Abstract
Background
Extracellular vesicles (EVs) are important mediators of cell-to-cell communication in healthy and pathological environments. Because EVs are present in a variety of biological fluids and contain molecular signatures of their cell or tissue of origin, they have great diagnostic and prognostic value. The ability of EVs to deliver biologically active proteins, RNAs and lipids to cells has generated interest in developing novel therapeutics. Despite their potential medical use, many of the mechanisms underlying EV biogenesis and secretion remain unknown.
Methods
Here, we characterized vesicle secretion across the NCI-60 panel of human cancer cells by nanoparticle tracking analysis. Using CellMiner, the quantity of EVs secreted by each cell line was compared to reference transcriptomics data to identify gene products associated with vesicle secretion.
Results
Gene products positively associated with the quantity of exosomal-sized vesicles included vesicular trafficking classes of proteins with Rab GTPase function and sphingolipid metabolism. Positive correlates of larger microvesicle-sized vesicle secretion included gene products involved in cytoskeletal dynamics and exocytosis, as well as Rab GTPase activation. One of the identified targets, CD63, was further evaluated for its role in vesicle secretion. Clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 knockout of the CD63 gene in HEK293 cells resulted in a decrease in small vesicle secretion, suggesting the importance of CD63 in exosome biogenesis.
Conclusion
These observations reveal new insights into genes involved in exosome and microvesicle formation, and may provide a means to distinguish EV sub-populations. This study offers a foundation for further exploration of targets involved in EV biogenesis and secretion.
To access the supplementary material to this article, please see Supplementary files under ‘Article Tools’.
Extracellular vesicles (EVs) are 40–1,000 nm membrane-bound delivery pods released from cells that have been implicated in many biological processes including the mediation of intercellular communication (Citation1, Citation2) and immune responses (Citation3–Citation5). These secreted vesicles have also been shown to play a role in tumour growth and metastasis (Citation6, Citation7), neurodegenerative diseases (Citation8, Citation9) and transfer of oncogenic material (Citation10–Citation13). Currently, EVs are divided into 2 major groups of interest: microvesicles (MVs) and exosomes, and are distinguished from one another by size, site of biogenesis and content. Recent research has focused on the role of EVs in pathological conditions and their potential use as biomarkers to diagnose cancer (Citation14–Citation16) and other diseases (Citation17). Evidence indicates that matrix metalloproteinases (MMPs) are selectively enriched in MVs (Citation18), suggesting the importance of these budding vesicles in extracellular matrix degradation. In the context of cancer, MVs have been reported to enable tumour invasion and metastasis through the activation of protease cargo (Citation19, Citation20). Exosomes have also been demonstrated to play a role in a variety of neoplastic processes through the selective packaging of oncoproteins into exosomes (Citation21). However, despite these important discoveries surrounding the roles of EV in disease, there is only limited understanding of the physiological functions of exosomes and MVs and the mechanisms governing their formation and release.
In general, exosomes and MVs are thought to have distinct mechanisms of formation within a cell. MVs, the larger of the 2 subtypes, are formed by outward budding of the plasma membrane directly into the extracellular environment (Citation22). It has been hypothesized that many of the mechanisms governing MV release from the cell surface resemble those used in retrovirus budding (Citation23). A growing body of research has suggested that these membrane fission events are likely mediated in part by lipid raft microdomain interactions and may be calcium dependent (Citation24, Citation25). The cytoskeleton is also likely to play a role in MV shedding; indeed, evidence shows that F-actin dynamics contribute to the morphological and structural reorganization within the cell that results in vesicle release (Citation26). Further involvement of contractile myosin proteins and small GTPases may facilitate membrane pinching and fission (Citation27).
Contrary to MVs, exosomes are generally believed to be smaller than 150 nm in size and generated at late endosomal membranes by inward budding events that produce intraluminal vesicles (ILVs) – the intracellular precursors of exosomes. Late endosomes containing ILVs are called multivesicular bodies (MVBs) (Citation28, Citation29). Protein, lipid and nucleic acid cargo of ILVs originates from the Golgi or endosomal pathway and is delivered to MVBs, which in turn fuse with the plasma membrane, releasing ILVs as exosomes into the extracellular space. Alternatively, MVBs that do not fuse with the plasma membrane can be divergently targeted to lysosomes for degradation (Citation30).
Several general mechanisms have been proposed to explain the regulation of protein-cargo trafficking and vesicle budding into the late endosome lumen. MVB regulation is classically thought to involve endosomal sorting complexes required for transport (ESCRT) proteins. The ESCRT multi-subunit machineries are evolutionarily conserved complexes that assemble on the endosome and guide inward budding of the late endosomal membrane into the lumen. The complexes are known to localize to endosomes by binding phosphatidylinositol-3-phosphate and ubiquitinated tails of transmembrane proteins, which directs trafficking to the ILV (Citation3, Citation31) (Citation32). Accessory proteins such as Alix can further interact with syndecan heparan sulphate residues and the adaptor protein syntenin to modulate cargo trafficking to vesicles and budding of late endosomal membranes (Citation33). Yet while the ESCRT machinery has been demonstrated to play a key role in endosomal cargo transport and MVB formation, it is not known how ESCRT protein interactions induce inward budding of ILVs.
There is also evidence that supports an ESCRT-independent mechanism of protein packaging that instead depends on ceramide-induced budding of ILVs through lipid raft microdomains (Citation34, Citation35). Tetraspanin proteins such as CD9, CD63 and CD81 that are enriched in membrane microdomains and in exosomes are often used as exosomal biomarkers and have been proposed to aid in ESCRT – dependent or independent – vesicle biogenesis (Citation36–Citation38). In particular, CD63 co-accumulates in vesicle populations with syndecan, syntenin and Alix, suggesting a potential role for these interactions in the formation of a unique population of exosomes (Citation33). Soluble N-ethlymaleimide-sensitive factor attachment protein receptor (SNARE) proteins also localize to lipid raft domains and exosomes with tetraspanins like CD63 (Citation39–Citation41). As SNARE proteins facilitate vesicle docking and fusion events in the endocytic pathway, these proteins are also likely involved in exosome biogenesis (Citation42).
In addition to SNARE proteins, several small GTPases are important in eukaryotic cargo transport between membrane-bound organelles, demonstrating roles in vesicle budding, uncoating, motility, tethering and fusion. Among these, Rab11, Rab35 and Rab27 stimulate tethering and fusion of MVBs to the plasma membrane (Citation43–Citation45). Additionally, Rab2, Rab4, Rab7 and Rab9 are proposed to be involved in vesicular trafficking and exosome release (Citation46, Citation47).
It is clear that multiple pathways likely orchestrate EV biogenesis. However, despite the fact that these vesicles likely have distinct functions (Citation48), many of the specific mechanisms guiding these events remain enigmatic. Further discoveries surrounding the biogenesis of exosomes and MVs will undoubtedly lead to a greater understanding of the diseases and intrinsic physiological processes to which they contribute. Therefore, it is crucial to determine the essential components involved in the origin and secretion of EVs.
In this study, we measured the size and quantity of EVs secreted from an array of human cancer cell lines. The panel of NCI-60 cells represents a diverse group of human cancers compiled by the Developmental Therapeutics Program of the U.S. National Cancer Institute. Originally assembled in the late 1980s for anticancer pharmaceutical research, the panel has more recently become a public high-throughput screening tool in the cancer research community. Since the open dissemination of these cells lines, several data-mining websites have been created, providing tools for the interpretation of genetic information associated with the NCI-60. Here, we used CellMiner (Citation49–Citation51), an online database created by the Genomics and Bioinformatics Group (DTB, LMP, CCR, National Cancer Institute), to examine the relationship between measured EV secretion and accessible gene expression data. The web application includes transcript expression data from 4 independent genome microarray platforms that were utilized in this study.
Our rationale for examining the EVs secreted by the panel of NCI-60 cells was 3 fold. First, evidence indicates that EVs play a role in cancer metastasis and tumour growth. Thus, we reasoned that a diverse group of cells harbouring various metastatic tendencies, patterns of aggressiveness and tumour grades would display varying patterns of EV secretion. Secondly, several publically available analytical tools have been designed for the panel of NCI-60 cells with the goal of high-throughput screening of pharmaceutical targets as well as genomic and proteomic analyses (Citation52–Citation54). And lastly, because these cells lines have been well described in the literature, many investigators are currently utilizing cell lines from the panel to study EV function and content. Therefore, our characterization of the EV secretion quantities across the NCI-60 panel provides valuable information when choosing cell lines to culture. For instance, in experiments requiring large amounts of vesicles, it might behove the researcher to identify and culture cell lines secreting higher levels of EVs, easing the burden of collecting sufficient vesicular material for innovative downstream analyses.
To characterize EV secretion across an extensive array of human cancer cells, we utilized a method of EV enrichment that yields a broad spectrum of vesicle populations for analysis. We quantified vesicles by nanoparticle tracking analysis (NTA), grouping particles by size to generate a set of quantities of exosomes and larger MVs. With the CellMiner analysis tools, we then performed a bioinformatics analysis to understand the associations between the quantity of EVs secreted by each cell type and gene transcript expression in the EV-producing cells. A list of candidate gene products involved in EV biogenesis and secretion was generated, and the utility of this screen was confirmed by functional analysis of CD63 knockout cells using a clustered regularly interspaced short palindromic repeat (CRISPR) and CRISPR-associated protein 9 (Cas9) gene editing technique (Citation55, Citation56). Further examination of the candidate effector proteins identified in the study will begin to answer the questions surrounding EV trafficking.
