Abstract
Cancer cells secrete small membranous extracellular vesicles (EVs) into their microenvironment and circulation. These contain biomolecules, including proteins and microRNAs (miRNAs). Both circulating EVs and miRNAs have received much attention as biomarker candidates for non-invasive diagnostics. Here we describe a sensitive analytical method for isolation and subsequent miRNA profiling of epithelial-derived EVs from blood samples of patients with colorectal cancer (CRC). The epithelial-derived EVs were isolated by immunoaffinity-capture using the epithelial cell adhesion molecule (EpCAM) as marker. This approach mitigates some of the specificity issues observed in earlier studies of circulating miRNAs, in particular the negative influence of miRNAs released by erythrocytes, platelets and non-epithelial cells. By applying this method to 2 small-scale patient cohorts, we showed that blood plasma isolated from CRC patients prior to surgery contained elevated levels of 13 EpCAM+-EV miRNAs compared with healthy individuals. Upon surgical tumour removal, the plasma levels of 8 of these were reduced (miR-16-5p, miR-23a-3p, miR-23b-3p, miR-27a-3p, miR-27b-3p, miR-30b-5p, miR-30c-5p and miR-222-3p). These findings indicate that the miRNAs are of tumour origin and may have potential as non-invasive biomarkers for detection of CRC. This work describes a non-invasive blood-based method for sensitive detection of cancer with potential for clinical use in relation to diagnosis and screening. We used the method to study CRC; however, it is not restricted to this disease. It may in principle be used to study any cancer that release epithelial-derived EVs into circulation.
To access the supplementary material to this article, please see Supplementary files under ‘Article Tools’.
Colorectal cancer (CRC) is the third most common cancer and the fourth most common cause of cancer deaths worldwide (Citation1). Detection and treatment of CRC at its early stages is a key to reduce the number of cancer deaths (Citation2). The time frame for development from premalignant to malignant disease is typically 10–15 years in individuals with sporadic disease, making CRC an ideal target for early detection and intervention (Citation3). At present, the faecal occult blood test and the faecal immune test are the only clinically implemented non-invasive diagnostic tests for CRC (Citation4). Unfortunately, these tests have low compliance rates and mediocre sensitivities (40–90%) (Citation5). Consequently, the clinical sensitivity of these tests ranges between 12 and 76% meaning that a significant number of neoplastic large bowel lesions are undetected by these approaches (Citation5). Therefore, current research in the field is focused on developing screening concepts that are accurate as well as acceptable by the screening population. Such concepts include blood-based procedures for which compliance rates >90% have been reported (Citation6).
Circulating microRNAs (miRNAs) have received much attention as biomarker candidates for non-invasive diagnostics but are not yet in clinical use. Recent reviews and meta-analyses of circulating miRNA studies in CRC highlight that while a few miRNAs, including miR-21-5p, miR-29a-5p and 92a-5p, are frequently found to be dysregulated, there are also multiple studies with conflicting findings (Citation7–Citation9). This may in part be due to unrecognized or underestimated influence of sample processing and other pre-analytical variables on the measured circulating miRNA levels. MiRNAs are released into circulation by nearly all cell types, and it has been reported that the majority of the cell-free miRNAs found in blood originate from blood cells (Citation10). Moreover, studies have reported that procedures for sample collection and storage dramatically affect the levels of circulating miRNAs (Citation7). In particular, platelet contamination and haemolysis have been reported to affect miRNA results (Citation11–Citation13). Indeed, the aforementioned miRNAs miR-21-5p, miR-29a-5p and 92a-5p are present in red blood cells, and their plasma levels have been shown to be strongly affected by haemolysis (Citation10, Citation13) (Citation14).
MiRNAs in circulation show a remarkable stability (Citation15, Citation16). It has been suggested that they are protected against RNase-mediated degradation because they are bound by proteins such as argonaute-2 and HDL or encapsulated by membranous extracellular vesicles (EVs) (Citation17–Citation20).
EVs have recently been recognized for their intercellular signalling role in normal physiology (Citation21). Their abundance in circulation is increased upon disease, including cancer (Citation22–Citation25). EVs have been reported to carry surface proteins characteristic of the cell and tissue of origin. These specific surface proteins have potential to be used as tags to identify and isolate EVs originating from specific tissues. The approach has been exemplified by the use of antibodies directed against the epithelial cell adhesion molecule (EpCAM) to isolate epithelial-derived EVs (EpCAM+-EVs) from serum of lung and ovarian cancer patients (Citation22, Citation23) (Citation25). By themselves, the EpCAM+-EVs are difficult to use as biomarkers as their low abundance makes detection and quantification non-trivial. However, some of the biomolecules they carry, including miRNAs, may be sufficiently abundant to be readily detected and quantified by sensitive methods such as quantitative reverse transcription polymerase chain reaction (qRT-PCR). MicroRNA profiling of epithelial-derived EVs potentially overcomes some of the specificity issues observed when profiling miRNAs directly from plasma and serum. Consequently, miRNAs from EpCAM+-EVs have potential as cancer-specific biomarkers (Citation22, Citation23).
In this study, we aimed to develop a method for miRNA profiling of EpCAM+-EVs isolated from plasma and serum. This method was applied to samples from CRC patients and healthy controls, and the results showed that specific miRNAs were more abundant in plasma and serum EpCAM+-EVs from CRC patients than healthy controls. Furthermore, post-operation the EpCAM+-EV miRNA levels were significantly reduced indicating that the EpCAM+-EVs originated from the CRC tumours. The method developed and the miRNAs identified have potential for non-invasive diagnosis of CRC and for post-operation patient monitoring.
Materials and methods
Cell lines and culture conditions
The human colorectal adenocarcinoma cell line SW620 was cultured in Dulbecco's Modified Eagle's Medium (DMEM) with 10% foetal bovine serum (FCS) and 100 µg/mL penicillin–streptomycin (Gibco Invitrogen, Carlsbad, CA, USA). SW620 cells were seeded at 25×106 cells in a volume of 15 mL into the lower cell chamber of CELLine Adhere 1000 (CLAD1000) bioreactors (INTEGRA Biosciences AG, Zizers, Switzerland) and maintained at high culture density at 37°C in a 5% CO2 humidified incubator for high-yield epithelial-derived EV harvest as previously described (Citation26, Citation27). For EV harvests, the medium in the lower cell compartment of the CLAD1000 bioreactors was replaced with serum-free Advanced DMEM (AdvDMEM, Gibco Invitrogen), whereas the upper compartment contained DMEM supplemented with d-glucose (4.5 g/L) and 10% FCS. EV-containing medium from the lower chamber was harvested every 48–96 hours. Cell line authentication using Short Tandem Repeat (STR) profiling (Cell ID System, Promega, Madison, WI, USA) confirmed cell line identity.
