Abstract
Introduction
The HIV Nef protein is a multifunctional virulence factor that perturbs intracellular membranes and signalling and is secreted into exosomes. While Nef-containing exosomes have a distinct proteomic profile, no comprehensive analysis of their miRNA cargo has been carried out. Since Nef functions as a viral suppressor of RNA interference and disturbs the distribution of RNA-induced silencing complex proteins between cells and exosomes, we hypothesized that it might also affect the export of miRNAs into exosomes.
Method
Exosomes were purified from human monocytic U937 cells that stably expressed HIV-1 Nef. The RNA from cells and exosomes was profiled for 667 miRNAs using a Taqman Low Density Array. Selected miRNAs and their mRNA targets were validated by quantitative RT-PCR. Bioinformatics analyses were used to identify targets and predict pathways.
Results
Nef expression affected a significant fraction of miRNAs in U937 cells. Our analysis showed 47 miRNAs to be selectively secreted into Nef exosomes and 2 miRNAs to be selectively retained in Nef-expressing cells. The exosomal miRNAs were predicted to target several cellular genes in inflammatory cytokine and other pathways important for HIV pathogenesis, and an overwhelming majority had targets within the HIV genome.
Conclusions
This is the first study to report miRnome analysis of HIV Nef expressing monocytes and exosomes. Our results demonstrate that Nef causes large-scale dysregulation of cellular miRNAs, including their secretion through exosomes. We suggest this to be a novel viral strategy to affect pathogenesis and to limit the effects of RNA interference on viral replication and persistence.
To access the supplementary material to this article, please see Supplementary files under Article Tools online.
The human immunodeficiency virus (HIV) encodes prototypic retroviral proteins (Gag, Pol, Env) as regulatory (Rev, Tat) and accessory (Nef, Vif, Vpr, Vpu/Vpx) proteins, the last group being dispensable for virus replication in vitro, but important for persistence and disease in an immunocompetent host (Citation1). The nef gene is located at the 3′-end of the viral genome, partially overlapping the env gene and the U3 region in the 3′ long terminal repeat (LTR). Nef is the largest of the four HIV accessory proteins, is expressed early in infection and at far higher levels than the other early proteins, Tat and Rev (Citation2). It is primarily localized at cellular membranes, which include endosomal membranes, the perinuclear region and the inner surface of plasma membrane. Nef is also released from cells, either in microvesicles (MVs) (Citation3, Citation4) or as a soluble protein, and has effects on bystander cells (Citation5–Citation7).
The extracellular MVs are called shedding vesicles or exosomes depending on their size and origin (Citation8). The shedding vesicles are 0.1–1 µm and are formed by outward budding and pinching of the plasma membrane. Exosomes are 30–100 nm and are formed by inward invagination of the multivesicular body (MVB) membrane forming intraluminal vesicles, which are released in the extracellular medium on fusion of the MVBs with the plasma membrane. These contain various mRNAs, miRNAs and proteins that vary depending on the producer cell type (Citation8, Citation9). Further, since these vesicles can be taken up by other cells, and the exosomal mRNAs and miRNAs are shown to be functional in recipient cells, the MVs are proposed as a novel mode of intercellular communication (Citation10). The modulatory effects of the exosomal cargo on cellular gene expression are likely to affect intracellular viral replication, persistence and pathogenesis.
The first evidence of extracellular Nef came from reports on the detection of anti-Nef antibodies in the sera of AIDS patients (Citation11). Later, Nef was also found in exosome-like vesicles released from HEK293 cells expressing a Nef-green fluorescent protein (Nef-GFP) fusion protein (Citation3). These vesicles were shown to be taken up by Jurkat CD4+ T cells, in which the Nef-GFP fusion protein localized mainly to the cytoplasm as punctate structures (Citation3). The vesicles are likely to enter recipient cells via endocytosis, which had been reported in other systems (Citation12). Importantly, Nef exosomes could also fuse with Nef-deficient HIV-1 virions and restore the infectivity of mature particles (Citation3).
The mammalian miRNA pathway restricts the replication of infecting viruses and promotes latency. While miR-323, miR-491 and miR-654 inhibit the replication of H1N1 influenza virus, miR-24 and miR-93 limit vesicular stomatitis virus (VSV) replication, and miR-32 restricts primate foamy virus type 1 (PFV-1) (Citation13). Human miR-28, miR-125b, miR-150, miR-223 and miR-382 target the 3′UTR of HIV-1 transcripts potentially taking productive infection towards latency (Citation14). Recent advances in deep sequencing technology have also led to the identification of small viral RNA species in HIV-1 infected cells (Citation15). Further, the knockdown of several miRNA processing pathway proteins, including Dicer, Drosha and DGCR8, leads to an increase in viral replication, and HIV-1 transcripts have been co-localized with RNAi effector proteins in P-bodies (Citation16). Thus, cellular miRNAs affect viral replication, either by targeting viral RNAs or cellular RNAs that encode host proteins necessary for viral replication (Citation16).
We have studied human monocytic U937 cells that stably expressed a Nef-EYFP fusion protein and found the protein in exosomes (Citation17). Transfer of exosomal miRNAs to other recipient cells is now recognized as a mode of communication between different cells and tissues (Citation10). Further, dysregulation of cellular miRNAs in diseased states also affects their repertoire in exosomes (Citation9). Nef localizes to MVBs, interacts with Argonaute-2, redistributes RNA-induced silencing complex (RISC) components between cells and exosomes, and acts as a viral suppressor of RNA interference (Citation17). We have profiled miRNAs in U937/Nef-EYFP cells and exosomes purified from these cells and have identified the pathways potentially regulated by these miRNAs. Our results suggest that Nef mediates the redistribution of miRNAs between cells and exosomes to aid in viral replication and persistence.