Materials and methods
Cell culture
HEK293 cells for preliminary growth and vesicle secretion experiments were cultured in DMEM medium (Lonza, 12-604Q). NCI-60 cell lines were grown in RPMI-1640 medium (Lonza, 12-702Q). Media were supplemented with a final concentration of 10% foetal bovine serum (FBS; Seradigm, 1400–500), 2 mM l-glutamine (Corning, 25-005-CI), 100 IU penicillin and streptomycin (Corning, 30-002-CI) and 100 IU: 100 µg/mL:0.25 µg/mL antiobiotic/antimycotic (Corning, 30-004-CI). Serum used for experiments was depleted of EVs by prior ultracentrifugation for 20 hours at 100,000 g (RCFavg). Cells were seeded based on a doubling time formula: C=S*2^(h/d), where C represents the number of cells at confluence, S represents the number of cells to be seeded, h represents the number of hours in culture and d represents the cell doubling time. Doubling times for each cell line were obtained from the NCI Developmental Therapeutics Program. To examine EV variation as a function of time and cell confluence, HEK293 cells were seeded at the same density (1.48×105 cells) for up to 6 days. Cell-conditioned media were collected from different plates at each time period. Thus, media collected on day 5, for example, represented EVs secreted by cells for the past 5 days. NCI-60 cells were seeded to achieve a confluent 9.62 cm2 well at the time of harvest, 96 hours after seeding, whereupon cell-conditioned media were collected and processed for EV enrichment. Live cell count, cell size and viability were measured at the time of harvest by staining cells with 0.2% trypan blue (Sigma, T8154) and analysing with an automated cell counter (Cellometer Vision, software version 2.1.4.2, Nexcelom Biosciences). For each cell line, three 9.62 cm2 wells were cultured, and media were enriched for EVs separately. To account for differences in cell number per surface area, particles measured by NTA after EV enrichment were divided by the total number of live cells counted at the time of harvest.
EV enrichment
Vesicles were enriched using an adaptable precipitation-based protocol developed in the laboratory using previously described techniques for virus isolation (Citation57). At higher concentrations of polymer (12%), this polyethylene glycol (PEG)-based method was demonstrated to effectively recover and concentrate all particles present in the cell-conditioned media before treatment. Similarly, levels of vesicular protein markers were highest with a final concentration of 12% PEG. As such, we determined this method as the most appropriate method for efficiently harvesting EVs from many cell lines and for ensuring the broadest spectrum of vesicle population recovery necessary for our later analyses. Briefly, after 4 days of culture, cell-conditioned media were centrifuged at 500 g for 5 minutes at 4°C to remove cells followed by 2,000 g for 30 minutes at 4°C to remove cell debris and vesicles larger than the expected size of exosomes and MVs. An equal volume of concentrated (2×) PEG Mn 6000 (Sigma-Aldrich 81260) with 1 M NaCl was added for a final PEG concentration of 12%. Samples were mixed by inversion and incubated for 16 hours at 4°C. Following the overnight incubation, samples were centrifuged in a bench-top centrifuge (Eppendorf 5810R) using a swing bucket rotor (S-4-104) at maximum speed (3,214 g) for 1 hour at 4°C. Supernatant was removed, and pellets were re-suspended in 200 µL of sterile filtered phosphate-buffered saline (PBS) (pH 7.4) and stored at −80°C until NTA.
Iodixanol density gradient purification
To confirm variation in particle secretion seen across the NCI-60 panel using PEG-based precipitation, EVs from cell lines secreting relatively high (SF268) and low (MCF7) quantities of vesicles were purified on an iodixanol (Optiprep) density gradient. For this experiment, EVs were enriched by PEG as described above. Following the 3,214 g centrifugation for 1 hour, EV pellets were suspended in 500 µL of particle-free PBS. Iodixanol solutions were diluted in a 0.25 M sucrose (10 mM Tris-HCl, pH 7.4) buffer to make 5, 10, 20 and 40% (w/v) iodixanol concentrations. Solutions were layered from bottom to top as follows: 3 mL of 40%, 3 mL of 20%, 3 mL of 10% and 2.5 mL of 5%. EV samples were layered on top of the gradient and ultracentrifuged at 100,000 g for 18 hours. Gradients were separated into 1 mL fractions, and refractive density was measured with a refractometer (Refracto 30PX). The fractions were then diluted in 10 mL of PBS and concentrated by a final ultracentrifugation of 100,000 g for 70 minutes. Pellets were suspended in 50 µL of particle-free PBS for NTA.
Nanoparticle tracking analysis
After vesicle enrichment from cancer cell lines, EVs were measured by NTA to quantify the mode and mean sizes and concentration of particles harvested. A Malvern NanoSight LM10 instrument with scientific CMOS sensor and a 488 nm laser was used. Samples were prepared by making dilutions between 1:250 and 1:1,000 in particle-free PBS to an acceptable concentration, according to the manufacturer's recommendations. For each biological replicate, 3 technical reads were measured. The instrument laser chamber was cleaned thoroughly between each sample. Camera shutter speed was set at 30.00 ms. NTA 3.1 software was used to process the 60-second video files of particles in suspension. Camera level was set to 13 and threshold to 3 for all samples.
Western blot analysis
Western blot analysis was performed to confirm exosome and MV protein markers. To facilitate lysis in a small volume for gel loading, PEG-precipitated samples were ultracentrifuged at 100,000 g (RCFavg) for 70 minutes at 4°C in a TLA120.2 rotor before immunoblot analysis, as previously described (Citation57). EVs were lysed by adding a strong lysis buffer containing 5% sodium dodecyl sulphate (SDS), 10 mM EDTA, 120 mM Tris-HCl, pH 6.8, 2.5% 2-mercaptoethanol and 8 M urea. Cell lysates were prepared in radioimmunoprecipitation assay (RIPA) buffer containing 20 mM Tris-Hcl, 150 mM NaCl, 1% NP-40, 0.1% SDS and 0.5% deoxycholate. Equal volume of EV lysate was loaded into an SDS 10% polyacrylamide gel and transferred onto a 0.45 µm pore size nitrocellulose membrane (GE Healthcare). Blots were blocked for 1 hour at room temperature in 5% (w/v) non-fat dry milk powder suspended in tris-buffered saline with tween-20 (TBS-T) and subsequently probed with the following primary antibodies, diluted as indicated in 5% milk/TBS-T solution: Alix (1:1,000; Q-19; Santa Cruz Biotechnology), TSG101 (1:1,000; C-2; Santa Cruz), HSC70 (1:6,000; B-6; Santa Cruz), Calnexin (1:1,000; H-70; Santa Cruz), MMP-2 (1:500; H-76; Santa Cruz), CD81 (1:500; H-121; Santa Cruz) and CD63 (1:3,000; TS63; Abcam). When probing for CD63, reducing agents were left out of lysis and sample buffers. After four 5-minute washes in TBS-T, blots were incubated with the following secondary antibodies, diluted as indicated in 5% milk/TBS-T: HRP-conjugated rabbit anti-goat IgG (1:10,000; Genetex), HRP-conjugated rabbit anti-mouse IgG (1:3,000; Genetex) and HRP-conjugated goat anti-rabbit IgG (Fab fragment) (1:3,000; Genetex). Chemiluminescent blot images were taken following four 5-minute washes in TBS-T and incubation with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, #1856136) or VisiGlo Select HRP Chemiluminescent Substrate (Amresco) using the ImageQuant LAS4000 instrument (GE Healthcare) and processed with ImageQuant TL v8.1.0.0 software and CorelDRAW Graphics Suite X5.
EXOCET exosome quantification
To further confirm variation seen in vesicle secretion, EV samples were measured using the EXOCET exosome quantification kit (SBI EXOCET96A-1). Samples were prepared following PEG precipitation according to kit instructions and analysed as relative secretion compared to MCF7 EV quantity.
Bioinformatics analysis of EV quantities
To generate a list of candidate genes involved in EV biogenesis and release, the number of vesicles secreted per cell across 58 cancer cell lines from the NCI-60 panel was analysed using CellMiner (www.discover.nci.nih.gov/cellminer/), a data-mining website compiled by the Genomics and Bioinformatics Group (DTB, LMP, CCR, National Cancer Institute). Genomic data from cell line MDA-MB-468 were not included in the CellMiner database and were therefore omitted from these analyses. Cell line RXF393 was also excluded from the study due to very low cell counts and inconsistent particle reads. Analyses were conducted with the intention of correlating transcriptomic data to secretion of specific EV populations. Because exosomes are generally classified as being smaller than 150 nm in diameter while MVs are predominately larger, particles per cell were partitioned into those ≤150 nm, and >150 nm in diameter, as measured by NTA. Small and large particles secreted per cell served as 2 distinct template files for molecular comparison using the CellMiner 60-element pattern comparison. Gene products significantly and positively correlating with the quantity of particles per cell across the cell lines were used as data sets for enrichment analysis. CellMiner output of gene transcripts was examined using FunRich 2.1.2 software (Citation58) containing the Vesiclepedia (Citation53) database of vesicle-associated gene products. Enrichment analyses of cellular component, molecular function and biological process were examined. Gene products previously found in exosomes and MVs were based on the Vesiclepedia database through selection of vesicle-specific content and the ExoCarta repository (Citation52, Citation59–Citation61). Gene product functions listed in Table and II were referenced from the UniProtKB/Swiss-Prot Knowledgebase. As negative (scrambled) controls, particles per cell in both vesicle size populations were randomized across the cell lines and used as template files for CellMiner analysis. Examination of enrichment categories revealed little to no overlap between gene products correlated with particles per cell and the respective scrambled control.