Patient samples
For miRNA profiling of circulating EpCAM+-EVs, 2 patient cohorts were included. Cohort I consisted of 6 patients diagnosed with CRC and 5 healthy control individuals (Table ). The healthy controls were all asymptomatic and all underwent colonoscopy, which showed no bowel lesions. From patients and controls, 54 mL whole blood was collected. For the generation of EDTA-plasma and serum, 27 mL of whole blood was processed for each. Within 2 hours of the phlebotomy, all blood samples were centrifuged at 3,000 g for 10 minutes at room temperature after which plasma and serum were isolated and stored at −80°C until use. Twelve mL plasma and 12 mL serum were collected from all patients and controls. For CRC patients, the samples were collected prior to surgery. Cohort II consisted of 7 stage III CRC patients who were all treated surgically with curative intent and who were recurrence free for at least 36 months (Table ). For all 7 patients, 9-mL EDTA blood was collected prior to surgery (median 1 day prior) and again 6 months after surgery. These samples were processed as described for cohort I. A total of 2 mL plasma was collected from each visit. The study was conducted in accordance with the Helsinki Declaration. Informed written consent was obtained from all participants according to ethical regulations. The research protocol for cohort I was approved by the Central Denmark Region Committees on Biomedical Research Ethics (J. no. 1999/4678) and the protocol for cohort II by the Danish National Ethics Committee (H-3-2009-110).
Table I. Patient cohort I (CRC cases and healthy controls)
Table II. Patient cohort II (pre- and post-operation analysis)
EV purification
Samples of conditioned culture medium and plasma or serum were diluted 1:10 in Phosphate-buffered saline (PBS) pH 7.4 (without Ca2+ and Mg2+) and subjected to centrifugation at 400×g for 10 minutes to remove whole cells, 2,000×g for 15 minutes to remove cellular debris and 16,000×g for 30 minutes (2 hours for plasma and serum samples to compensate for their higher viscosity and slower sedimentation) to remove large membrane vesicles and apoptotic bodies. The supernatant was transferred to new tubes and subjected to ultracentrifugation at 100,000×g for 70 minutes to pellet EVs. EVs were rinsed in a large volume of PBS followed by centrifugation at 100,000×g for 70 minutes. All steps of differential centrifugation were carried out at 4°C. Resuspension of the EVs in 3 mL PBS (pH 7.4, without Ca2+ and Mg2+) was performed over 20 hours at 4°C with gentle vortexing. Aliquots were used for total protein measurement (Bradford), proximity ligation assay (PLA), immunoaffinity magnetic bead isolation, immunoblotting or fixation in 2% (w/v) glutaraldehyde in 0.04 M phosphate buffer for transmission electron microscopy (TEM).
Immunoblotting
Immunoblotting of cellular and exosomal protein content was conducted using primary antibodies against CD63 (BD Pharmingen), CD81 (Santa Cruz), HSP90 (Santa Cruz) and EpCAM (E144, Abcam) followed by incubation with appropriate HRP-conjugated secondary antibodies (DAKO A/S, Glostrup, Denmark).
Transmission electron microscopy
For TEM, EV pellets were fixed using 2% (w/v) glutaraldehyde in 0.04 M phosphate buffer, dehydrated and embedded in Epon using a Leica EM TP tissue processor (Leica Microsystems GmbH, Wetzlar, Germany). Ultrathin sections (65 nm) were cut on a Leica Ultracut UCT ultramicrotome (Leica Microsystems GmbH, Wetzlar, Germany). Sections were stained with 3% uranyl acetate in water and Reynold's lead citrate and examined in a Phillips EM 208 transmission electron microscope (FEI, OR, USA). A total of 32 EVs from SW620 cells were analysed to estimate vesicle size.
Proximity ligation assay
PLA was used to evaluate the expression of EpCAM on isolated EVs quantitatively. PLA was performed as previously described (Citation28, Citation29). SW620 EVs were serially diluted into a plasma background and subjected to differential centrifugation. EV resuspensions were concentrated by speed-vac prior to analysis. The used EpCAM probe-conjugated oligo sequences were: 5′oligo: 5′—TCACGGTAGCATAAGGTGCAGTACCCAAATAACGGTTCAC—3′ and 3′ oligo: 5′—GGCCTCCTCCAATTAAAG AATCACGATGAGACTGGATGAA—3′. Primer sequences used for qPCR were: Forward: 5′—CGGCCTCCTCC AATTAAAGAA—3′, Reverse: 5′—CGTGAACCGTTATTTGGGTAC—3′. Quantitative RT-PCR was performed in a total sample volume of 25 µL and PCR cycle conditions of 1 cycle at 95°C for 10 minutes followed by 40 cycles of 15 seconds at 95°C and 1 minute at 60°C and finally 10 minutes at 46°C. The experiment was replicated twice.
Generation of the SW620 EV spike-in series
Conditioned SW620 culture medium was harvested after 3 days of culture and cleared for cells, cellular debris, membrane vesicles and apoptotic bodies by differential centrifugation as described above. Three independent harvests were collected and used to assess the concentration of EVs by nanoparticle tracking analysis (NTA). On average, the 3 supernatants contained 91.2±3.42×109 EVs per mL (Supplementary Table I). The plasma and serum spike-in series were generated by spiking SW620 supernatant (from a fourth independent harvest) into either plasma or serum. Each series consisted of 6 dilution points generated by spiking 1 mL of plasma (or serum) with incrementing (5-fold) volumes of SW620 supernatant, starting with 0.64 µl (~58×106 vesicles) and ending with 2,000 µl (~182×109 vesicles). Serum-free AdvDMEM was added to reach a total volume of 3 mL of each spiked sample. The 2 spike-in series were used to optimize and to evaluate the performance of the immunoaffinity-capture technique.
Nanoparticle tracking analysis
Concentration and size of vesicles present in conditioned medium were analysed by NTA using the NanoSight LM10 system (NanoSight, Malvern Instruments, Malvern, UK) configured with a 405 nm laser and a high-sensitivity sCMOS camera (OrcaFlash2.8, Hamamatsu C11440). Videos were collected and analysed using the NTA software (version 3.1), with the minimal expected particle size, minimum track length and blur setting, all set to automatic. Ambient temperature was recorded manually and did not exceed 25°C. Each sample was diluted 1:100 in particle-free PBS and had a final volume of 1.0 mL. For each sample, 5 videos of 60 seconds duration were recorded generating replicate histograms that were averaged.
Immunoaffinity magnetic bead capture of EVs
Immunoaffinity magnetic bead capture of EVs from the clinical as well as the EV-spiked dilution series was performed in 2 steps. In step 1, the total EV isolation was performed by differential centrifugation (described under EV purification). In step 2, the resuspended EV preparations were used for immunoaffinity-capture using beads coupled to an anti-EpCAM antibody (Dynabeads® Epithelial Enrich Cat. no. 161.02, Life Technologies). As negative control, anti-mouse IgG antibody-coupled magnetic beads were used (provided by Life Technologies). To deplete unspecific binding prior to anti-EpCAM bead incubation, beads conjugated with anti-Giardia IgG antibodies (provided by Life Technologies) were used for 1 hour. Except for the 2 following modifications, the beads were used according to the instructions supplied by the manufacturer (Dynabeads® Epithelial Enrich Whole blood and Buffy Coat). We used 4×107 beads per 3 mL EV suspension and incubated for 1 hour at 4°C before commencing the washing steps. The bead-bound EVs were used for total RNA isolation immediately after their isolation.