Materials and methods
Exosome isolation and characterization
The U937/Nef-EYFP and U937/EYFP cells and their culture conditions have been described previously (Citation17). Exosomes were prepared from culture supernatants (Citation18) and visualized by electron microscopy as described earlier (Citation17). Exosome quantification was done using Bradford reagent (BioRad, USA) as described elsewhere (Citation18). For flow cytometry, purified exosomes (~3–5 μg protein) were first incubated with 10 μl of aldehyde latex beads for 15 minutes at room temperature. Then PBS was added to a final volume of 1 ml and incubated on a rotator for 2 hour at room temperature, or overnight at 4°C. To this 110 μl of 1 M glycine (final concentration, 100 mM) was added and beads were incubated at room temperature for 30 minutes. The beads were then centrifuged for 3 minutes at 4,000 rpm at room temperature and washed twice with 1 ml PBS/0.5% BSA with final re-suspension in 0.5 ml PBS-BSA. For staining with antibodies, 10 μl of coated beads were incubated with 50 μl of the appropriate antibody diluted in PBS-BSA for 30 minutes at 4°C. The beads were then washed twice as described above followed by incubation with 50 μl secondary antibodies for 30 minutes at 4°C. The beads were again washed twice and were acquired on a Cyan-ADP flow cytometer (Beckman Coulter). Data were analysed using the Flow Jo software. For the labelling of exosomal membranes with PKH26, a fluorescent lipid dye (Sigma) the manufacturer's instructions were followed. About 10–50 µg of exosomes were used for each labelling reaction. The excess dye was removed by washing exosomes with 3 ml of PBS with ultracentrifugation at 100,000×g in a SW60 rotor at 4°C. These were visualized using a Nikon A1-R Confocal Microscope at 100× magnification. The labelled exosomes were also bound to aldehyde latex beads as described above and beads were observed under 60× magnification in the confocal microscope.
Isolation of cellular and exosomal RNAs
Total RNA was isolated from cells or exosomes using Trizol (Invitrogen) as per manufacturer's instructions. Approximately 10 million cells or 50–100 μg exosomes were re-suspended in 500 μl of Trizol and vortexed thoroughly. The mixture was kept at room temperature for 10 minutes. Then 200 μl of chloroform was added and mixed gently. This was left for 10 minutes at room temperature and then centrifuged at 13,000 rpm at 4°C for 10 minutes. The upper aqueous phase was mixed with 2 volumes of isopropanol and incubated overnight at −20°C. The mixture was then centrifuged at 13,000 rpm for 10 minutes at 4°C. The pellet was washed with 70% ethanol, air-dried and dissolved in DEPC-treated water. RNA preparations were treated with DNaseI (New England Biolabs, USA) according to manufacturer's instructions before further use. This RNA was used for messenger RNA analysis and for the Taqman miRNA array profiling. The miRNAs were isolated using the miRNeasy kit (Qiagen, Germany) following the supplier's protocol. Briefly, ~2–3 million cells or 80–100 μg exosomes were harvested and lysed in ~750 μl Qiazol. The cells were vortexed and ~200 μl chloroform was added. The mixture was vortexed for a few seconds and centrifuged at 13,000 rpm for 10 minutes at 4°C. An equal amount of ethanol was added to the supernatant and loaded on the columns, which were washed with the buffers provided in the kit, and eluted in ~50 μl nuclease-free water. The RNA amount and quality were estimated by absorbance values at 260 and 280 nm on a Nanodrop spectrophotometer.
Profiling of miRNAs in cells and exosomes
Total RNA was isolated as above using Trizol and the miRNA profiling was carried out at Labindia (Gurgaon, India) using the Applied Biosystems Taqman Low Density Array – Human MicroRNA Array Version 2.0, which covered 667 known human microRNAs, and included 6 endogenous miRNA controls – MammU6, RNU6B, RNU48, RNU44, RNU24 and RNU43. The small RNA fraction was purified from total RNA pool using the mirVana™ miRNA Isolation Kit (Qiagen), which was converted to cDNAs using Megaplex™ RT Primers and TaqMan MicroRNA Reverse Transcription kit. A pre-amplification step was carried out using Megaplex™ PreAmp Primers and TaqMan® PreAmp Master Mix. The cDNA was diluted, TaqMan universal PCR master mix was added and the mixture was loaded on a 384-well format Taqman Array. The arrays were run using Applied Biosystems 7900HT Fast Real-Time PCR System. Data were analysed using Data Assist Software v3.0.1 with the threshold Ct value as 35. For all analyses, the levels in control samples were set as 1, and the levels in the test sample were calculated with the software. The control miRNAs in the array were set as endogenous control for both samples.
Quantification of miRNAs and mRNAs
One μg total RNA from U937/Nef-EYFP and U937/EYFP cells, or 50–100 ng from exosomes was reverse transcribed with a specific stem-loop (SL) primer (Supplementary file). The reaction mix was prepared as described above and incubated as follows: 70°C for 5 minutes followed by 7 cycles of 16°C for 5 minutes and 42°C for 5 minutes. After heating at 85°C for 5 minutes, the mixture was either used immediately for quantitative PCR or stored at −70°C. An appropriate amount of cDNA was first used to optimize the RNU6B signal in each sample. Subsequently, amplifications of specific mature miRNAs were carried out using the primers shown in Supplementary file. For the quantification of mRNAs, total RNA was converted to cDNA using a kit (Promega, USA) according to the manufacturer's protocol. Briefly, a reaction mix was prepared using 1 μg total RNA, 500 ng oligo(dT)20 and DEPC water. The reaction mixture was incubated at 65°C for 5 minutes and chilled on ice for at least 1 minute. The following components were then added in the given order: 10 μl of 5X Promega RT buffer, 0.25 μl RNaseIN (Recombinant RNase Inhibitor; 40 U/μl), 0.5 μl of MULV RT enzyme (400 U/μl) and 0.5 μl of dNTPs (10 mM). The reaction was incubated at 42°C for 60 minutes, followed by heating at 70°C for 15 minutes and then chilled on ice. A 20-μl reaction mix was prepared using 2X EVAGreen dye, 1–2 μl of cDNA, and 5–10 pmoles each of forward and reverse primers. The amplification conditions were as follows: 94°C for 10 minutes, followed by 40 cycles of 94°C for 30 seconds, 60–62°C for 30 seconds, 72°C for 30 seconds, and a final extension at 72°C for 2 minutes. The PCR was carried out in a StepOne Plus thermocycler (Applied Biosystems). The Ct values of replicates were analysed to calculate relative fold change by the ΔΔCt method. Data Analysis was done using the Data Assist Software (Applied Biosystems). All primer sequences are listed in Supplementary file.
