance of P53 protein in cancer biology is undisputed. This protein is recognized as the guardian of the genome and is able to induce apoptosis. The P53 transcription factor is activated by potentially oncogenic stimuli, such as ribosomal stress, DNA damage, telomere erosion, nutrient deprivation and oncogene hyperactivation. This protein influences cell cycle arrest by changing the expression of CDKN1A gene encoding the P21 proteinCitation42,Citation44. Whereas compound 13 generated significant changes in the expression of TP53 gene (the increase in MCF-7 and C-32 cells but the decrease in SNB-19 cells); compound 6 did not show any alterations. Both compounds show significant increase of CDKN1A copies in MCF-7 and SNB-19 cells suggesting possibility of participation in cell cycle arrest and apoptosis.
The P53 protein can also stimulate the cell to changes in gene expression of proapoptopic BAX and antiapoptopic BCL-2 involved in mitochondrial pathway apoptosisCitation45–48. Both compounds reduced the expression of BCL-2 and BAX genes (with one exception for compound 13). Analysis of the gene expression ratio BAX/BCL-2 in the MCF-7 and SNB-19 cells showed activation of the mitochondrial apoptosis for compound 6 (in both cells) and 13 (only in SNB-19). Transcriptional activity of these genes in the C-32 cells suggests a different way of cell death.
Conclusion
We report here synthesis of 15 new 10-substituted 3,6-diazaphenothiazines. The parent compound, 10H-3,6-diazaphenothiazine, was obtained in cyclization reaction of 3-amino-3′-nitro-2,4′-dipyridinyl sulfide and in the reaction of sodium 3-amino-2-pyridinethiolate with 4-chloro-3-nitropyridine. This compound was transformed into 10-substituted derivatives with the alkyl, imidoalkyl, heteroaryl and dialkylaminoalkyl groups in the alkylation and heteroarylation reactions. The analysis of 2D NMR (ROESY, COSY, HSQC and HMBC) spectra of the N-methylated product showed that the thiazine ring formation proceeded through the Smiles rearrangement of the S–N type and the alkylation proceeded at the thiazine, not the azine nitrogen atom. This supposition was fully confirmed by X-ray analysis.
The parent compound 10H-3,6-diazaphenothizine was over 10 times more active (IC50 < 0.72 μg/mL) than a reference drug – cisplatin against glioblastoma SNB-19, melanoma C-32 and breast cancer MCF-7 cell lines. Two diazaphenothiazines with the 2-pyrimidinyl and dimethylaminopropyl substituents were very selectively antitumor active against breast cancer MCF-7 (10 times active) and melanoma C-32 (similarly active) cell lines, respectively, in relation to cisplatin. 3, 6 and 8 derivatives of 3,6-diazaphenothiazines turned out to be more active than monoazaphenothiazine drug, prothipendyl, against investigated cell lines. All 3,6-diazaphenothiazines were found to be non-toxic or almost non-toxic against normal human fibroblast (HFF-1) cell line in comparison with toxic cisplatin. The analysis of gene expressions (H3, TP53, CDKN1A, BCL-2, BAX) confirmed the antiproliferative activity of both compounds and indicated the activation of the p53 pathway in cancer cells, leading to the cell cycle arrest. The gene expression ratio BAX/BCL-2 suggested the mitochondrial apoptosis pathway activation in MCF-7 and SNB-19 cells.
Supplementary material available online
IENZ_1151014_Supplementary_File.pdf
Download PDF (1.4 MB)Declaration of interest
The work was supported by the Medical University of Silesia (grant KNW-1–004/K/4/0).
References
- Cancer Facts and Figures, American Cancer Society, 2013. Available from: http://www.cancer.org/acs/groups/content/@epidemiologysurveilance/documents/document/acspc-036845.pdf [last accessed 18 Aug 2015]
- Bray F, Ren J-S, Masuyer E, Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer 2013;32:1133–45.
- Bajaj S, Asati V, Singh J, Roy PP. 1,3,4-Oxadiazoles: an emerging scaffold to target growth factors, enzymes and kinases as anticancer agents. Eur J Med Chem 2015;97:124–41.