Table I. Gene products correlating with small vesicle quantity
Table II. Gene products correlating with large vesicle quantity
CRISPR/Cas9 CD63 knockout
The CD63 CRISPR was designed using the Zhang Lab protocol (Citation55) and MIT CRISPR design software (www.crispr.mit.edu/). The CRISPR guide oligos were annealed and ligated into the pLentiCRISPRv2 vector, a gift from Feng Zhang (Addgene plasmid # 52961). The plasmid was transformed using Endura E. coli (Lucigen) and sequenced to confirm its insertion. To generate lentivirus particles, HEK293-T cells were transfected with the CD63 CRISPR plasmid as well as pMD2.G, an envelope plasmid, and psPAX2, a lentiviral packaging plasmid (Didier Trono, Addgene plasmids #12259 and #12260, respectively). Resulting viral particles were harvested at 48 hours post-transfection and subsequently used to transduce HEK293 cells. The cells were incubated in viral medium containing 10 µg/mL polybrene (Sigma #107689) for 24 hours and then allowed to recover in complete growth medium (DMEM supplemented with 10% FBS). Cells were subcultured for 2 weeks in complete growth medium supplemented with 2 µg/mL puromycin (Amresco #J593) to select for transduced cells. Of these selected cells, single cell clones were generated by serial dilution in 96-well dishes and successful CD63 knockout was confirmed by western blot. Genomic DNA was isolated from the single cell population, cloned and prepared for sequencing analysis using the Guide-it Indel Identification Kit (Clontech, #631444). DNA sequencing analysis of cloned products confirmed that indels were generated at the targeted genomic site.
Statistical analysis
Statistical significance of results was evaluated by a repeated measures one-way analysis of variance (ANOVA) test with a post-hoc pairwise comparison (), a one-way ANOVA test ( and ) or Student's 2-sample t-tests () with confidence intervals of 95% using R statistical software version 3.2.2. Results in were analysed using linear regression. Data displayed in b–d were analysed using a forward selection multiple regression analysis in R to determine significant factors associated with the logarithm of the quantity of particles secreted per cell. This model was confirmed by a backwards elimination model. Both stepwise regression models were equivalent. Gene product compartmentalization and biological function () was determined in FunRich software using hypergeometric analysis where significance level was 0.01. Figures were produced using Microsoft Excel and CorelDraw X5 software.
Fig. 1. Extracellular vesicle levels are dependent upon cell culture time. HEK293 cells were grown in exosome-depleted 10% FBS-DMEM for 1–6 days. Cell-conditioned media were harvested and enriched for EVs at indicated time points. (a) Nanoparticle tracking analysis of the total number of particles after cumulative days in culture. (b) Number of live cells and the number of particles secreted per live cell over time in culture. Quantity of particles per cell on days 2–6 differed significantly from day 1, p<0.001. Live cell count reached steady state at day 4. Quantification of particles and cell counts were measured in triplicates for each day. Data are expressed as mean±s.d.
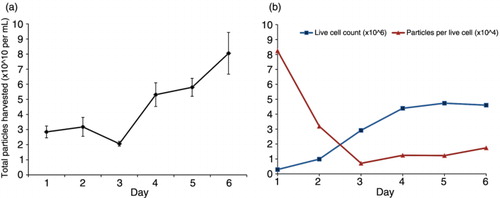
Fig. 2. Quantity of EVs secreted in vitro varies across different human cancer cell lines. Number of particles divided by live cell count at the time of harvest across (a) individual cell lines or (b) different tissue types. Data are expressed as mean±s.d. Significant variation in particle secretion among tissue types was determined by one-way analysis of variance, p<0.05. Leuk=leukaemia, Mel=melanoma.
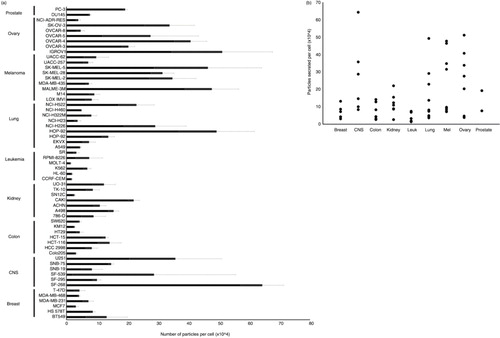
Fig. 3. Samples enriched for vesicles are the size of EVs. (a) Average mode size of EVs harvested from human cancer cell lines (NCI-60 panel) measured by NTA. Data expressed as mean±s.d. (b) Average mode and mean sizes of EV harvested by tissue type. Data expressed as mean±s.d. Significant variation in mode particle size across tissue types was determined by one-way analysis of variance, p<0.05.
Results
EV secretion varies by cell type
To determine whether cells from distinct cancers and tissues secrete different amounts or types of EVs, we compared the size and quantity of EVs across the NCI-60 cell panel. Since it has been suggested that EV abundance in cell culture medium varies with culture time and as a factor of confluence, we first investigated these culture conditions to optimize the yield of healthy vesicles. In HEK293 cultures, total particle numbers in the media were highest on days 4 through 6 (a). Interestingly, the number of particles per cell was highest on day 1 and decreased over time (b), suggesting that EV concentration in culture medium likely varies as a function of cell growth phase and confluence. At 4 days, when the cells reached confluency, live cell count and particles per cell appeared to reach equilibrium. We concluded that this time point was the most suitable to compare relative rates of EV secretion and uptake across a variety of cells.
Using this method, cancer cells were seeded based upon individual doubling times and cultured for 4 days to reach confluency. We found variation in the number of EVs secreted across the NCI-60 panel (a). The average number of EVs secreted was dependent upon tissue type (p<0.05) (b). Notably, all cell lines produced between 1×104 and 7×105 particles per cell. MOLT-4, a leukaemia cancer, produced the fewest number of EVs in culture, while SF268, a CNS cancer line, secreted the greatest number of EV.
In addition to particle counts across the NCI-60 panel, particle size was measured by NTA. Mode sizes of vesicles in our enrichment ranged from 95 to 157 nm, consistent with the reported size of EVs (a). Particle size was also tissue dependent (p<0.05) (b), suggesting tissue type is overall an important indicator of vesicle production and morphology in cancer cells.
To confirm the relative differences in vesicle quantity found among cells secreting various levels of EVs, we employed a highly pure method of EV enrichment. Vesicles from cell lines secreting high (SF268) and low (MCF7) quantities of vesicles were purified on an iodixanol density gradient following PEG-based precipitation. Evidence from our lab and others (Citation62–Citation65) has shown small EV enrichment at densities of 1.08–1.13 g/mL on a discontinuous iodixanol gradient, corresponding to fractions 5–7 by decreasing density. NTA of these combined fractions demonstrated similar relative levels of vesicle secretion following the density gradient purification step (a–b). However, fewer vesicles were recovered after subsequent iodixanol gradient purification compared to PEG purification alone. These absolute differences are likely due to the inefficiency in vesicle recovery following multiple steps of purification and the limited vesicle densities isolated. It should be noted that in this study, we did not extensively compare PEG-based isolation methods to other methods of purifying EVs across the NCI-60 panel, which may result in different quantities or sub-populations of vesicles enriched. Nevertheless, the relative differences in vesicle secretion were comparable regardless of the 2 enrichment technique used here.
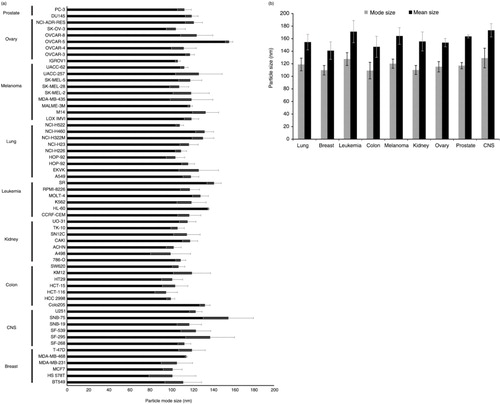
Fig. 4. Biochemical analyses of extracellular vesicles (EVs) demonstrate relative secretion levels across cancer cells. Quantity of particles per cell from SF268 and MCF7 cells following (a) PEG precipitation and (b) iodixanol gradient purification. (c) HEK293 cell lysate (CL) and representative EV lysates from cancer cells in the NCI-60 panel loaded by equal volume. EV lysates are enriched for exosomal markers Alix, TSG101, CD63 and CD81; microvesicle marker MMP-2; and positive for EV marker HSC70. Calnexin is found predominately in CL while minimal to undetectable levels are found in EV samples. (d) Quantification of relative EV levels from MCF7, SK-MEL-5 and OVCAR-5 cells by esterase activity.