Bioinformatic analysis
The miRWalk database (http://www.umm.uniheidelberg.de/apps/zmf/mirwalk/) (Citation19) was searched to identify validated target genes of the selected miRNAs, followed by pathway analysis using the DAVID Bioinformatics Database (http://david.abcc.ncifcrf.gov/gene2gene.jsp) (Citation20, Citation21). Microinspector (http://bioinfo.uni-plovdiv.bg/microinspector/) (Citation22) and RNA Hybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/) (Citation23) algorithms were used to identify miRNAs targeting HIV-1 strains Indie-C1 (Accession No. AB023804) and NL4-3 (Accession No. AF324493). All of the 2,578 mature human miRNAs available in miRBase version 20 were analysed using a free-energy cut-off of –14 kcal/mole. We also compared the list of validated targets of miRNAs upregulated or downregulated in U937/Nef-EYFP cells with genes that are reported in literature to be upregulated or downregulated by Nef; the latter information is part of the HIV, Human Protein Interaction Database and is available at http://www.ncbi.nlm.nih.gov/projects/RefSeq/HIVInteractions/nef.html.
Western blotting
Cells were washed twice in cold 1X PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 18 mM KH2PO4) and lysed in 1X RIPA Buffer (20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% NP-40, 1% sodium deoxycholate, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 μg/ml leupeptin). Cell lysis was carried out for 45 minutes at 4°C with gentle vortexing every 10 minutes. The lysate was clarified by centrifugation at 13,000×g for 20 minutes at 4°C. The protein concentration of the lysate was estimated using the Bradford reagent (Bio-Rad, USA). For western blotting, 100 microgram of cell lysate was boiled in Laemmli buffer, and the proteins were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE). The separated proteins were transferred to a nitrocellulose membrane (Hybond ECL; Amersham Biosciences) at constant voltage of 65 V for 60 minutes. The membrane was then blocked with Tris-buffered saline (TBS) containing 3% Blotto (Bio-Rad) for 1 hour at room temperature and washed with TBST (TBS containing 0.1% Tween 20). The membrane was then incubated overnight at 4°C with the primary antibody appropriately diluted in TBST–3% Blotto, washed 4 times for 10 minutes each with TBST, and incubated with horseradish peroxidase-linked secondary antibodies diluted in TBST–5% Blotto for 1 hour at room temperature. After the membrane was washed as described above, chemiluminescent detection of proteins was carried out using Luminol reagent (Santa Cruz Biotechnology, USA) according to the supplier's protocol. Primary antibodies anti-IL6 (sc7920), anti-IL1β (sc1250), anti-Actin and HRP-linked secondary antibodies were from Santa Cruz Biotechnology (USA).
Results
Characterization of exosomes secreted from U937 human monocytic cells
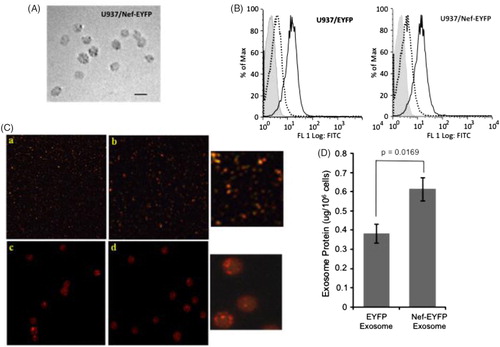
Exosomes isolated from U937/EYFP and U937/Nef-EYFP cells using a differential centrifugation protocol were found by electron microscopy to be of the expected size of 30–90 nm (A). These were also tested for the exosomal marker proteins Alix, Tsg101 and CD81, which were present in purified exosomes as well as in both types of cells. However, unlike cell lysates we did not detect voltage-dependent anion channel (VDAC), Calnexin or Cytochrome C in the exosome preparation, indicating that it was not contaminated with mitochondria, endoplasmic reticulum or apoptotic bodies (Citation17). We further detected CD81 on the surface of exosomes prepared from both cell lines by flow cytometry (B). The exosomes were also labelled with PKH26, a red fluorescent lipid dye, and were visualized by confocal microscopy either directly (C, panels a and b) or after binding to aldehyde latex beads (C panels c and d). This confirmed that exosomes were isolated with an intact limiting membrane. We also quantified exosome secretion from the two cell lines by normalizing total exosomal protein with the number of cells harvested for exosome preparation. The average exosome yields were 0.384 μg/million U937/EYFP cells and 0.615 μg/million U937/Nef-ETFP cells (D). Thus, the Nef-expressing U937 cells secreted on an average about 60% more exosomes than control cells.
Fig. 1 Characterization of exosomes from U937/Nef-EYFP and U937/EYFP cells. (A) Electron microscopy of exosomes from U937/Nef-EYFP cells; bar 100 nm. (B) Flow cytometry of exosomes for the marker protein CD81. (C) Exosomes were labelled with lipid dye PKH26; these were (a, b) observed directly at 100X magnification, or (c, d) first bound to 4 μm latex beads and then observed at 100X magnification. The insets on the right show enlarged views of the images. (D) Amount of exosomes secreted by U937/EYFP and U937/Nef-EYFP cell lines presented as protein concentration from 1×106 cells. Data is presented as mean±SD from 3 independent experiments. The p-value calculated using Students t-test is shown.