Isolation of total RNA
Total RNA was isolated according to the manufacturer's protocol (miRNeasy Mini Kit, Qiagen) with the exception that 1 µg MS2 carrier RNA (Roche) was added to the QIAzol Lysis Reagent to increase the extracted RNA yield for each sample and to minimize variation in purification efficiency. QIAzol Lysis Reagent was added (at 6:1 volume ratio) to the bead suspension containing bound vesicles. The lysate was collected while sample tubes were attached to a magnet (DynaMag™, Life Technologies). To enable assessment of the RNA purification efficiency, 3 RNA species (UniSp2, UniSp4 and UniSp5 from the RNA spike-in kit, Exiqon) were spiked into the Lysis Reagent according to the manufacturer's protocol. RNA yield and purification efficiency was assessed by qRT-PCR. The large abundance of MS2 carrier in the extracted RNA prohibited assessment of RNA concentration and quality by spectrometry and fluorescence-based approaches, such as Qubit (Thermo-Fisher) and Bioanalyzer (Agilent).
miRCURY LNA™ Universal RT microRNA PCR analysis
The miRCURY LNA™ Universal RT microRNA PCR system (Exiqon) was used for cDNA synthesis and qRT-PCR analysis of miRNA abundance as this platform was previously reported to have superior sensitivity for miRNAs with low abundance (Citation30). The approach included adding fixed quantities of 2 RNAs to control the quality of the cDNA-synthesis (UniSp6 and cel-miR-39-3p from the RNA spike-in kit, Exiqon). RNAs extracted from the spike-in series and from the plasma and serum samples of the clinical cohorts were profiled for miRNA. From clinical cohort I, we had available 12 mL plasma/serum; and from cohort II, we had available 2 mL plasma. For each sample, all the RNA extracted (50 µl eluate) was used as input for cDNA synthesis, using the Universal cDNA synthesis kit II (Exiqon). The spike-in series and cohort I samples were profiled using the human miRNome panels I and II (Ver.3) (Exiqon), which together screens 721 distinct human miRNAs and a number of controls (including the 3 RNA purification controls, the 2 cDNA synthesis controls and a plate-to-plate variation control, UniSp3). For cohort I samples, the plasma/serum input per qPCR reaction was ~17 µl. Cohort II samples were profiled using a custom Pick & Mix microRNA PCR Panel querying 43 selected miRNAs and 5 internal controls (Supplementary Table II) corresponding to ~42 µl plasma per qPCR reaction. Accordingly, the reported data are relative to the plasma input volume. They were not normalized further. qPCR was performed on a LightCycler 480 Real-Time PCR system (Roche) using the thermal-cycling parameters recommended by Exiqon.
Data analysis and statistical testing
Spike-in series
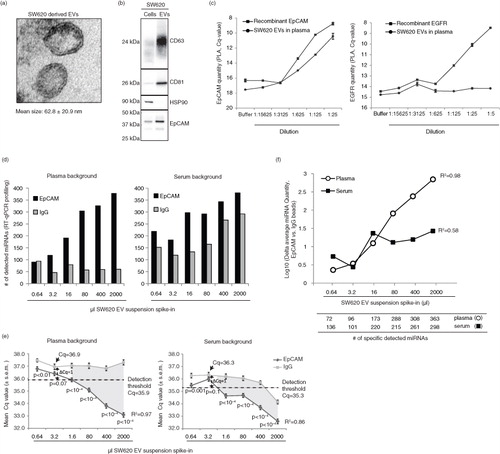
For the plasma and serum SW620 EV spike-in series, the number of identified miRNAs upon EpCAM or IgG immunoaffinity-capture of EVs was calculated as the number of miRNA assays with Cq<40. False positive miRNA assays were defined as assays detected with Cq<40 in all IgG (negative control) data points of the spike-in series for plasma and serum, respectively. Conservative miRNA detection thresholds, to be applied on the clinical samples, were defined for both plasma and serum. These were based on assessment of the signal-to-noise ratio ΔCq (EpCAM vs. IgG) at the individual points in the spike-in series. From 3.2 µl of SW620 EV suspension spike-in and upwards acceptable signal versus noise values, ΔCq (EpCAM vs. IgG) of ~0.5 or higher was observed (). Accordingly, the detection thresholds were defined based on the IgG background signals observed at the spike-in volume of 3.2 µl. For plasma and serum, the average IgG miRNA-assay Cq-values at 3.2 µl were 36.9 and 36.3, respectively (). Conservatively, the detection thresholds were set 1 Cq-value below the average IgG Cq-values. Accordingly, in the clinical samples, miRNA assays were only called as detected if Cq<35.9 for plasma and Cq<35.3 for serum.
Fig. 1. Characterization of isolated CRC cell-derived extracellular vesicles. (a) TEM analysis of EVs isolated from the bioreactor-cultivated SW620 CRC cell line. The mean size and standard deviation of 32 EVs is indicated. (b) SW620 cells and their secreted EVs were analysed for the expression of exosomal markers CD63, CD81, and Hsp90, as well as epithelial-specific EpCAM by immunoblotting using 3.6 µg protein. (c) Evaluation of the quantity of EpCAM and EGFR (negative control) on SW620 EVs by PLA. Serial dilutions of isolated SW620 EVs spiked into plasma from a healthy individual were performed and followed by PLA analysis. As positive controls serial dilutions of recombinant EpCAM and EGFR spiked into PLA buffer were used. Shown are mean values and standard deviations of 2 experiments. (d) Evaluation of the EpCAM immunoaffinity approach's ability to recover known levels of circulating EVs. As a measure of recovery, we used the number of miRNAs detected (Cq<40) in the RNA extracted from the vesicles isolated by applying the immunoaffinity-capture to series of plasma and serum samples spiked with incrementing amounts of SW620 EVs. IgG beads were used as a negative control. (e) The mean miRNA-assay Cq-values±S.E.M. measured after EpCAM or IgG immunoaffinity-capture of the plasma (left panel) and serum (right panel) SW620 EV spike-in series. From 3.2 µl of EV spike-in and upwards a linear relationship between the mean Cq value and the EV input level was observed. The detection thresholds for EpCAM were defined as 1 Cq-value below the IgG level of the 3.2 µl SW620 EV spike-in point. The dashed horizontal lines define the detection thresholds. Below the detection thresholds the signal (EpCAM) is significantly higher than the noise (IgG) as indicated by the grey-shaded area of the graph. P-values represent Mann–Whitney U tests (H0: no difference between EpCAM and IgG Cq-values). (f) Recovery was also evaluated as the difference in average miRNA quantity (EpCAM vs. IgG beads) at each dilution point. Only miRNAs detected with Cq<40 in the EpCAM analysis were included. MicroRNAs with a signal (Cq<40) in all of the IgG spike-in series points were excluded as technical false positive assays. Depicted below the graph, is the number of miRNAs detected at each dilution point, excluding the false positive.