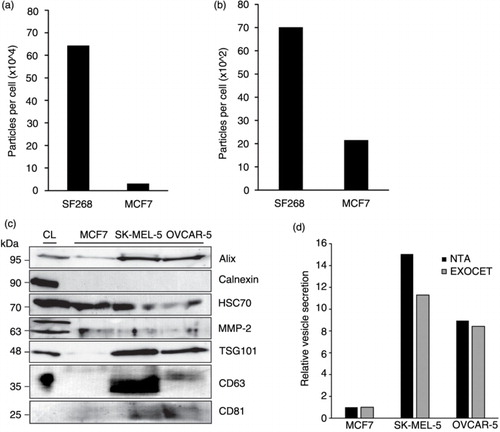
To identify the presence of vesicle populations, western blot analysis of EVs from cell lines exhibiting a range of secretion quantities and EV sizes was conducted. Vesicle lysates were collectively positive for the exosomal markers Alix, TSG101, CD63, CD81 and EV marker HSC70, as well as a MV marker, MMP-2. EV lysates were negative for calnexin, an ER/Golgi protein marker that is generally not associated with exosomes and microvesicles (c). When EV lysates were loaded by equal volume, levels of TSG101 and Alix reflected the number of particles per cell secreted by the respective cell lines; MCF7 exhibited the lowest levels of particles and corresponding protein markers, while higher particle secretion and protein signal were found in SK-MEL-5 samples. We also noted that the pro-peptide of MMP-2 seemed to be enriched in the HEK293 cell lysate, while the cleaved 63 kDa peptide was enriched specifically in the vesicle fraction. It has previously been suggested that MMPs are secreted as zymogens and cleaved outside the cell to undergo activation (Citation66). This observation suggests that EVs may be sites of activation for MMPs. Altogether, the varying levels of protein markers among the EV samples suggest that some cell lines secrete more of a particular vesicle subtype than others. However, many of these protein markers have been found in various vesicle types, and therefore, it remains difficult to guide conclusions about the quantity of EV sub-populations (Citation67). Regardless of these limitations, western blot analysis confirmed that the isolation method successfully yielded a range of vesicles containing exosomes and MVs.
To further verify the relative differences in vesicle secretion seen through nanoparticle tracking before and after iodixanol gradient purification, and through western blot analysis, we measured EVs secreted from a subset of cell lines by quantifying enzymatic esterase activity within vesicles. EVs have previously been shown to be enriched in acetylcholinesterase (Citation68); increasing levels of enzymatic activity can be correlated with vesicle number. Using this assay, we again observed similar trends in vesicle secretion between cell lines (d). EVs from MCF7 cells exhibited the lowest levels of esterase activity, while OVCAR-5 and SK-MEL-5 samples contained increasing levels of activity.
Cell size correlates with the amount of EVs secreted
In quantifying EVs, we controlled for the number of live cells at the time of harvest. However, a few cell lines exhibited viabilities <85% regardless of how the cells were maintained. As the presence of dying cells has previously been suggested to impact the populations of EVs secreted (Citation69), we wanted to ensure that differences in particle size were independent of any variation in cell viability. We reasoned that cells with lower viabilities may lead to enrichment of larger particle sizes, indicating the presence of larger vesicles such as apoptotic bodies. To test this, cell viability at the time of harvest was compared to particle size (a). The results of this comparison revealed no significant correlation between viability and EV size (p>0.05), suggesting that cell viability was not a determining factor in the sizes of vesicle populations in our samples.
Fig. 5. Cell size is a predictor of the number of EVs secreted per cancer cell. (a) EV mode size is not associated with cell viability measured at the time of harvest, p>0.05. Simple linear regression analysis was performed to examine the relationship between cell viability and particle size. (b) Cell viability does not correlate with particles per cell secreted, p>0.05. (c) An association between cell doubling time, in hours, and EV quantity was seen, p<0.01. (d) A multiple regression model was further used to determine the most significant factors associated with EV quantity. In this model, no significant association was seen between doubling time and EV number, p>0.05. However, cell size was positively associated with the number of particles per cell, p<0.001, adjusted R-squared=0.34. Vesicle quantity is expressed as the logarithm of the number of particles secreted per cell (× 104).
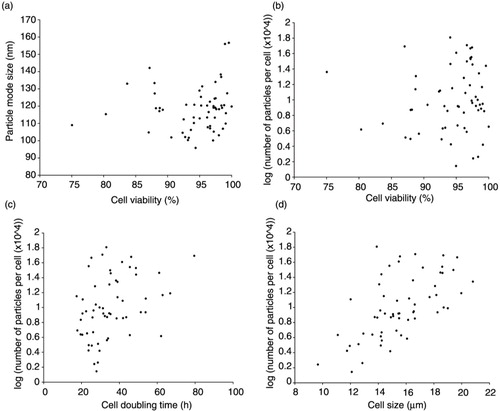
While cell viability was not associated with EV size, we further sought to ascertain whether viability may be correlated with the number of EV secreted. Moreover, because EVs are known to play a role in cell-to-cell communication and enhance growth of target cells, we similarly wanted to determine whether there was an association between the growth rate of cells (i.e. doubling time) and EV production. We also assessed the association between cell size and number of EV secreted per cell. In these analyses, no significant correlation was seen between the quantity of particles secreted per cell and cell viability (p>0.05) (b). Significant associations were seen between particle quantity and doubling time (p<0.01) (c) and between particle quantity and cell size (p<0.0001) (d). To further understand the impact of these variables (cell viability, doubling time and cell size) on particle quantity, a stepwise multiple regression analysis was conducted. The analysis suggested that cell size had the most significant impact on the number of particles secreted (p<0.001, adjusted R-squared=0.34). Cell doubling time and viability did not contribute significantly to the overall model (p>0.05). These findings suggest that cell size is a more important predictor than viability or growth rate of the number of vesicles secreted by a given cell. We hypothesize that larger cells likely contain more endosomal machinery and greater plasma membrane surface area and that these factors could result in increases in exosome and MV secretion, respectively.
CellMiner analysis of EV quantity suggests different genes are involved in exosome and MV biogenesis
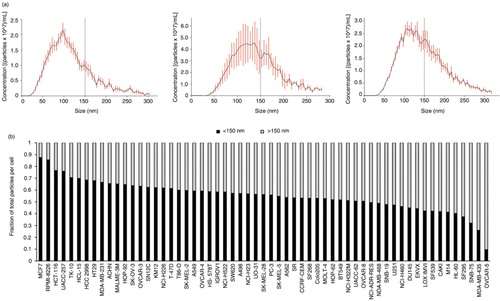
To discover potential genes regulating exosome and MV biogenesis, a bioinformatics approach was employed to examine the correlation of highly active genes with vesicle secretion. CellMiner was used to relate gene expression data on the NCI-60 panel to EVs secreted across the respective cancer cells. In order to determine which genes influence exosome versus MV formation, we first characterized the vesicles we harvested from each cell line. Because exosomes and MVs reportedly originate from different cellular locations, it has been hypothesized that mechanisms governing their budding and secretion may also be different. Our nanoparticle tracking data across the NCI-60 panel supported the presence of a variety of vesicle sub-populations. As exosomes are generally believed to be 40–150 nm in diameter (Citation59, Citation67) (Citation70), we counted the particles each cell line produced that were ≤150 nm to represent small exosomal-sized vesicles. Larger MV-sized vesicles were classified as >150 nm in size. Interestingly, the proportion of smaller and larger vesicles varied across different cell types (a–b), suggesting that some cells may preferentially secrete more of a particular EV population. For example, MCF7 cells mostly release exosome-sized vesicles, whereas OVCAR-5 cells shed predominately larger MV-sized vesicles.
Fig. 6. Cancer cells secrete distinct vesicle populations. (a) Representative NTA graphs of EV analysis from MCF7 (left), SK-MEL-5 (middle) and MD-MB-435 (right) show differences in the quantity of vesicles <150 nm and >150 nm. (b) Proportion of small (<150 nm) and large (>150 nm) vesicles to total number of EV secreted by each cell type varies across the NCI-60 panel.
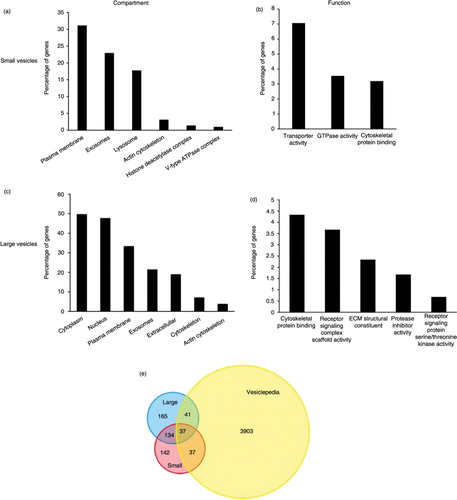
Analysis of particles per cell <150 nm in diameter using CellMiner revealed 350 genes for which transcript expression levels significantly and positively correlated with small vesicle quantity (Supplementary Fig. 1). To confirm the enrichment of gene products known to be involved in vesicular trafficking pathways and to assess candidate genes that may be involved in exosome biogenesis and regulation of secretion, positively correlating genes were cross-referenced with the Vesiclepedia database of vesicle-associated gene products (Supplementary Fig. 2). Of the 350 genes, 74 were previously identified as associated with EVs, and 69 of those have been shown to be found in exosomes specifically. Notably, enrichment analysis revealed that gene products were most significantly localized to the plasma membrane (p<0.01), exosomes (p<0.001) and lysosomes (p<0.01) (a). Molecular function analysis indicated enrichment of gene products involved in transporter activity (p<0.001), GTPase activity (p<0.01) and cytoskeletal protein binding (p<0.01) (b). A list of correlating genes to a scrambled control data set (see Materials and methods) generated by CellMiner (Supplementary Fig. 3) showed almost no commonality between enrichment of these categories (Supplementary Fig. 4). This negative control supports the enrichment of vesicular-related genes in our analysis.
Fig. 7. FunRich enrichment analysis shows that different genes are involved in exosome and microvesicle biogenesis and secretion. Enrichment of cellular components (a) and molecular function (b) corresponding to genes involved in small vesicle secretion. Cell compartment (c) and molecular function (d) enrichment of gene products mapped to large vesicle secretion. Reported p-values are based on hypergeometric tests calculated in FunRich software. Percentage of genes reflects the number of gene products within each category divided by the total significant genes generated by CellMiner analysis for each data set. (e) Venn diagram of overlapping genes found in the Vesiclepedia database.