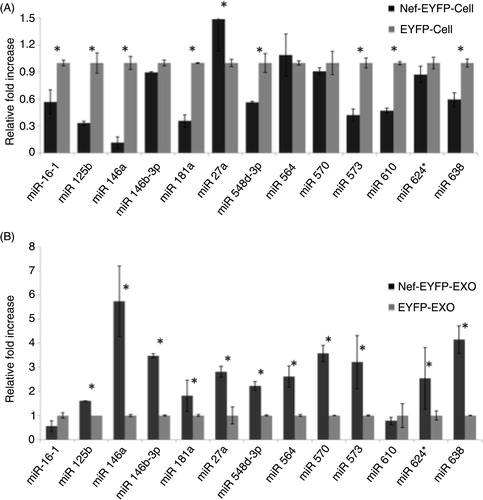
Differential miRNA expression and secretion in exosomes
The RNA isolated from both cell lines and their exosomes was analysed on an Agilent Bioanayzer. The yield for cellular RNA was 1.177 µg/µl for U937/EYFP cells and 2.055 µg/µl for U937/Nef-EYFP cells with RIN of 9.4 and 9.1, respectively (Supplementary file). The high RIN values indicate the extracted RNA to be of high quality. The yields for exosomal RNAs were 53 and 52 ng/µl for control and Nef exosomes, respectively. The bioanalyzer profiles showed 2 distinct peaks of ribosomal RNAs in the cellular RNA pool, which is characteristic of the 18S and 28S species that were absent in the exosomal RNA pool (Supplementary file). On the contrary, the exosomal RNAs showed a major peak around 25 nt demonstrating the enrichment of small RNAs in exosomes. The miRNA population was then isolated from both cellular and exosomal RNA pools using the miRVana miRNA isolation kit and profiling was carried out using Taqman low density arrays. The miRNAs detected in U937/Nef-EYFP cellular and exosomal pools were then compared to their respective control pools to determine differential cellular expression and exosomal secretion of miRNAs in Nef-expressing U937 monocytic cells. The results from cellular and exosomal datasets were then compared to identify the miRNAs that were preferentially packaged in or excluded from Nef-containing exosomes. In any sample, only ~50% of the 667 profiled miRNAs were detected and the rest remained undetected for each pool. This showed that not all miRNAs were produced in U937 cells, confirming the cell-specific expression of miRNAs. The profiling data also suggested that cellular and exosomal miRNA pools of U937/Nef-EYFP cells vary considerably from that of control U937/EYFP cells.
Of the 667 miRNAs analysed, 320 miRNAs (i.e. 48%) were detected in U937/Nef-EYFP and U937/EYFP cells, of which 87 (27%) were expressed at 1.4-fold or higher levels (miNCup) and 67 (21%) were expressed at lower than 0.8-fold (miNCdown) in U937/Nef-EYFP compared to U937/EYFP cells; the remaining 166 (52%) miRNAs were expressed at similar levels in both cell lines (A and Supplementary file). Analogous to the cellular pool, ~50% of the profiled miRNAs were detected in any of the exosome samples and the rest remained undetected. Of the 349 miRNAs detected in Nef exosomes, 311 (89%) were present at 1.4-fold or higher levels (miNEup) and 28 (8%) were present at lower that 0.8 fold (miNEdown) in exosomes from U937/Nef-EYFP compared to U937/EYFP cells; the remaining 10 (3%) were present at similar levels in exosomes from both cell lines (B and Supplementary file). While the miRNA profile of Nef-expressing cells showed almost similar percentages of upregulated and downregulated miRNAs, the exosomes from these cells contained increased levels of most of the detected miRNAs.
Fig. 2 Nef expression modulates cellular and exosomal miRNAs profiles. The miRNAs in (A) cells and (B) exosomes were profiled and relative fold-changes calculated as described in Methods. (C) The Venn diagrams show the intersection of detected miRNAs as follows: (i) Upregulated miRNAs in both U937/Nef-EYFP cells and exosomes; (ii) Downregulated miRNAs in U937/Nef-EYFP cells and upregulated miRNAs in U937/Nef-EYFP exosomes; (iii) Upregulated miRNAs in U937/Nef-EYFP cells and downregulated miRNAs in U937/Nef-EYFP exosomes.
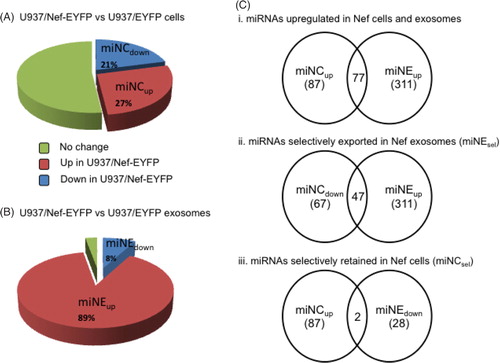
We then examined miRNAs upregulated in both cellular and exosomal pools of U937/Nef-EYFP compared to U937/EYFP cells (miNCup∩miNEup). There were 87 and 311 in Nef-expressing cells and exosomes, respectively, of which 77 miRNAs were common to these groups (Ci). These 77 miRNAs are likely to be packaged more in Nef exosomes because their levels are also higher in Nef-expressing cells. To identify miRNAs selectively secreted in Nef exosomes (miNEsel) we compared the miRNAs down regulated in Nef-expressing cells with those that were upregulated in Nef exosomes (miNCdown∩nmiNEup) and found 47 miRNAs in this category (Cii, Supplementary file). Finally, we analysed miRNAs that are selectively retained in Nef-expressing cells (miNCsel) and are not exported out through exosomes. For this, we compared the group of miRNAs upregulated in Nef-expressing cells with those that were downregulated in Nef exosomes (miNCup∩miNEdown), and found only 2 such miRNAs (Ciii, Supplementary file). Thus, Nef expression leads to the selective secretion of many more miRNAs in exosomes compared to those that are selectively retained in Nef-expressing cells.