Patient cohort I
Data points exceeding the detection thresholds were filtered out, and miRNA assays defined as technical false positive assays from the IgG immunoaffinity spike-in series were excluded. Subsequently, the number of detected miRNAs in each sample was calculated, and unsupervised hierarchical clustering (similarity metric: Spearman rank correlation; linkage method: average linkage clustering) was performed in Multi experiment Viewer (MeV) version 4.9.0 (Citation31). Principal component analysis (PCA) was conducted using Gene Cluster 3.0 and Java TreeView v1.1.6r4 (Citation32). The cluster and PCA analyses were conducted on the miRNA assays that were detected in all cancer samples (plasma n=31, serum n=7). Statistical testing for differences in the number of detected miRNAs was conducted by Student's t-test.
Patient cohort II
In this cohort of pre- and post-operation plasma samples from 7 stage III CRC patients, 43 miRNAs were profiled using a custom Pick & Mix panel. The miRNA detection threshold established for the plasma spike-in series (Cq<35.9) was applied. Only miRNA assays detected with Cq values below the threshold in all 7 pre-operation samples were included for the analysis (n=26). The average ΔCq (pre/post) for each individual miRNA, and for all miRNAs together, was calculated. Significance of the Cq difference between the pre- and post-operation measurements was evaluated by paired Student's t-test. For visualization of the measured Cq-values pre- and post-operation for the top 10 identified miRNA biomarkers, Cq>35.9 was depicted when relevant in the post-operation samples.
Results
In order to evaluate the feasibility of isolating circulating tumour-derived EVs from CRC patient plasma for miRNA biomarker identification, we first evaluated methodological efficacy and accuracy using an EV spike-in system. The CRC cell line SW620 was cultivated in bioreactors for harvest of epithelial-derived EVs. The EVs had a mean size of ~63 nm in diameter and showed enrichment of the exosomal markers CD63 and CD81 relative to their cellular expression (a and b). EpCAM, which has previously been detected on epithelial-derived EVs and used for affinity-capture of EVs (Citation23), was also enriched (b). On average, 1-mL conditioned medium harvested after 3 days of culture contained 91.2×109 EVs (Supplementary Table I).
We next investigated if plasma samples spiked with increasing levels of SW620 EVs also showed increasing amounts of EpCAM protein (as measured by PLA) since this would indicate that an EpCAM-based immunoaffinity-capture approach for isolation of epithelial-derived EVs would be feasible. Indeed, a dose-dependent EpCAM signal was found in the serial plasma samples and likewise in a series of positive control samples spiked with increasing amounts of recombinant EpCAM protein (c, left panel). PLA did not detect EGFR expression on SW620 EVs (c, right panel), which is consistent with a previous report stating that EGF receptors were undetectable on SW620 cells (Citation33).
We then tested the feasibility and accuracy of miRNA profiling of EVs isolated from plasma and serum using EpCAM immunoaffinity-capture, that is, by the use of anti-EpCAM antibodies conjugated with magnetic beads or anti-IgG beads as negative control. Initially, immunoaffinity-capture was conducted directly on the plasma samples spiked with increasing amounts of SW620 EVs. However, a non-linear relationship between the spike-in level and the measured miRNA quantities was observed (data not shown), indicating that constituents in the plasma affected the specificity of the immunoaffinity-capture. This issue was overcome by isolation of EVs by ultracentrifugation prior to bead incubation. Following ultracentrifugation, proper dissociation and resuspension of EVs was critical to obtain linearity. We found that 20 hours of resuspension at 4°C with gentle vortexing was needed. From plasma and serum samples spiked with incrementing amounts of SW620 EVs, EpCAM+-EVs were isolated. RNA was extracted, and 721 human miRNAs were profiled using the miRCURY LNA™ qRT-PCR platform (Exiqon). MicroRNA assays yielding false positive signals, defined as assays detected in all IgG (negative control) points of the spike-in series, were excluded from analysis. In total, 21 and 88 miRNAs were excluded from the plasma and serum analyses, respectively (Supplementary Table III). As expected, EpCAM but not IgG-capture led to an increased number of miRNAs being detected in the plasma samples spiked with increasing volumes of SW620 EV suspension (d). A similar observation was made in serum although here an increasing number of miRNAs were also detected upon IgG capture (d). EpCAM-capture recovered the increasing EV numbers of the spike-in series as illustrated by a linear relationship between the mean miRNA Cq-values and the spiked EV numbers for both plasma (R2=0.97) and serum (R2=0.86) (e). The signal-to-noise relationship at varying EV levels was assessed by calculating the average ▵miRNA quantity (EpCAM vs. IgG) at each point in the EV spike-in series. A dose-dependent correlation between vesicle input and the signal-to-noise ratio was observed in plasma (R2=0.98), but not in serum (R2=0.58) (f). The latter because in serum the noise (IgG-capture) increased with increasing SV620 EV input (e). The linearity and high signal-to-noise ratio observed for plasma were replicated twice on independent spike-in series (Supplementary Fig. 1). In conclusion, EpCAM-immunoaffinity-capture of EVs from plasma (and to a lesser extent from serum) is a feasible and reliable isolation strategy for EV-derived miRNA quantification.
CRC patients have increased abundance of EpCAM+-EV associated miRNAs
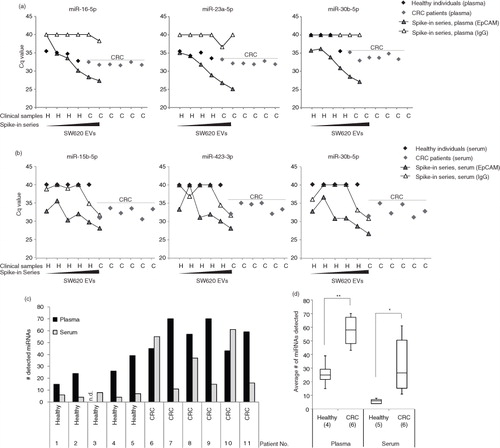
We next addressed whether plasma and serum samples collected from CRC patients prior to surgery contained elevated levels of EpCAM+-EV miRNAs compared with healthy individuals. We applied the EpCAM immunoaffinity approach to 12 mL of sample material (plasma and serum) and subsequently profiled 721 human miRNAs on the RNA extracted from the isolated EVs. Indeed, many miRNAs were detected at markedly lower Cq value (indicating increased abundance) in CRC patients compared with healthy individuals in both plasma and serum (). Next, the Cq-values of the clinical samples were compared with the Cq-values obtained from the SW620 spike-in series. This revealed for plasma that the Cq values of the CRC samples were within the linear range of the spike-in series, while the values obtained from the healthy controls were at or below the EpCAM detection level for plasma (a). Similar results were obtained for serum (b). The higher miRNA abundance in CRC patients was also reflected in a significantly higher number of miRNAs being detected in EpCAM+-EVs from CRC patients than from healthy controls (c and d). Generally, the results from serum varied more than from plasma, which may be due to the poorer performance of the immunoaffinity-capture on serum.