Examination of these proteins revealed several novel candidates for exosome biogenesis (Table ) that warrant further exploration. Several proteins have been previously hypothesized to play a role in endocytosis and vesicular trafficking, including CD63, vacuolar protein sorting 41 (VPS41), sorting nexin 9 (SNX9), syntaxin 7 (STX7) and Golgi autoantigen (GOLGA4) (Citation71–Citation75). Among the significantly correlating targets, many gene products have been previously demonstrated to play a role in GTP binding and regulation of GTPase activity, including Rab17, Rab9A, Rab5B, E-Ras, M-Ras, ADP-ribosylation factor-like 8A (ARL8A), Ras protein activator like 2 (RASAL2), cytohesin 3 (CYTH3), amyotrophic lateral sclerosis 2 (ALS2) and rho GDP dissociation inhibitor alpha (ARHGDIA) (Citation47, Citation76–Citation80). These proteins may be directly involved in protein trafficking to endosomal compartments and mediate MVB fusion at the plasma membrane.
Other gene products significantly correlated with small vesicle secretion include hepatocyte growth factor-regulated tyrosine kinase substrate (HGS), a protein involved in ESCRT-0 sorting of cargo into MVBs (Citation32), and ATP6V0D2, the V0 subunit of a hydrogen ion transporting ATPase, a family of proteins which have been implicated in exosome secretion (Citation81). Phosphatidylinositol-3-phosphate-binding proteins such as SNX24, ubiquitinating proteins MARCH2 and SMURF1, and AP1S2, a protein implicated in recognizing cytosolic tail signals of transmembrane cargo molecules, suggest a more specific network by which many of these proteins may participate in cargo-specific endocytosis and trafficking to MVBs (Citation82). Interestingly, genes including NPC2 and PSAP are known to be involved in glycosphingolipid transport and cleavage, suggesting a role in ceramide-dependent MVB formation (Citation83–Citation85).
Moreover, the biological process of cell growth and maintenance was significantly enriched (p<0.01) with regard to genes present in this data set (Supplementary Fig. 2). This may suggest that gene transcripts positively correlating with exosome secretion may regulate processes of cell growth, including neoplastic development. Altogether, this analysis highlights many genes positively correlating with exosome quantity across the NCI-60 panel that have been found to associate with endosomal and lysosomal pathways.
Next, we evaluated candidate genes involved in larger vesicle budding, predominately MVs. The quantity of particles per cell >150 nm was analysed using CellMiner to generate a list of positively correlated gene transcripts (Supplementary Fig. 5). A total of 377 genes were determined to be significantly correlated with this data set. CellMiner output was again cross-referenced with Vesiclepedia to quantitatively evaluate gene products overlapping with those previously found in EVs. FunRich software revealed 78 corresponding proteins that have been found in EV, 40 of which have been specifically found in MVs. Enrichment analysis revealed that a majority of the gene protein products were located within the cytoplasm, (p<0.001), nucleus (p<0.01), plasma membrane (p<0.001) or cytoskeletal components (p<0.001) with some overlap in exosomal compartments (p<0.001) (c). Molecular function analysis showed significant enrichment of gene products involved in cytoskeletal protein binding (p<0.001). Receptor signalling activity (p=0.02), extracellular matrix structure (p=0.02) and protease inhibitor activity (p=0.02) were also enriched in genes correlating with large vesicle quantity, suggesting a closer tie to the plasma membrane from which MVs bud (d). Again, genes involved in cell growth and maintenance (p<0.001), as well as signal transduction (p<0.01) and cell communication (p<0.01), were highly enriched in these data set as well (Supplementary Fig. 6). Importantly, a scrambled negative control data set (Supplementary Fig. 7) failed to show any enrichment in the above-mentioned classes (Supplementary Fig. 8).
A multitude of the identified gene products have previously been implicated in general vesicle biogenesis and secretion (Table ). Here, our data indicate a more specific participation of these genes in MV formation. Many of the gene products were associated with cytoskeletal dynamics such as actin-binding proteins actinin, syntrophin and coactosin-like 1, or dynein and myosin proteins, suggesting the involvement of microfilaments and microtubules in the release of budding MVs from the cell surface. Interestingly, several of the gene products with GTPase functions such as cytohesin 3, ARHGD1A and ALS2 were found to be also associated with large vesicle secretion. These findings similarly imply a potential role of guanine nucleotide exchange in MV biogenesis.
Finally, the overlap of genes correlating with small and large vesicle quantity was compared (e). Of the 74 genes correlating with small EV secretion and found in Vesiclepedia, 37 were commonly associated with large vesicle quantity, while 37 were unique to small vesicle levels. Similarly, of the 78 genes correlated with larger vesicle quantity and found in Vesiclepedia, 41 were specific to EV secretion. Many more genes (134) were commonly associated with large and small vesicle levels that were not found in the Vesiclepedia database. These gene products likely regulate events within the cell required for EV formation and secretion. The commonality of genes correlating with exosome and MV secretion may reflect both overlapping vesicle size populations in our study, as well as similar mechanisms of vesicle budding and membrane fusion that guide formation of multiple vesicle populations. Overall, these analyses demonstrate the usefulness of this bioinformatics approach to screen for potential genes involved in exosome and MV secretion.
CD63 is important for small vesicle secretion
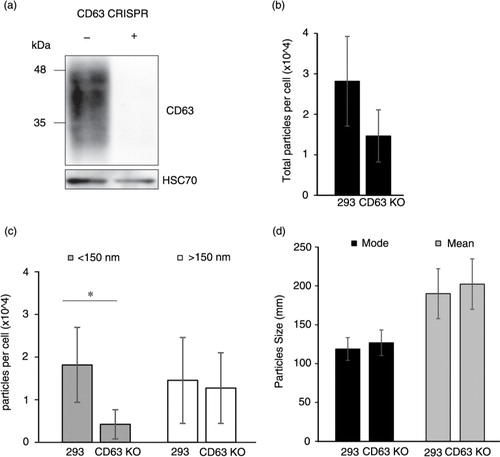
Our analysis clearly shows an enrichment of vesicle-related genes that we propose are involved in EV biogenesis and secretion. One of the candidate genes associated with small exosome-sized vesicle quantity, CD63, is a well-known exosomal marker. However, to our knowledge, the effect of CD63 knockout on exosome secretion has not been examined. To confirm the role of CD63 in exosome biogenesis, we designed a stable CD63 knockout cell line using the CRISPR/Cas9 system. Endogenous CD63 protein was effectively eliminated in HEK293 cells (a). We observed a reduction in particles secreted per cell by CD63 knockout cells compared with wild-type cells (b). Specifically, vesicles smaller than 150 nm in size were decreased in the medium of CD63 knockout cells (p<0.05) (c). Interestingly, no reduction in vesicles >150 nm was noted (p>0.05), supporting the observation that CD63 is important for exosomal-sized vesicle secretion. No overall change in mode (p>0.05) or mean (p>0.05) size of particles was noted (d). Cell viability and doubling time was not affected by CD63 knockout (Supplemental Fig. 9). These findings confirm the utility of our NTA and provide new insight into the significance of CD63 in exosomal biogenesis and secretion.
Fig. 8. CD63 CRISPR knockout results in decreased EV secretion. (a) Western blot demonstrating knockout (KO) of CD63 protein in cells with CRISPR/Cas9 targeting. (b) Nanoparticle tracking analysis of EV harvested from HEK293 and HEK293 CD63-KO cells showed a reduction in total particles per cell. (c) Specifically, particles per cell smaller than 150 nm were significantly decreased in CD63-KO cell-conditioned medium, p < 0.05. Larger particles (>150 nm) did not differ between cells. (d) Mode and mean sizes of EV harvested from HEK293 and HEK293 CD63-KO cells were not significantly different, p>0.05. Significant reduction in particle secretion was determined by a Student's t-test. Data are expressed as mean±s.d. and *denotes p<0.05.
Discussion
In this study, we characterized the quantity of EVs secreted by an extensive panel of human cancer cells originating from a variety of tissues and found large variations in the number of EVs harvested. This observation has multiple implications. First, the quantification of vesicles provides an extensive comparison of these valuable reference cell lines that will direct future investigation. For instance, MCF7, a well-known and commonly used cell line for exosome characterization, secretes among the lowest levels of EVs across the NCI-60 panel. These data explain, in part, why some cell lines require harvesting significantly more media in order to purify sufficient quantities of vesicular protein and can guide more informative decisions regarding the most efficient cells to culture for EV enrichment.
Secondly, the variation seen in EVs secreted may relate to the level of aggression of the cancer cell. The NCI-60 panel originates from primary or metastasized tumours of oncological patients. The cells express a diverse pattern of chromosomal alterations, tumour suppressor gene mutations, oncogene activation, and tumorigenic and invasive properties, representing an assortment of biological phenotypes. As cancer patients have been observed to have higher levels of circulating exosomes that facilitate metastasis (Citation86), we hypothesize that the level of aggression of a neoplastic cell may be associated with the number of EVs secreted. This phenomenon may facilitate cell-to-cell communication as well as tumour growth and metastasis in vivo. Although this study does not directly address the correlation between cancer cell aggressiveness and the number of EVs secreted, as comparative clinical data on these cell lines is limited, genes involved in cell growth and maintenance were enriched in our analyses, providing support for this hypothesis. Further exploration of the heterogeneity of EV secretion as it relates to cancer aggression across neoplastic cell lines is warranted.