Validation of miRNAs in cells and exosomes
To confirm the profiling data, we selected 13 miRNAs for validation by stem-loop quantitative RT-PCR in multiple replicates of cells and exosomes. These include miR-573 and miR-638 (upregulated in Nef cells), miR-548d-3p and 564 (same levels in Nef and control cells), and miR-16-1, miR-146a, miR-125b, miR-146b-3p, miR-181a, miR-27a, miR-570, miR-610 and miR-624* (down regulated in Nef cells). All these miRNAs, except miR-146a, were upregulated in exosomes in the profiling array data. As there is no well documented control small RNA for exosomes, we quantified RNU6 levels in both Nef and control exosomes, and found these to be almost similar in multiple replicates of the 2 populations. The average Ct values obtained were 23.3 in control exosomes and 23.7 in Nef exosomes (Supplementary file). Therefore, the exosomal miRNA data was normalized to RNU6. The results showed most of the miRNA levels to correlate with the profiling data. In the cells we found 4 miRNAs to show a trend opposite to the profiling data. These include miR-27a, which is upregulated, and miR-548d-3p, miR-573 and miR-638, which are down regulated in Nef-expressing cells based on qRT-PCR analysis (A). All the analysed miRNAs showed higher levels in Nef exosomes, which is consistent with the profiling data (B). Thus, the qRT-PCR results broadly validate our profiling data.
Nef exosomes are enriched in miRNAs that potentially target innate immune responses and the HIV-1 genome
Since each miRNA can potentially target hundreds of transcripts, changes in their abundance can have significant effects on cellular pathways. The validated targets of miRNAs that were selectively secreted from Nef-expressing cells were extracted using the miRwalk software. A majority of these miRNAs were found to target the expression of cytokines/chemokines (e.g. interleukins), proteins involved in innate immunity (e.g. interferon, NFKB, STATs), tumour suppressors (PTEN, RB1), cell survival factors (BCL2, MCL1) and the HIV restriction factor APOBEC3G. In monocytes, Nef induces the synthesis of proinflammatory cytokines and chemokines. This effect is observed with Nef expressed intracellularly as well as that added exogenously to monocytes (Citation24). We therefore selected 6 of these genes – IL1α, IL1β, IL6, TNFα, MIP1α, MIP1β, which are induced in the presence of Nef, and observed that many of the miRNAs that target these cytokines, such as miR-146a, miR-146b-3p, miR-125b and miR-181a, are downregulated in Nef-expressing cells, but are selectively secreted in exosomes from these cells. These 4 miRNAs regulate proinflammatory cytokine levels, antigen processing and presentation pathways (Citation25).
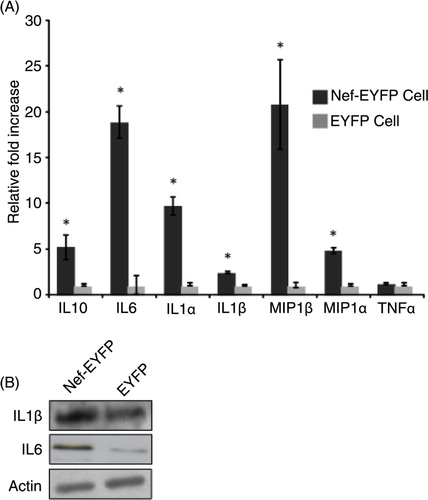
We quantified the expression levels of IL1α, IL1β, IL6, TNFα, MIP1α, MIP1β, and IL10 mRNAs in U937/Nef-EYFP and U937/EYFP cells. Except for TNFα, which did not show any significant change, all other mRNAs were present at much higher levels in Nef-expressing cells (A). The results showed >10-fold increase for IL1β, IL6 and MIP1β mRNAs, and about 5-fold increase for IL10 and MIP1α mRNAs. For 2 of these, IL1β and IL6, the protein expression levels were also estimated by western blotting; these were also higher in Nef-expressing cells (B).
Fig. 4 Nef induces the expression of proinflammatory cytokines in U937 cells. (A) RNA isolated from U937/Nef-EYFP and U937/EYFP cells were tested for the expression of selected cytokine genes by qRT-PCR; relative fold-changes were calculated from raw Ct values using the Data Assist Software (ABI). Error bars represent standard errors from 3 experiments, each with triplicate measurements; *p<0.05. (B) Western blots of lysates from U937/Nef-EYFP and U937/EYFP cells for IL1β and IL6 levels. Actin was used as a loading control.
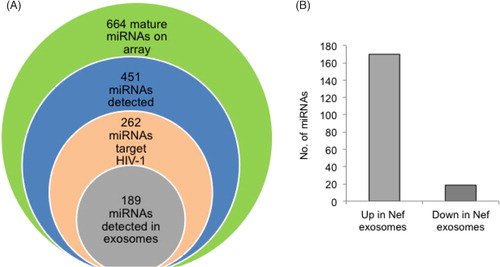
Host miRNAs can also target HIV transcripts leading to a reduced level of viral replication. We then analysed our profiling data to identify the status of miRNAs that can potentially target the HIV genome. In silico analysis was performed with the 451 detected miRNAs that were matched against 2 HIV-1 genomes – NL4-3 (clade B) and IndieC1 (clade C), using two separate software tools – Microinspector and RNAHybrid. Of the 262 potential HIV-1 targeting miRNAs identified through this process, 189 were also present in exosomes from the 2 cell lines. These are listed in Supplementary file. Interestingly, 170 of these miRNAs were upregulated and only 19 were downregulated in exosomes from U937/Nef-EYFP compared to U937/EYFP cells (). This overwhelming preference for the secretion of potentially HIV-targeting miRNAs suggests that Nef-induced vesicle secretion might be a unique viral strategy to selectively export virus targeting host miRNAs.
Fig. 5 HIV targeting host miRNAs are preferentially secreted into U937/Nef-EYFP exosomes. (A) Diagram showing the correlation between HIV targeting miRNAs and Nef exosomal miRNAs. All the detected miRNAs were screened for potential binding sites on 2 different HIV-1 genomes (NL4-3 and IndieC1) using the MicroInspector algorithm (http://bioinfo.uni-plovdiv.bg/microinspector/); the identified miRNAs were verified using the RNAHybrid algorithm (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/). (B) miRNAs present in exosomes that can potentially target the HIV-1 genome are separated based on whether their levels are higher or lower in Nef compared to control exosomes.