- Yugandhar D, Nayak VL, Archana S, et al. Design, synthesis and anticancer properties of novel oxa/azaspiro[4,5]trienones as potent apoptosis inducers through mitochondrial disruption. Eur J Med Chem 2015;101:348–57.
- Gupta RR, Kumar M. Synthesis, properties and reactions of phenothiazines. In: Gupta RR, ed. Phenothiazine and 1,4-benzothiazines – chemical and biological aspect. Amsterdam: Elsevier; 1988:1–161.
- Motohashi N, Kawase M, Saito S, Sakagami H. Antitumor potential and possible targets of phenothiazine-related compounds. Curr Drug Targets 2000;1:237–45.
- Motohashi N, Kawase M, Satoh K, Sakagami H. Cytotoxic potential of phenothiazines. Curr Drug Targets 2006;7:1055–66.
- Mitchell SC. Phenothiazine: the parent molecule. Curr Drug Targets 2006;7:1181–9.
- Dasgupta A, Dastridara SG, Shirataki Y, Motohashi N. Antibacterial activity of artificial phenothiazines and isoflavones from plants. Top Heterocycl Chem 2008;1567–132.
- Aaron JJ, Gaye Seye MD, Trajkovska S, Motohashi N. Bioactive phenothiazines and benzo[a]phenothiazines: spectroscopic studies and biological and biomedical properties and applications. Top Heterocycl Chem 2009;16:153–231.
- Sudeshna G, Parimal K. Multiple non-psychiatric effects of phenothiazines: a review. Eur J Pharmacol 2010;648:6–14.
- Pluta K, Morak-Młodawska B, Jeleń M. Recent progress in biological activities of synthesized phenothiazines. Eur J Med Chem 2011;46:3179–89.
- Wesołowska O. Interaction of phenothiazines, stilbenes and flavonoids with multidrug resistance-associated transporters, P-glycoprotein and MRP1. Acta Biochim Polon 2011;58:433–48.
- Jaszczyszyn A, Gąsiorowski K, Świątek P, et al. Chemical structure of phenothiazines and their biological activity. Pharmacol Rep Rep 2012;64:16–23.
- Viveiros M, Martins M, Couto I, et al. The in vitro activity of phenothiazines against Mycobacterium avium: potential of thioridazine for therapy of the co-infected AIDS patient. In Vivo 2005;19:733–6.
- Mosnaim AD, Ranade VV, Wolf ME, et al. Phenothiazine molecule provides the basic chemical structure for various classes of pharmacotherapeutic agents. Am J Therapeut 2006;13:261–73.
- González-Muñoz GC, Arce MP, López B, et al. Acylamino-phenothiazines: neuroprotective agents displaying multifunctional activities for a potential treatment of Alzheimer’s disease. Eur J Med Chem 2011;46:2224–35.
- Pohjala L, Utt A, Varjak M, et al. Inhibitors of alphavirus entry and replication identified with a stable chikungunya replicon cell line and virus-based assays. PLos One 2011;6:e28923.
- Kaur P, Chu JJH. Chikungunya virus: an update on antiviral development and challenges. Drug Discovery Today 2013;18:969–83.
- Pluta K, Jeleń M, Morak-Młodawska B, et al. Anticancer activity of newly synthesized azaphenothiazines from NCI's anticancer screening bank. NCI’s screening. Pharmacol Rep 2010;62:319–32.
- Morak-Młodawska B, Pluta K, Zimecki M, et al. Synthesis and selected immunological properties of 10-substituted 1,8-diazaphenothiazines. Med Chem Res 2015;24:1408–18.
- Zimecki M, Artym J, Kocięba M, et al. The immunosuppressive activities of newly synthesized azaphenothiazines in human and mouse models. Cell Mol Biol Lett 2009;14:622–35.
- Morak-Młodawska B, Pluta K, Matralis AN, Kourounakis AP. Antioxidant activity of newly synthesized 2,7-diazaphenothiazines. Arch Pharm (Weinheim) Pharm Life 2010;343:268–73.
- Morak-Młodawska B, Pluta K, Latocha M, et al. Synthesis and anticancer and lipophilic properties of 10-dialkylaminobutynyl derivatives of 1,8- and 2,7-diazaphenothiazines. J Enzyme Inhib Med Chem 2015. [Epub ahead of print]. 1–7. doi: 10.3109/14756366.2015.1101092.