Fig. 2. Increased miRNA abundance associated with EpCAM+ extracellular vesicles in plasma and serum of CRC patients. Representative examples of miRNA amounts detected in EpCAM+-EVs in (a) plasma or in (b) serum from healthy individuals and CRC patients compared to the amount detected when SW620 EVs were spiked into (a) plasma or (b) serum from a healthy individual. The miRNA abundance in CRC plasma samples represented an acceptable signal as compared to the signal-to-noise window of EpCAM (signal) s. IgG (noise) bead spike-in series. (c) The number of miRNAs detected in EpCAM+-EVs isolated from plasma and serum of healthy individuals and CRC patients (Patient cohort I). Following criteria were used: i) All false positive miRNAs assays were excluded, ii) Only miRNAs detected in clinical samples at Cq<35.9 (plasma) and Cq<35.3 (serum) were included. Plasma from healthy individual 3 was excluded based on an extreme outlier profile in the initial quality check. (d) Box plot of the number of miRNAs detected from isolated EpCAM+-EVs (Student's t-test, p<0.05, **p<0.01).
To further investigate whether the EpCAM+-EV miRNA profiles reflected whether the samples were collected from patients or controls, unsupervised PCA and hierarchical clustering analyses were conducted (). Prior to analysis, false positive miRNA-assays (positive in all IgG spike-in samples) and assays that were not detected in all cancer samples were filtered out resulting in 31 and 7 miRNA-assays for plasma and serum, respectively. Unsupervised PCA analysis based on these miRNAs demonstrated near complete separation of healthy and diseased individuals in both plasma (a) and serum (b). More variation was observed between healthy individuals than between CRC patients. Similar to the PCA analysis, unsupervised hierarchical clustering showed near complete separation of healthy individuals and CRC patients (c and d). A total of 13 (plasma, Table ) and 7 (serum, Table ) miRNAs were significantly more abundant in the CRC than the healthy control samples (p<0.05, Student's t-test). Three miRNAs (miR-16-5p, miR-23a-3p and miR30b-5p) were significant for both plasma and serum (e). In conclusion, the miRNA profiles of EpCAM+-EVs distinguish CRC from healthy controls. Both the number of miRNAs detected and their abundance was increased in the EpCAM+-EVs of the CRC patients compared with the controls.
Fig. 3. Analysis of altered miRNA profiles in EpCAM+-EVs from CRC patients. (a, b) Unsupervised Principal Component Analysis (PCA) demonstrating separation of samples based on their origin from CRC patients or healthy individuals from miRNA profiling of circulating EpCAM+-EVs. (a) Plasma, a total of 31 miRNAs were included in the analysis using the following selection criteria: i) miRNA expression in 6/6 cancer samples, ii) detected at Cq<35.9, and iii) exclusion of false positive miRNA assays. (b) Serum, a total of 7 miRNAs were included in the analysis using selection criteria: i) miRNA expression in 6/6 cancer samples, ii) detected at Cq<35.3, iii) exclusion of false positive assays. The patient ID number is depicted at each data point. (c, d) Unsupervised hierarchical clustering analysis based on the miRNAs from (a, b) (yellow-high expression, blue-low expression, n.d- not detected). (e) miRNAs significantly more abundant in EpCAM+-EVs from CRC patients than healthy controls (p≤0.05, Student's T-test). Shown are data for both plasma and serum.
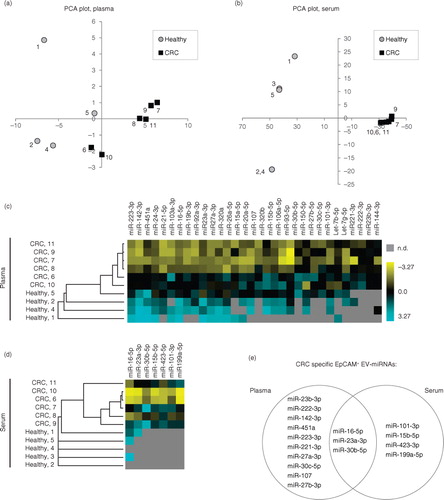
Fig. 4. Circulating EpCAM+-EV-derived miRNAs are reduced following CRC surgery. (a) The number of miRNAs detected before and after surgery. EpCAM+-EVs were isolated from matched pre- and post-operation plasma from 7 stage III patients. From these, the abundance of 43 miRNAs associated with EpCAM+-EVs were profiled using a custom-designed Pick & Mix qRT-PCR panel. A miRNA was considered detected if it was below the previously defined threshold of Cq<35.9. Individual patient ID numbers are displayed. (b) 26 miRNAs were detected in all 7 pre-operation samples and shown are average abundance of these in pre- and post-operation samples. The increased Cq values observed in the post-operation samples relative to the pre-operation samples indicate reduced abundance. The data are depicted as the average abundance in all CRC samples. (c) A volcano plot of the 26 individual miRNAs showing the relationship between statistical significance (paired Student's t-test) and the fold-change between pre- and post-operation samples (▵Cq (pre – post)). The dashed line indicates p=0.05. MicroRNAs with distinct abundance in EpCAM+-EVs in plasma samples from CRC patients and healthy controls are marked in red. (d) Shown are paired pre- and post-OP Cq-values for 10 selected EpCAM+-EV miRNAs with significant difference in pre- and post-operation abundance.
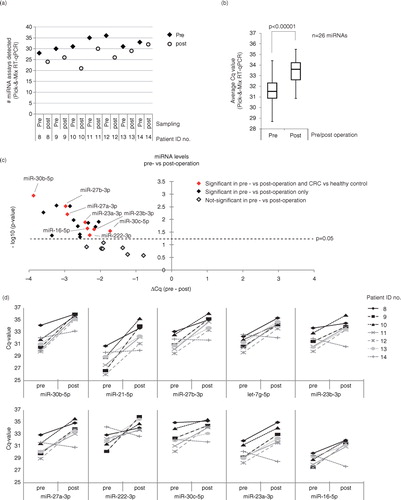
Table III. MicroRNAs with significantly different abundance in EpCAM+-EVs from plasma of CRC patients and healthy controls
Table IV. MicroRNAs with significantly different abundance in EpCAM+-EVs from serum of CRC patients and healthy controls
EpCAM+-EV-associated miRNA levels in circulation are reduced upon surgical tumour removal
We next analysed whether surgical removal of the primary CRC tumour affected the levels of circulating EpCAM+-EV miRNAs. Circulating miRNAs were examined pre- and post-operation in a patient cohort comprising 7 stage III CRC patients (Table ). All patients were considered surgically cured based on >3 years of disease-free follow up. Only plasma samples were analysed due to the superior assay specificity observed in plasma. EpCAM+-EVs were isolated from 2 mL plasma using the same EpCAM bead affinity-capture approach applied to patient cohort I. The EpCAM+-EVs were subsequently profiled using a 43 miRNA custom-designed miRCURY LNA™ qRT-PCR Pick & Mix panel. The 43 miRNAs in the panel were selected because they were reliably detected in the CRC samples of cohort I (Supplementary Table II), that is, they were detected in either all CRC plasma or all serum samples. Given the reduced number of miRNAs profiled (43 vs. 721 miRNAs) on cohort II compared with cohort I, it was possible to reduce the plasma input volume from 12 to 2 mL and still ensure that the amount of input EpCAM+-EV cDNA available per miRNA qPCR reaction was ~3× higher for cohort II than for cohort I.