Pathway analysis of selectively deregulated miRNAs in Nef expressing cells and exosomes
Using miRWalk, we obtained 1,565 validated target genes for the 47 miRNAs that were selectively secreted in Nef exosomes (miNEsel; Supplementary file) and 107 validated target genes of the 2 miRNAs that were selectively retained in Nef-expressing cells (miNCsel; Supplementary file). These were then subjected to pathway analysis using the DAVID (Database for Annotation, Visualization and Integrated Discovery) Bioinformatics Resources. We identified 63 KEGG pathways (p<0.05) corresponding to the 1,565 validated targets of the 47 miRNAs in miNEsel. Several pathways known to be involved in HIV pathogenesis came up in this analysis. These include TGF-beta signalling, toll-like receptor signalling, apoptosis, cell cycle, T-cell receptor signalling, p53 signalling, mitogen activated protein kinase (MAPK) signalling, cytokine–cytokine receptor interaction, Janus kinase-signal transducer and activator of transcription (Jak-STAT) signalling, B-cell receptor signalling, mTOR signalling and chemokine signalling (Supplementary file). We also identified 26 KEGG pathways (p<0.05) corresponding to the 107 validated targets of the miRNAs in miNCsel. Some important pathways here include cell cycle, MAPK signalling, p53 signalling, T-cell receptor signalling, toll-like receptor signalling, cytokine–cytokine receptor interaction and regulation of actin cytoskeleton (Supplementary file). We further compared the entries reported in the HIV, Human Protein Interaction Database to be upregulated and downregulated by Nef with the miRNA targets identified in this study. This identified 19 proteins (genes) reported to be upregulated by Nef to also be present in our set of 1,565 upregulated genes; similarly, 2 proteins (genes) were in common with our set of 107 downregulated genes. A list of these is provided in Supplementary file.
Discussion
The Nef protein of HIV is expressed early during viral infection and is associated with cellular membranes, including endosomal membranes (Citation2). In addition to perturbing intracellular membranes and signalling, Nef is secreted in exosomes (Citation4). Interestingly, Nef increases the formation of MVBs (Citation26), which are intracellular sites for exosome biogenesis, the secretion of these vesicles (Citation27), and interacts with the AIP1/Alix protein that is involved in MVB biogenesis (Citation28). Consequently, Nef promotes its own export through exosomes from different cell types and can induce apoptosis in bystander CD4+ T cells (Citation3, Citation4). Exosomes contain different proteins, mRNAs and miRNAs depending on their cell of origin (Citation9). They are taken up by recipient cells, in which the exosomal mRNA is translated and the miRNA can post-transcriptionally regulate gene expression (Citation10), thus supporting the view that exosomes function as intercellular messengers. Modulation of exosomal miRNAs is reported in several cancers as well as viral infections (Citation29).
We have carried out the first miRNome analysis of Nef-expressing human monocytic cells and their exosomes. We show that Nef exosomes are enriched in miRNAs that can target proinflammatory cytokines and other genes involved in key pathways like JAK-STAT signalling, MAPK signalling and apoptosis. Further, an overwhelming percentage of miRNAs that can potentially target HIV-1 are secreted out of Nef-expressing cells into exosomes. As reported earlier for other cell types, U937 cells expressing Nef also showed increased exosome secretion. There was differential expression of about 50% of detected miRNAs under the influence of Nef. It was shown recently that reducing GW182, which is an important component of Glycine-Tryptophan (GW) bodies, also reduced miRNA secretion through exosomes (Citation30). This agrees with our recent findings that Nef-expressing U937 cells have higher levels of GW182 (Citation17) and display increased secretion of exosomes and miRNAs reported here.
We observed significant changes in the levels of several miRNAs that regulate innate immune responses, especially the proinflammatory cytokines. These include miR-16, miR-125b, miR-146a, miR-146b-3p and miR-181a, which are reduced in Nef-expressing U937 cells. Of these, miR-16 is required for the degradation of transcripts that contain AU-rich elements in their 3′UTR, which includes mRNAs for most of the inflammatory mediators. It regulates IL6, TNFα and IL8, and high levels of miR-16 restrict the production of inflammatory mediators under non-stimulated conditions (Citation25). Interestingly, miR-16 levels were ~50% lower in the Nef cellular pool, suggesting that Nef might regulate proinflammatory cytokines through this pathway. MiR-125b inhibits HIV replication by targeting viral transcripts (Citation31). Our profiling data showed about 3-fold reduction in miR-125b levels in Nef-expressing U937 cells and a corresponding increase in their exosomes, suggesting that Nef might also render cells more permissive for viral replication through miR-125b modulation.
The miR-146 family comprises 2 members – miR146a and miR146b, of which the former is also a well-known inhibitor of innate immune responses (Citation32). Its levels increase in myeloid cells in response to TLR2, TLR4 or TLR5, and exposure to inflammatory cytokines like TNFα or IL1β (Citation33, Citation34); in alveolar epithelial cells it reduces IL1β levels (Citation33, Citation34). We found miR-146a levels to be about 5-fold lower in U937/Nef-EYFP cells compared to U937/EYFP cells, and observed a corresponding increase in Nef exosomes. In THP-1 monocytic cells, miR-146a regulates inflammatory cytokines/chemokines, including IL1β, IL6, TNFα, IL8, IP10, and MCP-1 (Citation34). High levels of miR-146a correlated with increased numbers of GW bodies in THP-1 cells, suggesting that GW bodies are markers for miRNA activity during innate immune signalling (Citation34). Our data showing reduced levels of miR-146a in Nef-expressing monocytes, suggests that Nef modulates key miRNAs that regulate innate immune responses. Further, miR-181a levels are also ~50% lower in U937/Nef-EYFP cells compared to U937/EYFP cells, and show a corresponding increase in Nef exosomes. This miRNA regulates T-cell signalling by down regulating multiple phosphatases, and is thought to act as a rheostat that regulates protein phosphorylation levels (Citation25). Thus, several miRNAs that affect inflammatory cytokines and innate immune responses are present at reduced levels in Nef-expressing monocytes and show a corresponding increase in exosomes secreted from these cells. Correspondingly, the mRNAs and expression of these cytokines/chemokines are at higher levels in Nef-expressing cells.