Despite the potential implications of our findings, a major question still remains regarding the dynamic kinetics of cellular uptake and secretion of vesicles. In this study, EVs were purified at a single time point from each cell line, utilizing a method that isolates a broad spectrum of EV populations. Fluctuations in EV quantity between different cells were considered a measure of cellular vesicle secretion. However, higher levels of vesicles accumulating in the media may reflect either greater secretion of vesicles or decreased cellular uptake of vesicles. Our initial observations () suggest that cell growth conditions such as confluence and time in culture may influence the number of vesicles in the medium at any given time. To minimize the current limited understanding of vesicle dynamics, we harvested cell-conditioned media at a steady state of cell growth that demonstrated stable EV secretion. We further normalized particle numbers to live cell counts to account for differences in cell numbers at the same level of confluence in the culture well. Thus while much remains to be discovered about the rates of vesicle release and uptake, we believe our methods in this study overcome some of these limitations and provide valuable information about the variation in EV quantity across cells that can help to further understand these mechanisms in the future.
Historically, the inconsistency of EV isolation methods has hampered the reliable separation of vesicle populations to study these mechanisms. Here, we provide new and valuable information about genes involved in the formation of different vesicle populations, independent of the variable EV enrichment protocols encountered in community compendiums. We chose a method of EV enrichment that has been recently demonstrated to concentrate the vast majority of vesicles in cell-conditioned medium (Citation57). One limitation of PEG-based EV isolation is the presence of contaminating serum protein precipitation. To minimize the impact of potential protein aggregates in our bioinformatics analysis, identical cell culture conditions were maintained across the panel of cells, and relative differences in vesicle secretion were analysed by CellMiner. These variations in particle count were also seen by measuring esterase activity known to be contained within EVs. If an overwhelming amount of protein aggregates contributed to the vesicle enrichments, then similar relative differences in EV secretion and size would likely not have been observed across methods. Further purification of PEG-enriched vesicles on iodixanol density gradients revealed similar profiles of the relative differences in EV quantity between cell lines that were found to secrete low and higher levels of vesicles. However, the gradient purification, fractionation and subsequent ultracentrifugation wash step resulted in a loss of particles of up to 2 orders of magnitude. Furthermore, little has been described about the relative densities of different EV populations. Thus, while gradient purification remains one of the most pure methods of vesicle enrichment and allows separation of different EV populations, it also has limitations.
For the purpose of this study, we used a rapid and efficient precipitation method to analyse a broad spectrum of vesicles secreted by a large set of cancer cells, providing a wealth of information for future researchers in the field. Our screening approach has highlighted several candidate genes that may be involved in exosome and MV release. After differentiation of smaller and larger vesicle quantities per cell, analyses revealed subsets of genes positively correlating with exosome and MV abundance, respectively. Gene products associated with the number of smaller vesicles per cell appeared to be most significantly enriched in plasma membrane, lysosomal and endosomal structural compartments and functional pathways, consistent with the presumed cellular location for the biogenesis of these vesicles. Larger vesicle quantity was correlated with gene products found in the cytoplasm, on the plasma membrane, in vesicles or within the cellular cytoskeleton. Genes correlated with larger vesicle quantity were not enriched in lysosomal compartments, suggesting a pathway independent from that of exosomal biogenesis. Overall, these findings suggest enrichment of vesicular trafficking proteins in our study and provide insight into the mechanistic regulation of exosome and MV release or uptake.
While it is likely that there are different mechanisms orchestrating exosome and MV release from cells, in this study we also found genes commonly involved in small and large vesicle release. This may not be surprising, considering enveloped viruses that bud from the plasma membrane or at internal, endosomal-derived membranes, have been found to utilize similar ESCRT machinery (Citation11). In both small and large vesicles, enrichment of genes involved in endocytosis was found which could explain, in part, molecular mechanisms governing cargo trafficking to the site of vesicle formation. Specifically, the enrichment of multiple small Rab GTPases in our analyses highlights the importance of these factors in both exosome and MV biogenesis alike. Rab proteins are thought to orchestrate vesicular trafficking within a cell by acting as molecular switches recruiting effector molecules to guide vesicle movement throughout the endocytic pathway (Citation87). In this study, Rab5B, Rab9A and Rab17 expression levels correlated with small, exosome-sized vesicle secretion, whereas Rab32 was found to be specific for larger MV release. As upregulation of Rab proteins in cancer cells has been associated with increasing tumour aggressiveness and metastatic potential (Citation88, Citation89), a broad connection between vesicle formation and cancer growth therefore may result from the cellular activity of Rab proteins and the dysregulation of physiological exosome or MV functions.
Notably, many gene products previously implicated in exosome and MV biogenesis, including SNARE proteins, other ESCRT machinery components and additional tetraspanin proteins, were not found to be correlated with EV secretion in this study. For instance, Rab27, Rab11 and Rab35 are among the few proteins that have been directly linked with the quantity of vesicles secreted by cells (Citation44, Citation45) (Citation68, Citation90). However, many of these experiments have been limited to single cell lines and often involve inhibition of the GTPase protein function rather than reductions in transcript levels, as analysed in this study. Our data certainly does not negate the role of these proteins in EV biogenesis and secretion. Rather, here we present evidence supporting new players contributing to vesicle secretion among a wide variety of cells.
Gene expression of one of these players, CD63, was found to be significantly associated with small vesicle secretion. To confirm the utility of our nanoparticle screening assay in determining genes involved in small and large vesicle secretion, we generated a CD63 knockout cell line. CD63 is a highly conserved ubiquitous tetraspanin protein enriched in exosomal membranes (Citation36) and commonly used as an exosomal marker. Synthesized in the endoplasmic reticulum, CD63 undergoes post-translational palmitoylation in the Golgi and is trafficked to the plasma membrane, where further glycosylation occurs (Citation91). These modifications enable organization of tetraspanin-enriched microdomains (TEMs) on the cell membrane that allow interaction with integrin molecules (Citation92). While CD63 can localize to TEMs on the cell surface, the majority of the protein accumulates in the late endosome and lysosomal compartments. Though the cellular function of CD63 is unclear, recruitment of CD63 to immature exosomes and interaction between the tetraspanin and other membrane proteins suggest a role of the protein within the endocytic pathway (Citation71). Furthermore, a decrease in cell surface CD63 expression has been correlated with increased tumour aggressiveness in late stage cancer (Citation93–Citation96). Combined with an increase in circulating CD63-positive exosomes throughout cancer development, we hypothesize that trafficking of CD63 to ILVs from the cell surface may be involved in exosome biogenesis as well as tumour progression and metastasis.
Consistent with these results, exosome levels have been previously demonstrated to decrease following CD63 knockdown by shRNA in HEK293 cells (Citation97). In contrast, an independent study showed an increase in exosome secretion following CD63 knockdown by shRNA in dendritic cells (Citation98). However, in the latter study, exosome secretion was measured only by total protein content, while EM results did not confirm this increase in secretion. Here, we demonstrate a decrease in small vesicle secretion following complete CD63 knockout, providing evidence for the importance of this protein in EV biogenesis, especially in the context of oncogenic environments. The residual presence of EVs following removal of CD63 suggests possible competing or compensatory pathways of exosome or MV biogenesis. However, the unique reduction in small vesicle release following CD63 knockout supports a role for this protein specific to small EV or exosome biogenesis. It will now be important to determine the precise role of CD63 in small vesicle formation and the composition of CD63 containing vesicles.
Altogether, our data further provide a foundation from which many future studies can continue to explore the role of exosomes and MVs in both physiological conditions and in the context of cancer cell-to-cell communication. Future manipulation of the genes revealed in this study, as described for CD63, will begin to clarify the mechanisms orchestrating the biogenesis of EV populations and their functions in tumour progression.
Conflict of interest and funding
The authors declare no conflict of interest.
Supplementary figure 1
Download MS Excel (43.3 KB)Supplementary figure 2
Download MS Excel (12.2 KB)Supplementary figure 3
Download MS Excel (32.7 KB)Supplementary figure 4
Download MS Excel (11.1 KB)Supplementary figure 5
Download MS Excel (44.1 KB)Supplementary figure 6
Download MS Excel (13.8 KB)Supplementary figure 7
Download MS Excel (29.8 KB)Supplementary figure 8
Download MS Excel (20.1 KB)Supplementary figure 9
Download TIFF Image (8.1 MB)Acknowledgements
This study was supported by grants from the Florida Department of Health Bankhead-Coley Cancer Research Program (4BB05) and the National Institutes of Health (CA204621 and CA188941) awarded to D.G.M. Special thanks to Xia Lui for confirming the indels created in the CD63 CRISPR cell line by PCR and sequencing.
Notes
To access the supplementary material to this article, please see Supplementary files under ‘Article Tools’.
References
- Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010; 73: 1907–20. [PubMed Abstract].
- Théry C. Exosomes: secreted vesicles and intercellular communications. F1000 Biol Rep. 2011; 3: 15.
- Bobrie A, Colombo M, Raposo G, Théry C. Exosome secretion: molecular mechanisms and roles in immune responses. Traffic. 2011; 12: 1659–68. [PubMed Abstract].
- Graner MW, Alzate O, Dechkovskaia AM, Keene JD, Sampson JH, Mitchell DA, etal. Proteomic and immunologic analyses of brain tumor exosomes. FASEB J. 2009; 23: 1541–57. [PubMed Abstract] [PubMed CentralFull Text].
- Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002; 2: 569–79.
- Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK, etal. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015; 17: 816–26. [PubMed Abstract].
- Park JE, Tan HS, Datta A, Lai RC, Zhang H, Meng W, etal. Hypoxic tumor cell modulates its microenvironment to enhance angiogenic and metastatic potential by secretion of proteins and exosomes. Mol Cell Proteomics. 2010; 9: 1085–99. [PubMed Abstract] [PubMed CentralFull Text].