The results showed that the overall number of miRNAs detected in the EpCAM+-EVs decreased upon surgical removal of the primary tumour (a). A comparison of the average miRNA abundance before and after surgery for the 26 miRNAs detected in all the pre-operation samples revealed a significant reduction (p<0.00001, paired Student's t-test) in miRNA abundance after surgery (b). Nine of the 26 miRNAs were significantly less abundant in the post-operation samples (p<0.05, paired Student's t-test, c and Supplementary Table IV). In support of these miRNAs originating from the CRC tumours, 8 of them were among the miRNAs significantly more abundant in plasma from CRC patients compared with healthy controls (e and 4c, and Supplementary Table IV). In d, we show the pre- and post-operation Cq values for all 7 patients for 10 of the 19 significant miRNAs. The post-operation miRNA levels were reduced for 6 of 7 patients (d). But for the last patient (ID no. 14), no-change or an increase was often seen post-operation. Clinically, this patient was distinct from the rest of the patients. Over a period of more than 14 years, including the period with treatment for CRC, the patient was diagnosed with, and treated for, numerous recurrent Ta-bladder papillomas. These lesions may release EpCAM+-EVs into the circulation potentially explaining why no reduction in miRNA levels was seen post-CRC-operation.
Discussion
For studies investigating the diagnostic potential of circulating miRNAs in blood, there is an unmet need for a method that is unaffected by miRNAs released to the circulation during pre-analytical handling of the samples, for example, through lysis of erythrocytes and platelets. Here, we have addressed this need and devised a method for miRNA profiling of EpCAM+-EVs isolated from plasma and serum. This approach is, in principle, unaffected by miRNAs released by non-epithelial cells as these are not associated with EpCAM.
The performance of the approach was tested using EpCAM+-EV spike-in series with up to 3 orders of magnitude difference in EpCAM+-EV content. A linear relationship between the measured miRNA quantities and the EV input levels was obtained in both plasma and serum. Furthermore, the miRNA quantities measured from the EpCAM+-EVs were significantly higher than the background miRNA quantities obtained by immunoaffinity-capture using an unspecific IgG-antibody. This was particularly evident for the analysis of plasma where the miRNA background signals remained negligible across the whole range of EpCAM+-EV inputs. Also for serum, the miRNA signals from the EpCAM immune-capture were higher than the IgG background although the background level increased with increasing input amounts of EpCAM+-EVs, indicating that our approach is less specific in serum than plasma. The reason for this discrepancy between plasma and serum remains unknown. Implementation of a pre-clearing step using anti-Giardia IgG beads prior to EpCAM beads did not improve the signal-to-noise ratio in the spike-in series at low EV amounts (data not shown).
Importantly, when we applied the approach to plasma and serum samples from CRC patients and compared the obtained miRNA signals with those obtained from the spike-in series, we found that they were generally above the detection limit and within the linear range of the approach. This indicates that the plasma/serum input we use per qPCR-reaction (≥17 µl) is sufficient for reliable miRNA quantification. Given that EpCAM+-EVs are not expected to be abundant in the blood of healthy individuals, we did not expect prominent miRNA signal in the healthy control samples, and indeed, the majority of the queried miRNAs were either undetectable or detected at the limit of sensitivity in the control samples. In agreement with this, PCA as well as unsupervised hierarchical clustering analysis based on the miRNAs detected in the CRC samples also separated CRC and control samples. Moreover, these analyses indicated that miRNA profiles of the healthy individuals varied more than that of the CRC patients. This may reflect that the level of EpCAM+-EVs is lower in healthy individuals and that the organ of origin for the EVs (and consequently, their miRNA content) varies more between healthy individuals than CRC patients.
We generally observed more variation in the results obtained from serum than from plasma, which probably relates to the poorer performance of the immunoaffinity-capture approach on serum. We also detected fewer differentially expressed miRNAs (cases vs. controls) in serum than plasma (7 vs. 13) consistent with serum being a poorer starting material for the developed approach. Nevertheless, 3 of the 7 differentially expressed serum miRNAs (43%) were also differentially expressed in plasma, indicating that some of the signal seen in serum may be informative, despite of the higher noise. Due to the lower variation and superior specificity, we focused on the plasma results.
Our finding that miRNA profiling of epithelial-derived EVs can distinguish between plasma from CRC patients and healthy controls was corroborated when we compared plasma samples collected pre- and post-CRC-operation. Consistent with the reduced tumour burden, the miRNA signals measured in the post-operation samples were significantly lower than in the pre-operation samples for 6 out of 7 patients. The 7th patient, like the other patients, was clinically relapse free for 36 months and as such likely cured of CRC. However, in contrast to the others, this patient had recurrent bladder neoplasia both concomitant and after the CRC surgery. The bladder lesions could potentially be the source of the detected post-CRC-operation EpCAM+-EV miRNAs. While an increase in EpCAM+-EV miRNA abundancy distinguishes healthy individuals from cancer patients, it will likely not allow distinction between different types of epithelial-derived cancers. In principle, it should be possible to develop the method further and perform the immuno-capture with cancer subtype- or organ-specific markers. However, this necessitates the availability and identification of specific surface marker proteins. Additional studies are needed to address this.