Host miRNAs also target viral transcripts and limit replication. Naturally then, modulation of host miRNAs is emerging as a viral counter-strategy for successful infection (Citation13, Citation35). HIV-1 infection of Jurkat cells downregulates the polycistronic miRNA cluster miR-17/92, which includes miR-17/-18/-20a, miR-19a/-19b and miR-92a (Citation16). The downregulation of miR-21, miR-26a, miR-29a, miR-29b and miR-29c was also observed in HIV patients (Citation31). We found miR-18, miR-19a, miR-20a, miR-21 and miR-29b to be downregulated in Nef expressing monocytes. A comparison of miRNA expression patterns in resting and activated CD4+ T cells and use of specific antagomirs concluded that miR-28, miR-125b, miR-150, miR-223 and miR-382 target the nef/3′LTR region and contribute to HIV latency in resting CD4+ T cells; similar data were also reported for monocytes and macrophages (Citation14, Citation16). Our profiling demonstrated reduced levels of miR-125b and miR-223 in Nef-expressing monocytes, whereas miR-382 was not detected. Based on in silico analyses, miR-29a, miR-29b, miR-149, miR-324-5p and miR-378 were reported to target conserved regions of the HIV-1 genome, including the nef gene (Citation16). Of these, we found miR-29b to be downregulated in Nef-expressing cells. A majority of miRNAs that inhibit HIV replication, including miR-17, miR-19a, miR-19b, miR-20a, miR-26a, miR-28, miR-29a, miR-29b, miR-29c, miR-92a, miR-125b, miR-149, miR-150, miR-223, miR-324-5p, miR-378 and miR-382 were present at 1.5-folds or higher levels in Nef exosomes. Further, when correlated with our in silico analysis of miRNA target sites in the HIV genome, an overwhelming majority of miRNAs that can potentially target HIV-1 genomes were present at increased levels in exosomes secreted by Nef-expressing cells. We also found 47 miRNAs to be present at increased levels in Nef exosomes despite being present at reduced levels in Nef-expressing cells. Of these selectively secreted miRNAs, 21 also had target sites on HIV-1 genomes. Thus, Nef expression reduces the cellular levels of several host miRNAs that target innate immune responses and viral transcripts by exosome-mediated export. This is likely to modify the host cell environment to favour virus replication.
The increased formation of MVB membranes (Citation26, Citation28) and enhanced vesicle secretion (Citation4, Citation27) in the presence of Nef can favour virus egress and spread. However, recent reports also show MVBs to be sites for miRNA assembly and activity (Citation36, Citation37). Thus, the host can limit HIV replication by targeting viral transcripts to membranes, which it evolved to exploit. We have recently shown that Nef attenuates the activity of let-7a, which silences target genes primarily at MVBs (Citation17). This is attributed to the ability of Nef to interact with Ago2 through 2 highly conserved GW motifs (Citation17). We now show that many miRNAs whose levels are reduced in Nef-expressing cells accumulate in exosomes secreted from these cells. Thus, besides employing endosomal membranes for Ago2 interaction, HIV Nef further exploits the exosomal pathway to alter the cellular and exosomal distribution of miRNAs. Based on these findings, we propose that Nef has evolved a 2-pronged suppressor function to inhibit miRNA-mediated viral silencing and to modulate exosome-mediated cell-to-cell communication. How Nef influences the selective expression and export of miRNAs, and how the exosomal cargo influences uninfected cells remain important issues to explore in future studies.
Conflict of interest and funding
This work was funded by grants from the Department of Biotechnology (DBT), Government of India and the Indian Council of Medical Research (ICMR). A Senior Research Fellowship from the Council for Scientific and Industrial Research (CSIR), India, supported MA.
Supplementary Tables and figures
Download PDF (699 KB)Notes
To access the supplementary material to this article, please see Supplementary files under Article Tools online.
References
- Das SR, Jameel S. Biology of the HIV Nef protein. Indian J Med Res. 2005; 121: 315–32.
- Landi A, Iannucci V, Nuffel AV, Meuwissen P, Verhasselt B. One protein to rule them all: modulation of cell surface receptors and molecules by HIV Nef. Curr HIV Res. 2011; 9: 496–504.
- Campbell TD, Khan M, Huang MB, Bond VC, Powell MD. HIV-1 Nef protein is secreted into vesicles that can fuse with target cells and virions. Ethn Dis. 2008; 18: S2–14-9.
- Lenassi M, Cagney G, Liao M, Vaupotic T, Bartholomeeusen K, Cheng Y et al. HIV Nef is secreted in exosomes and triggers apoptosis in bystander CD4+ T cells. Traffic. 2010; 11: 110–22.
- Raymond AD, Campbell-Sims TC, Khan M, Lang M, Huang MB, Bond VC et al. HIV type 1 Nef is released from infected cells in CD45(+) microvesicles and is present in the plasma of HIV-infected individuals. AIDS Res Hum Retroviruses. 2011; 27: 167–78.
- Fujii Y, Otake K, Tashiro M, Adachi A. Soluble Nef antigen of HIV-1 is cytotoxic for human CD4+ T cells. FEBS Lett. 1996; 393: 93–6.
- Campbell PE, Isayev O, Ali SA, Roth WW, Huang MB, Powell MD et al. Validation of a novel secretion modification region (SMR) of HIV-1 Nef using cohort sequence analysis and molecular modeling. J Mol Model. 2012; 18: 4603–13.