- Rajendran L, Honsho M, Zahn TR, Keller P, Geiger KD, Verkade P, etal. Alzheimer's disease beta-amyloid peptides are released in association with exosomes. Proc Natl Acad Sci USA. 2006; 103: 11172–7. [PubMed Abstract] [PubMed CentralFull Text].
- Pegtel DM, Peferoen L, Amor S. Extracellular vesicles as modulators of cell-to-cell communication in the healthy and diseased brain. Philos Trans R Soc Lond B Biol Sci. 2014; 369: 20130516
- Meckes DG, Shair KH, Marquitz AR, Kung CP, Edwards RH, Raab-Traub N. Human tumor virus utilizes exosomes for intercellular communication. Proc Natl Acad Sci USA. 2010; 107: 20370–5. [PubMed Abstract] [PubMed CentralFull Text].
- Meckes DG, Raab-Traub N. Microvesicles and viral infection. J Virol. 2011; 85: 12844–54. [PubMed Abstract] [PubMed CentralFull Text].
- Meckes DG. Exosomal communication goes viral. J Virol. 2015; 89: 5200–3. [PubMed Abstract] [PubMed CentralFull Text].
- Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, etal. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci USA. 2010; 107: 6328–33. [PubMed Abstract].
- Rabinowits G, Gerçel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009; 10: 42–6. [PubMed Abstract].
- Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics. 2009; 6: 267–83. [PubMed Abstract].
- Théry C. Cancer: diagnosis by extracellular vesicles. Nature. 2015; 523: 161–2.
- Boukouris S, Mathivanan S. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteomics Clin Appl. 2015; 9: 358–67. [PubMed Abstract].
- Taraboletti G, D'Ascenzo S, Borsotti P, Giavazzi R, Pavan A, Dolo V. Shedding of the matrix metalloproteinases MMP-2, MMP-9, and MT1-MMP as membrane vesicle-associated components by endothelial cells. Am J Pathol. 2002; 160: 673–80. [PubMed Abstract] [PubMed CentralFull Text].
- Dolo V, D'Ascenzo S, Violini S, Pompucci L, Festuccia C, Ginestra A, etal. Matrix-degrading proteinases are shed in membrane vesicles by ovarian cancer cells in vivo and in vitro. Clin Exp Metastasis. 1999; 17: 131–40. [PubMed Abstract].
- Ginestra A, La Placa MD, Saladino F, Cassarà D, Nagase H, Vittorelli ML. The amount and proteolytic content of vesicles shed by human cancer cell lines correlates with their in vitro invasiveness. Anticancer Res. 1998; 18: 3433–7. [PubMed Abstract].
- Keerthikumar S, Gangoda L, Liem M, Fonseka P, Atukorala I, Ozcitti C, etal. Proteogenomic analysis reveals exosomes are more oncogenic than ectosomes. Oncotarget. 2015; 6: 15375–96. [PubMed Abstract] [PubMed CentralFull Text].
- Heijnen HF, Schiel AE, Fijnheer R, Geuze HJ, Sixma JJ. Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood. 1999; 94: 3791–9. [PubMed Abstract].
- Gan X, Gould SJ. Identification of an inhibitory budding signal that blocks the release of HIV particles and exosome/microvesicle proteins. Mol Biol Cell. 2011; 22: 817–30. [PubMed Abstract] [PubMed CentralFull Text].
- Salzer U, Hinterdorfer P, Hunger U, Borken C, Prohaska R. Ca(++)-dependent vesicle release from erythrocytes involves stomatin-specific lipid rafts, synexin (annexin VII), and sorcin. Blood. 2002; 99: 2569–77. [PubMed Abstract].
- Muralidharan-Chari V, Clancy JW, Sedgwick A, D'Souza-Schorey C. Microvesicles: mediators of extracellular communication during cancer progression. J Cell Sci. 2010; 123: 1603–11. [PubMed Abstract] [PubMed CentralFull Text].
- Masi G, Mercati D, Vannuccini E, Paccagnini E, Riparbelli MG, Lupetti P, etal. p66Shc regulates vesicle-mediated secretion in mast cells by affecting F-actin dynamics. J Leukoc Biol. 2014; 95: 285–92. [PubMed Abstract].
- Muralidharan-Chari V, Clancy J, Plou C, Romao M, Chavrier P, Raposo G, etal. ARF6-regulated shedding of tumor cell-derived plasma membrane microvesicles. Curr Biol. 2009; 19: 1875–85. [PubMed Abstract] [PubMed CentralFull Text].
- Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983; 33: 967–78. [PubMed Abstract].
- Pan BT, Teng K, Wu C, Adam M, Johnstone RM. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J Cell Biol. 1985; 101: 942–8. [PubMed Abstract].
- Futter CE, Pearse A, Hewlett LJ, Hopkins CR. Multivesicular endosomes containing internalized EGF-EGF receptor complexes mature and then fuse directly with lysosomes. J Cell Biol. 1996; 132: 1011–23. [PubMed Abstract].
- Prag G, Watson H, Kim YC, Beach BM, Ghirlando R, Hummer G, etal. The Vps27/Hse1 complex is a GAT domain-based scaffold for ubiquitin-dependent sorting. Dev Cell. 2007; 12: 973–86. [PubMed Abstract] [PubMed CentralFull Text].
- Hurley JH. ESCRT complexes and the biogenesis of multivesicular bodies. Curr Opin Cell Biol. 2008; 20: 4–11. [PubMed Abstract] [PubMed CentralFull Text].
- Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, etal. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012; 14: 677–85. [PubMed Abstract].
- Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, etal. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008; 319: 1244–7. [PubMed Abstract].
- Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, etal. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci. 2013; 126: 5553–65. [PubMed Abstract].
- Escola JM, Kleijmeer MJ, Stoorvogel W, Griffith JM, Yoshie O, Geuze HJ. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J Biol Chem. 1998; 273: 20121–7. [PubMed Abstract].
- van Niel G, Charrin S, Simoes S, Romao M, Rochin L, Saftig P, etal. The tetraspanin CD63 regulates ESCRT-independent and-dependent endosomal sorting during melanogenesis. Dev Cell. 2011; 21: 708–21. [PubMed Abstract] [PubMed CentralFull Text].
- Andreu Z, Yáñez-Mó M. Tetraspanins in extracellular vesicle formation and function. Front Immunol. 2014; 5: 442. [PubMed Abstract] [PubMed CentralFull Text].
- Perez-Hernandez D, Gutiérrez-Vázquez C, Jorge I, López-Martín S, Ursa A, Sánchez-Madrid F, etal. The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J Biol Chem. 2013; 288: 11649–61. [PubMed Abstract] [PubMed CentralFull Text].
- Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014; 30: 255–89. [PubMed Abstract].
- Salaün C, Gould GW, Chamberlain LH. Lipid raft association of SNARE proteins regulates exocytosis in PC12 cells. J Biol Chem. 2005; 280: 19449–53.
- Hyenne V, Apaydin A, Rodriguez D, Spiegelhalter C, Hoff-Yoessle S, Diem M, etal. RAL-1 controls multivesicular body biogenesis and exosome secretion. J Cell Biol. 2015; 211: 27–37. [PubMed Abstract] [PubMed CentralFull Text].
- Savina A, Fader CM, Damiani MT, Colombo MI. Rab11 promotes docking and fusion of multivesicular bodies in a calcium-dependent manner. Traffic. 2005; 6: 131–43. [PubMed Abstract].
- Hsu C, Morohashi Y, Yoshimura S, Manrique-Hoyos N, Jung S, Lauterbach MA, etal. Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A-C. J Cell Biol. 2010; 189: 223–32. [PubMed Abstract] [PubMed CentralFull Text].
- Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, etal. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010; 12: 19–30; sup pp 1–13. [PubMed Abstract].
- Gillingham AK, Sinka R, Torres IL, Lilley KS, Munro S. Toward a comprehensive map of the effectors of rab GTPases. Dev Cell. 2014; 31: 358–73. [PubMed Abstract] [PubMed CentralFull Text].
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009; 10: 513–25. [PubMed Abstract].
- Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013; 200: 373–83. [PubMed Abstract] [PubMed CentralFull Text].
- Shankavaram UT, Varma S, Kane D, Sunshine M, Chary KK, Reinhold WC, etal. CellMiner: a relational database and query tool for the NCI-60 cancer cell lines. BMC Genomics. 2009; 10: 277. [PubMed Abstract] [PubMed CentralFull Text].
- Reinhold WC, Sunshine M, Liu H, Varma S, Kohn KW, Morris J, etal. CellMiner: a web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res. 2012; 72: 3499–511. [PubMed Abstract] [PubMed CentralFull Text].
- Genomics & Bioinformatics Group L, CCR, NCI. CellMiner. 2009. [Database Version:1.6.1] [Cited 2015 Nov 23]. Available from: http://discover.nci.nih.gov/cellminer/analysis.do.
- Simpson RJ, Kalra H, Mathivanan S. ExoCarta as a resource for exosomal research. J Extracell Vesicles. 2012; 1: 18374.
- Kalra H, Simpson RJ, Ji H, Aikawa E, Altevogt P, Askenase P, etal. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012; 10: e1001450. [PubMed Abstract] [PubMed CentralFull Text].
- Benito-Martin A, Peinado H. FunRich proteomics software analysis, let the fun begin!. Proteomics. 2015; 15: 2555–6. [PubMed Abstract].
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013; 8: 2281–308. [PubMed Abstract] [PubMed CentralFull Text].