- Okafor CO. Studies in the heterocyclic series. I. A novel diazaphenothiazine system. J Org Chem 1967;32:2006–7.
- Rodig OR, Collier RE, Schlatzer RK. Pyridine chemistry. I. The Smiles rearrangement of the 3-amino-2,2′-dipyridyl sulfide system. J Org Chem 1964;29:2652–8.
- Sheldrick G. SHELXS-97. Program for crystal structure solution. Germany: University of Göttingen; 1990.
- Sheldrick GM. SHELXL-2013. Program for structure refinement. Germany: University of Göttingen; 2013.
- Flack HD. On enantiomorph-polarity estimation. Acta Cryst 1983;A39:876–81.
- Pluta K, Morak-Młodawska B, Jeleń M. Synthesis and properties of diaza-, triaza- and tetraazaphenothiazines. J Heterocycl Chem 2009;46:355–91.
- Silberg IA, Cormos G, Oniciu DC. Retrosynthetic approach to the synthesis of phenothiazines. In: Katritzky AR, ed. Advances in heterocyclic chemistry. New York: Elsevier; 2006:205
- Morak B, Pluta K, Suwińska K. Unexpected simple route to novel dipyrido-1,4-thiazines. Heterocyclic Commun 2002;8:331–4.
- Morak-Młodawska B, Suwińska K, Pluta K, et al. 8-diazaphenothiazine as the double Smiles rearrangement. J Mol Struct 2012;1015:94–8.
- Clarke FH, Silverman GB, Watnick CM, Sperber N. 3-Azaphenothiazine and dialkylaminoalkyl derivatives. J Org Chem 1961;26:1126–232.
- Werle E, Kopp E, Leysath G. Die Antihistaminwirkung von 2,7-diazaphenothiazin und einiger seiner derivate. Arzneim-Forsch 1962;4:443–4.
- Pappalardo G, Vittorio F, Ronsisvalle G. Investigation on 2,3-diazaphenothiazine. Quaternization reactions. Ann Chim 1973;63:255–67.
- Carter S, Cheeseman G. Some aspects of 1,4-diazaphenothiazine chemistry. Tetrahedron 1977;33:827–32.
- Saari W, Cochran D, Lee Y, et al. Preparation of some 10-[3-(dimethylamino)-1-propyl]-10H-pyrazino[2,3-b][1,4] benzothiazines as potential neuroleptics. J Med Chem 1983;26:564–9.
- Cambridge Structural Database System, Release v5.36. Cambridge, UK: Cambridge Crystallographic Data Centre; [last accessed 30 Oct 2015].
- Morak-Młodawska B, Suwińska K, Pluta K, Jeleń M. Alkylations of 10H-2,7-diazaphenothiazine to alkyl-2,7-diazaphenothiazinium salts and 7-alkyl-2,7-diazaphen-othiazines. Heterocycles 2010;81:2511–22.
- Morak-Młodawska B, Suwińska K, Pluta K, Jeleń M. 10-(Prop-2-yn-1-yl)-2,7-diazaphenothiazine. Acta Crystallogr Sect E Struct Rep Online 2012;68:o1590–1.
- Allen MA, Andrysik Z, Dengler VL, et al. Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms. eLife 2014;3:1–29.
- Giui CY, Ngo L, Xu WS, et al. Histone deacetylase (HDAC) inhibitor activation of p21WAF1 involves changes in promoter-associated proteins, including HDAC1. PNAS 2004;101:1241–6.
- Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004;303:844–9.
- Hemann MT, Lowe SW. The p53-Bcl-2 connection. Cell Death Differ 2006;13:1256–9.
- Antonsson B, Montessuit S, Sanchez B, Martinou JC. Bax is present as a high molecular weight oligomer/complex in the mitochondrial membrane of apoptotic cells. J Biol Chem 2001;276:11615–23.
- Scorrano L, Korsmeyer SJ. Mechanisms of cytochrome c release by proapoptotic BCL-2 family members. Biochem Biophys Res Commun 2003;304:437–44.
- Antonsson B, Martinou JC. The Bcl-2 protein family. Exp Cell Res 2000;256:50–7.