We found 8 miRNAs with higher EpCAM+-EV abundance in plasma from CRC patients than healthy controls and lower abundance in post- versus pre-operation plasma (c). We speculate that these are of tumour origin and may hold potential as non-invasive biomarkers. One should note though that these encouraging findings were based on small sample numbers and need to be confirmed in larger cohorts. All 8 miRNAs have previously been reported to be functionally related to the development of CRC, metastasis and/or prognosis. Comparison of normal mucosa and tumour tissue indicates that 5 of the 8 are downregulated (miR-16-5p (Citation34), miR-23b-3p (Citation34), miR-27b-3p (Citation34), miR-30b-5p (Citation34) and miR-30c-5p (Citation34)) and 3 upregulated in tumours (miR-23a-3p (Citation35, Citation36), miR-27a-3p (Citation34–Citation37) and miR-222-3p (Citation37, Citation38)). Both down- and upregulation may be consistent with our observation of increased levels in blood. We have recently published a study indicating that exosome-mediated secretion of miRNAs (including miR-23b-3p) from cancer cells may represent a cellular disposal route of tumour- or metastasis-suppressor miRNAs (Citation39). In this study, the cellular export of miR-23b was found to be of advantage to the primary tumour since its high cellular presence would otherwise restrain proliferation, invasion, anoikis and capacity of cancer cell lung colonization in vivo (Citation39). On the contrary, secretion of tumour-derived EVs may also be a way to prime distant sites into pre-metastatic niches to allow for micrometastases to establish (Citation40). There is, thus, a complex interplay between deregulated miRNAs in the primary tumour tissue, the selection of miRNAs being secreted through EVs, and their impact on distant organs to prime for metastases. The expression of 2 of the 8 miRNAs (miR-23b-3p and miR-30-5p) has, furthermore, been shown to correlate with survival of CRC patients (Citation41). Consistent with our findings, increased plasma and/or serum levels have previously been reported in CRC for 3 of the 8 miRNAs (miR-23a-3p (Citation15, Citation42) (Citation43), miR-30b-5p (Citation44) and miR-222-3p (Citation45)). It is worth noting that other miRNAs have also been reported to be deregulated in crude plasma/serum samples from CRC patients but were undetected or inconsistently detected in our EpCAM+-EVs. Among the most frequently reported are miR-18a-5p, miR-21-5p, miR-29a-5p, miR-92a-5p, miR-143-5p and miR-378-5p (reviewed in (Citation7)). Of these, miR-21-5p, miR-29a-5p and miR-92a-5p are known to be expressed by red blood cells, and it has been suggested that this cell type may be the major source for these miRNA in plasma/serum (Citation7). In situ hybridization studies of colorectal adenocarcinomas have furthermore revealed that miR-21-5p and miR-143-5p are expressed primarily by mesenchymal cells and not cancer cells (Citation46, Citation47). To the best of our knowledge, the cellular origin of miR-18a-5p and miR-378-5p are currently unknown. Of the 6 miRNAs, only miR-21-5p was detected in EpCAM+-EVs, and this inconsistently, as it was only observed in cohort II. Accordingly, our findings are consistent with the notion that the primary cellular origin of these miRNAs is not colonic epithelia and moreover indicate that EpCAM+-EV profiling, with its improved specificity, may be superior to crude plasma/serum profiling.
Normalization remains a major challenge in the analysis of circulating miRNAs (Citation7, Citation48). In this study, we report the EpCAM+-EV miRNA Cq-values relative to the plasma input volume used per qPCR-reaction (17 µl plasma for cohort I and 43 µl plasma for cohort II). This was done because we wanted the miRNA differences (between cases and controls for cohort I and between pre- and post-operation plasma for cohort II) to reflect changes in both EV abundancy and EV miRNA content. Accordingly, we cannot determine if the observed quantity changes are due to changes in EV abundance or in EV miRNA abundance, or both. To the best of our knowledge, no reference marker besides plasma input has yet been reported for normalizing circulating EV miRNA data (Citation7, Citation48). It has been reported that the input plasma volume may affect miRNA purification efficiency (Citation49). Since our input was 12 mL for cohort I and 2 mL for cohort II, variable purification efficiencies may have affected the absolute Cq values obtained. However, we expect that it has only minor influence on the ΔCq values (CRC vs. controls and pre- vs. post-operation) we use to compare the 2 cohorts, at least for the miRNAs that are detectable in both cohorts. Nevertheless, it may be part of the explanation why the consistency between the cohorts is not complete.
A few studies have previously reported on miRNA profiling of EpCAM+-EVs isolated from plasma/serum of lung- and ovarian-cancer patients (Citation22, Citation23), but none so far from CRC patients. While each of these studies used unique approaches for the isolation of the EpCAM+-EVs, they identified a number of miRNAs with promising diagnostic potential, similar to this study. This indicates that the suggested approach is feasible and generally applicable to epithelial-derived tumour types.
Importantly, the approach we report is not inherently restricted to analysis of miRNAs. All epithelial-derived biomolecules present in or on EVs could in principle be investigated for their biomarker potential, for example, mRNAs, long non-coding RNAs, proteins and lipids. Further studies are needed to investigate the biomarker potential of these biomolecules.
In summary, we report on the development of a non-invasive approach for miRNA profiling of epithelial-derived EVs isolated from blood. This approach mitigates many of the specificity issues observed in earlier studies of circulating miRNAs and their biomarker potential in relation to epithelial cancers. Using the approach, we identified 8 miRNAs that were more abundant in the blood circulation of CRC patients than healthy controls and whose abundance dropped significantly upon surgical removal of the primary tumour. While the reported results are promising, the approach and miRNA markers need to be validated in larger patient cohorts before their clinical value can be confidently affirmed.
Conflict of interest and funding
The authors have not received any funding or benefits from industry or elsewhere to conduct this study.
Supplementary figure 1
Download (1.3 MB)Supplementary Tables
Download MS Word (40.2 KB)Acknowledgements
The authors thank Martin Rasmussen (Department of Clinical Pathology, Odense University Hospital) for technical assistance and Life Technologies for providing the anti-IgG-beads. This work was supported by the Novo Nordisk Foundation, The Toyota Foundation, The John and Birthe Meyer Foundation, The Danish Council for Strategic Research, The Danish Council for Independent Research – Medical Sciences, The Lundbeck Foundation, The Danish Cancer Biobank, and the Danish Cancer Society.
Notes
To access the supplementary material to this article, please see Supplementary files under ‘Article Tools’.
References
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, etal. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015; 136: E359–86.
- Etzioni R, Urban N, Ramsey S, McIntosh M, Schwartz S, Reid B, etal. The case for early detection. Nat Rev Cancer. 2003; 3: 243–52.
- Levin TR, Jamieson L, Burley DA, Reyes J, Oehrli M, Caldwell C. Organized colorectal cancer screening in integrated health care systems. Epidemiol Rev. 2011; 33: 101–10.
- Kuipers EJ, Rosch T, Bretthauer M. Colorectal cancer screening – optimizing current strategies and new directions. Nat Rev Clin Oncol. 2013; 10: 130–42.
- Nielsen HJ, Jakobsen KV, Christensen IJ, Brunner N. Danish Study Group on Early Detection of Colorectal Cancer. Screening for colorectal cancer: possible improvements by risk assessment evaluation?. Scand J Gastroenterol. 2011; 46: 1283–94.
- Sanford KW, McPherson RA. Fecal occult blood testing. Clin Lab Med. 2009; 29: 523–41.
- Clancy C, Joyce MR, Kerin MJ. The use of circulating microRNAs as diagnostic biomarkers in colorectal cancer. Cancer Biomark. 2015; 15: 103–13.
- Du M, Liu S, Gu D, Wang Q, Zhu L, Kang M, etal. Clinical potential role of circulating microRNAs in early diagnosis of colorectal cancer patients. Carcinogenesis. 2014; 35: 2723–30.
- Rokkas T, Kothonas F, Rokka A, Koukoulis G, Symvoulakis E. The role of circulating microRNAs as novel biomarkers in diagnosing colorectal cancer: a meta-analysis. Eur J Gastroenterol Hepatol. 2015; 27: 819–25.
- Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, Miyaji MM, etal. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res. 2012; 5: 492–7.
- Cheng HH, Yi HS, Kim Y, Kroh EM, Chien JW, Eaton KD, etal. Plasma processing conditions substantially influence circulating microRNA biomarker levels. PLoS One. 2013; 8: e64795.