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, etal. RNA-guided human genome engineering via Cas9. Science. 2013; 339: 823–6. [PubMed Abstract] [PubMed CentralFull Text].
- Rider MA, Hurwitz SN, Meckes DG. ExtraPEG: a polyethylene glycol-based method for enrichment of extracellular vesicles. Sci Rep. 2016; 6: 23978. [PubMed Abstract] [PubMed CentralFull Text].
- Pathan M, Keerthikumar S, Ang CS, Gangoda L, Quek CY, Williamson NA, etal. FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015; 15: 2597–601. [PubMed Abstract].
- Keerthikumar S, Chisanga D, Ariyaratne D, Al Saffar H, Anand S, Zhao K, etal. ExoCarta: a web-based compendium of exosomal cargo. J Mol Biol. 2015; 428:688–92
- Mathivanan S, Simpson RJ. ExoCarta: a compendium of exosomal proteins and RNA. Proteomics. 2009; 9: 4997–5000. [PubMed Abstract].
- Mathivanan S, Fahner CJ, Reid GE, Simpson RJ. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012; 40: D1241–4. [PubMed Abstract].
- Sung BH, Ketova T, Hoshino D, Zijlstra A, Weaver AM. Directional cell movement through tissues is controlled by exosome secretion. Nat Commun. 2015; 6: 7164. [PubMed Abstract] [PubMed CentralFull Text].
- Tauro BJ, Greening DW, Mathias RA, Ji H, Mathivanan S, Scott AM, etal. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods. 2012; 56: 293–304. [PubMed Abstract].
- Van Deun J, Mestdagh P, Sormunen R, Cocquyt V, Vermaelen K, Vandesompele J, etal. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J Extracell Vesicles. 2014; 3: 24858. http://dx.doi.org/10.3402/jev.v3.24858.
- Ji H, Greening DW, Barnes TW, Lim JW, Tauro BJ, Rai A, etal. Proteome profiling of exosomes derived from human primary and metastatic colorectal cancer cells reveal differential expression of key metastatic factors and signal transduction components. Proteomics. 2013; 13: 1672–86. [PubMed Abstract].
- Page-McCaw A, Ewald AJ, Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Biol. 2007; 8: 221–33. [PubMed Abstract] [PubMed CentralFull Text].
- Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, etal. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci USA. 2016; 113: E968–77. [PubMed Abstract].
- Savina A, Vidal M, Colombo MI. The exosome pathway in K562 cells is regulated by Rab11. J Cell Sci. 2002; 115: 2505–15. [PubMed Abstract].
- Théry C, Boussac M, Véron P, Ricciardi-Castagnoli P, Raposo G, Garin J, etal. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001; 166: 7309–18.
- Greening DW, Xu R, Ji H, Tauro BJ, Simpson RJ. A protocol for exosome isolation and characterization: evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Mol Biol. 2015; 1295: 179–209. [PubMed Abstract].
- Pols MS, Klumperman J. Trafficking and function of the tetraspanin CD63. Exp Cell Res. 2009; 315: 1584–92. [PubMed Abstract].
- Asensio CS, Sirkis DW, Maas JW, Egami K, To TL, Brodsky FM, etal. Self-assembly of VPS41 promotes sorting required for biogenesis of the regulated secretory pathway. Dev Cell. 2013; 27: 425–37. [PubMed Abstract] [PubMed CentralFull Text].
- Lundmark R, Carlsson SR. SNX9 – a prelude to vesicle release. J Cell Sci. 2009; 122(Pt 1): 5–11. [PubMed Abstract].
- Pryor PR, Mullock BM, Bright NA, Lindsay MR, Gray SR, Richardson SC, etal. Combinatorial SNARE complexes with VAMP7 or VAMP8 define different late endocytic fusion events. EMBO Rep. 2004; 5: 590–5. [PubMed Abstract] [PubMed CentralFull Text].
- Wong M, Munro S. Membrane trafficking. The specificity of vesicle traffic to the Golgi is encoded in the golgin coiled-coil proteins. Science. 2014; 346: 1256898. [PubMed Abstract] [PubMed CentralFull Text].
- Takahashi K, Mitsui K, Yamanaka S. Role of ERas in promoting tumour-like properties in mouse embryonic stem cells. Nature. 2003; 423: 541–5. [PubMed Abstract].
- Matsumoto K, Asano T, Endo T. Novel small GTPase M-Ras participates in reorganization of actin cytoskeleton. Oncogene. 1997; 15: 2409–17. [PubMed Abstract].
- Chavrier P, Goud B. The role of ARF and Rab GTPases in membrane transport. Curr Opin Cell Biol. 1999; 11: 466–75. [PubMed Abstract].
- Donaldson JG, Jackson CL. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Biol. 2011; 12: 362–75. [PubMed Abstract] [PubMed CentralFull Text].
- Lai C, Xie C, Shim H, Chandran J, Howell BW, Cai H. Regulation of endosomal motility and degradation by amyotrophic lateral sclerosis 2/alsin. Mol Brain. 2009; 2: 23. [PubMed Abstract] [PubMed CentralFull Text].
- Liégeois S, Benedetto A, Garnier JM, Schwab Y, Labouesse M. The V0-ATPase mediates apical secretion of exosomes containing Hedgehog-related proteins in Caenorhabditis elegans . J Cell Biol. 2006; 173: 949–61.
- Cullen PJ. Endosomal sorting and signalling: an emerging role for sorting nexins. Nat Rev Mol Cell Biol. 2008; 9: 574–82. [PubMed Abstract].
- Abdul-Hammed M, Breiden B, Adebayo MA, Babalola JO, Schwarzmann G, Sandhoff K. Role of endosomal membrane lipids and NPC2 in cholesterol transfer and membrane fusion. J Lipid Res. 2010; 51: 1747–60. [PubMed Abstract] [PubMed CentralFull Text].
- Gallala HD, Breiden B, Sandhoff K. Regulation of the NPC2 protein-mediated cholesterol trafficking by membrane lipids. J Neurochem. 2011; 116: 702–7. [PubMed Abstract].
- Carvelli L, Libin Y, Morales CR. Prosaposin: a protein with differential sorting and multiple functions. Histol Histopathol. 2015; 30: 647–60. [PubMed Abstract].
- Silva J, Garcia V, Rodriguez M, Compte M, Cisneros E, Veguillas P, etal. Analysis of exosome release and its prognostic value in human colorectal cancer. Genes Chromosomes Cancer. 2012; 51: 409–18. [PubMed Abstract].
- Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev. 2011; 91: 119–49. [PubMed Abstract] [PubMed CentralFull Text].
- Li Y, Meng X, Feng H, Zhang G, Liu C, Li P. Over-expression of the RAB5 gene in human lung adenocarcinoma cells with high metastatic potential. Chin Med Sci J. 1999; 14: 96–101. [PubMed Abstract].
- Fukui K, Tamura S, Wada A, Kamada Y, Igura T, Kiso S, etal. Expression of Rab5a in hepatocellular carcinoma: possible involvement in epidermal growth factor signaling. Hepatol Res. 2007; 37: 957–65. [PubMed Abstract].
- Li W, Hu Y, Jiang T, Han Y, Han G, Chen J, etal. Rab27A regulates exosome secretion from lung adenocarcinoma cells A549: involvement of EPI64. APMIS. 2014; 122: 1080–7. [PubMed Abstract].
- Yang X, Claas C, Kraeft SK, Chen LB, Wang Z, Kreidberg JA, etal. Palmitoylation of tetraspanin proteins: modulation of CD151 lateral interactions, subcellular distribution, and integrin-dependent cell morphology. Mol Biol Cell. 2002; 13: 767–81. [PubMed Abstract] [PubMed CentralFull Text].
- Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol. 2005; 6: 801–11. [PubMed Abstract].
- Zhijun X, Shulan Z, Zhuo Z. Expression and significance of the protein and mRNA of metastasis suppressor gene ME491/CD63 and integrin alpha5 in ovarian cancer tissues. Eur J Gynaecol Oncol. 2007; 28: 179–83. [PubMed Abstract].
- Kwon MS, Shin SH, Yim SH, Lee KY, Kang HM, Kim TM, etal. CD63 as a biomarker for predicting the clinical outcomes in adenocarcinoma of lung. Lung Cancer. 2007; 57: 46–53. [PubMed Abstract].
- Sauer G, Kurzeder C, Grundmann R, Kreienberg R, Zeillinger R, Deissler H. Expression of tetraspanin adaptor proteins below defined threshold values is associated with in vitro invasiveness of mammary carcinoma cells. Oncol Rep. 2003; 10: 405–10. [PubMed Abstract].
- Sordat I, Decraene C, Silvestre T, Petermann O, Auffray C, Piétu G, etal. Complementary DNA arrays identify CD63 tetraspanin and alpha3 integrin chain as differentially expressed in low and high metastatic human colon carcinoma cells. Lab Invest. 2002; 82: 1715–24. [PubMed Abstract].
- Verweij FJ, van Eijndhoven MA, Hopmans ES, Vendrig T, Wurdinger T, Cahir-McFarland E, etal. LMP1 association with CD63 in endosomes and secretion via exosomes limits constitutive NF-κB activation. EMBO J. 2011; 30: 2115–29. [PubMed Abstract] [PubMed CentralFull Text].
- Petersen SH, Odintsova E, Haigh TA, Rickinson AB, Taylor GS, Berditchevski F. The role of tetraspanin CD63 in antigen presentation via MHC class II. Eur J Immunol. 2011; 41: 2556–61. [PubMed Abstract].