- Zomer A, Vendrig T, Hopmans ES, van Eijndhoven M, Middeldorp JM, Pegtel DM. Exosomes: fit to deliver small RNA. Commun Integr Biol. 2010; 3: 447–50.
- de Jong OG, Verhaar MC, Chen Y, Vader P, Gremmels H, Posthuma G et al. Cellular stress conditions are reflected in the protein and RNA content of endothelial cell-derived exosomes. J Extracell Vesicles. 2012; 1 18396, doi: http://dx.doi.org/10.3402/jev.v1i0.18396.
- Ekström K, Valadi H, Sjöstrand M, Malmhäll C, Bossios A, Eldh M et al. Characterization of mRNA and microRNA in human mast cell-derived exosomes and their transfer to other mast cells and blood CD34 progenitor cells. J Extracell Vesicles. 2012; 1 18389, doi: http://dx.doi.org/10.3402/jev.v1i0.18389.
- Ameisen JC, Guy B, Chamaret S, Loche M, Mach B, Tartar A et al. Persistent antibody response to the HIV-1-negative regulatory factor in HIV-1-infected seronegative persons. N Engl J Med. 1989; 320: 251–2.
- Svensson KJ, Christianson HC, Wittrup A, Bourseau-Guilmain E, Lindqvist E, Svensson LM et al. Exosome uptake depends on ERK1/2-heat shock protein 27 signaling and lipid Raft-mediated endocytosis negatively regulated by caveolin-1. J Biol Chem. 2013; 288: 17713–24.
- Song L, Gao S, Jiang W, Chen S, Liu Y, Zhou L et al. Silencing suppressors: viral weapons for countering host cell defenses. Protein Cell. 2011; 2: 273–81.
- Wang X, Ye L, Hou W, Zhou Y, Wang YJ, Metzger DS et al. Cellular microRNA expression correlates with susceptibility of monocytes/macrophages to HIV-1 infection. Blood. 2009; 113: 671–4.
- Yeung ML, Bennasser Y, Watashi K, Le SY, Houzet L, Jeang KT. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res. 2009; 37: 6575–86.
- Sun G, Li H, Wu X, Covarrubias M, Scherer L, Meinking K et al. Interplay between HIV-1 infection and host microRNAs. Nucleic Acids Res. 2012; 40: 2181–96.
- Aqil M, Naqvi AR, Bano AS, Jameel S. The HIV-1 Nef protein binds argonaute-2 and functions as a viral suppressor of RNA interference. PLoS One. 2013; 8: e74472.
- Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006; Chapter 3 Unit 3 22.
- Dweep H, Sticht C, Pandey P, Gretz N. miRWalk – database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011; 44: 839–47.
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009; 37: 1–13.
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009; 4: 44–57.
- Rusinov V, Baev V, Minkov IN, Tabler M. MicroInspector: a web tool for detection of miRNA binding sites in an RNA sequence. Nucleic Acids Res. 2005; 33: W696–700.
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004; 10: 1507–17.
- Olivetta E, Percario Z, Fiorucci G, Mattia G, Schiavoni I, Dennis C et al. HIV-1 Nef induces the release of inflammatory factors from human monocyte/macrophages: involvement of Nef endocytotic signals and NF-kappa B activation. J Immunol. 2003; 170: 1716–27.
- Lindsay MA. microRNAs and the immune response. Trends Immunol. 2008; 29: 343–51.
- Stumptner-Cuvelette P, Jouve M, Helft J, Dugast M, Glouzman AS, Jooss K et al. Human immunodeficiency virus-1 Nef expression induces intracellular accumulation of multivesicular bodies and major histocompatibility complex class II complexes: potential role of phosphatidylinositol 3-kinase. Mol Biol Cell. 2003; 14: 4857–70.
- Muratori C, Cavallin LE, Kratzel K, Tinari A, De Milito A, Fais S et al. Massive secretion by T cells is caused by HIV Nef in infected cells and by Nef transfer to bystander cells. Cell Host Microbe. 2009; 6: 218–30.
- Costa LJ, Chen N, Lopes A, Aguiar RS, Tanuri A, Plemenitas A et al. Interactions between Nef and AIP1 proliferate multivesicular bodies and facilitate egress of HIV-1. Retrovirology. 2006; 3: 33.
- Turchinovich A, Samatov TR, Tonevitsky AG, Burwinkel B. Circulating miRNAs: cell–cell communication function?. Front Genet. 2013; 4: 119.
- Yao B, La LB, Chen YC, Chang LJ, Chan EK. Defining a new role of GW182 in maintaining miRNA stability. EMBO Rep. 2012; 13: 1102–8.
- Witwer KW, Watson AK, Blankson JN, Clements JE. Relationships of PBMC microRNA expression, plasma viral load, and CD4+ T-cell count in HIV-1-infected elite suppressors and viremic patients. Retrovirology. 2012; 9: 5.
- Chan EK, Ceribelli A, Satoh M. MicroRNA-146a in autoimmunity and innate immune responses. Ann Rheum Dis. 2013; 72: ii90–5.
- Nahid MA, Pauley KM, Satoh M, Chan EK. miR-146a is critical for endotoxin-induced tolerance: implication in innate immunity. J Biol Chem. 2009; 284: 34590–9.
- Pauley KM, Satoh M, Pauley BA, Dominguez-Gutierrez PR, Wallet SM, Holliday LS et al. Formation of GW/P bodies as marker for microRNA-mediated regulation of innate immune signaling in THP-1 cells. Immunol Cell Biol. 2010; 88: 205–12.
- Tan Gana NH, Onuki T, Victoriano AF, Okamoto T. MicroRNAs in HIV-1 infection: an integration of viral and cellular interaction at the genomic level. Front Microbiol. 2012; 3: 306.
- Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009; 11: 1143–9.
- Lee YS, Pressman S, Andress AP, Kim K, White JL, Cassidy JJ et al. Silencing by small RNAs is linked to endosomal trafficking. Nat Cell Biol. 2009; 11: 1150–6.