- MacLellan SA, MacAulay C, Lam S, Garnis C. Pre-profiling factors influencing serum microRNA levels. BMC Clin Pathol. 2014; 14: 27.
- Yamada A, Cox MA, Gaffney KA, Moreland A, Boland CR, Goel A. Technical factors involved in the measurement of circulating microRNA biomarkers for the detection of colorectal neoplasia. PLoS One. 2014; 9: e112481.
- Zanutto S, Pizzamiglio S, Ghilotti M, Bertan C, Ravagnani F, Perrone F, etal. Circulating miR-378 in plasma: a reliable, haemolysis-independent biomarker for colorectal cancer. Br J Cancer. 2014; 110: 1001–7.
- Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006..
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, etal. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008; 105: 10513–8.
- Li L, Zhu D, Huang L, Zhang J, Bian Z, Chen X, etal. Argonaute 2 complexes selectively protect the circulating microRNAs in cell-secreted microvesicles. PLoS One. 2012; 7: e46957.
- Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, etal. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011; 108: 5003–8.
- Koberle V, Pleli T, Schmithals C, Augusto Alonso E, Haupenthal J, Bonig H, etal. Differential stability of cell-free circulating microRNAs: implications for their utilization as biomarkers. PLoS One. 2013; 8: e75184.
- Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011; 13: 423–33.
- Yanez-Mo M, Siljander PR, Andreu Z, Zavec AB, Borras FE, Buzas EI, etal. Biological properties of EVs and their physiological functions. J Extracell Vesicles. 2015; 4: 27066, doi: http://dx.doi.org/10.3402/jev.v4.27066.
- Rabinowits G, Gercel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009; 10: 42–6.
- Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008; 110: 13–21.
- Lazaro-Ibanez E, Sanz-Garcia A, Visakorpi T, Escobedo-Lucea C, Siljander P, Ayuso-Sacido A, etal. Different gDNA content in the subpopulations of prostate cancer extracellular vesicles: apoptotic bodies, microvesicles, and exosomes. Prostate. 2014; 74: 1379–90.
- Yoshioka Y, Kosaka N, Konishi Y, Ohta H, Okamoto H, Sonoda H, etal. Ultra-sensitive liquid biopsy of circulating extracellular vesicles using ExoScreen. Nat Commun. 2014; 5: 3591.
- Jeppesen DK, Nawrocki A, Jensen SG, Thorsen K, Whitehead B, Howard KA, etal. Quantitative proteomics of fractionated membrane and lumen exosome proteins from isogenic metastatic and nonmetastatic bladder cancer cells reveal differential expression of EMT factors. Proteomics. 2014; 14: 699–712.
- Mitchell JP, Court J, Mason MD, Tabi Z, Clayton A. Increased exosome production from tumour cell cultures using the Integra CELLine Culture System. J Immunol Methods. 2008; 335: 98–105.
- Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gustafsdottir SM, etal. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol. 2002; 20: 473–7.
- Lundberg M, Thorsen SB, Assarsson E, Villablanca A, Tran B, Gee N, etal. Multiplexed homogeneous proximity ligation assays for high-throughput protein biomarker research in serological material. Mol Cell Proteomics. 2011; 10: M110 004978.
- Jensen SG, Lamy P, Rasmussen MH, Ostenfeld MS, Dyrskjot L, Orntoft TF, etal. Evaluation of two commercial global miRNA expression profiling platforms for detection of less abundant miRNAs. BMC Genomics. 2011; 12: 435.
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, etal. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003; 34: 374–8.
- de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004; 20: 1453–4.
- Coffey RJ Jr , Shipley GD, Moses HL. Production of transforming growth factors by human colon cancer lines. Cancer Res. 1986; 46: 1164–9.
- Christensen LL, Holm A, Rantala J, Kallioniemi O, Rasmussen MH, Ostenfeld MS, etal. Functional screening identifies miRNAs influencing apoptosis and proliferation in colorectal cancer. PLoS One. 2014; 9: E96767.
- Jahid S, Sun J, Edwards RA, Dizon D, Panarelli NC, Milsom JW, etal. miR-23a promotes the transition from indolent to invasive colorectal cancer. Cancer Discov. 2012; 2: 540–53.
- Slattery ML, Wolff E, Hoffman MD, Pellatt DF, Milash B, Wolff RK. MicroRNAs and colon and rectal cancer: differential expression by tumor location and subtype. Genes Chromosomes Cancer. 2011; 50: 196–206.
- Monzo M, Navarro A, Bandres E, Artells R, Moreno I, Gel B, etal. Overlapping expression of microRNAs in human embryonic colon and colorectal cancer. Cell Res. 2008; 18: 823–33.
- Liu S, Sun X, Wang M, Hou Y, Zhan Y, Jiang Y, etal. A microRNA 221- and 222-mediated feedback loop maintains constitutive activation of NFkappaB and STAT3 in colorectal cancer cells. Gastroenterology. 2014; 147: 847–59. e11.
- Ostenfeld MS, Jeppesen DK, Laurberg JR, Boysen AT, Bramsen JB, Primdal-Bengtson B, etal. Cellular disposal of miR23b by RAB27-dependent exosome release is linked to acquisition of metastatic properties. Cancer Res. 2014; 74: 5758–71.
- Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, etal. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012; 18: 883–91.
- Zhou X, Xu X, Wang J, Lin J, Chen W. Identifying miRNA/mRNA negative regulation pairs in colorectal cancer. Sci Rep. 2015; 5: 12995.
- Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K, etal. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One. 2014; 9: e92921.
- Yong FL, Law CW, Wang CW. Potentiality of a triple microRNA classifier: miR-193a-3p, miR-23a and miR-338-5p for early detection of colorectal cancer. BMC Cancer. 2013; 13: 280.
- Ho GY, Jung HJ, Schoen RE, Wang T, Lin J, Williams Z, etal. Differential expression of circulating microRNAs according to severity of colorectal neoplasia. Transl Res. 2015; 166: 225–32.
- Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, etal. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009; 58: 1375–81.
- Kent OA, McCall MN, Cornish TC, Halushka MK. Lessons from miR-143/145: the importance of cell-type localization of miRNAs. Nucleic Acids Res. 2014; 42: 7528–38.
- Nielsen BS, Jorgensen S, Fog JU, Sokilde R, Christensen IJ, Hansen U, etal. High levels of microRNA-21 in the stroma of colorectal cancers predict short disease-free survival in stage II colon cancer patients. Clin Exp Metastasis. 2011; 28: 27–38.
- Toiyama Y, Tanaka K, Inoue Y, Mohri Y, Kusunoki M. Circulating cell-free microRNAs as biomarkers for colorectal cancer. Surg Today. 2016; 46: 13–24.
- El-Khoury V, Pierson S, Kaoma T, Bernardin F, Berchem G. Assessing cellular and circulating miRNA recovery: the impact of the RNA isolation method and the quantity of input material. Sci Rep. 2016; 6: 19529.
