Abstract
Nasopharyngeal carcinoma (NPC) is one of the most common head and neck malignancies and exhibits regional differences in incidence. Because many fusion genes have been discovered in different types of tumors over the past few years, we aimed to investigate the existence of a fusion gene in primary NPC patients using RNA-seq. In this study, for the first time, we found that fibroblast growth factor receptor 3-transforming acidic coiled-coil-containing protein 3 (FGFR3-TACC3) fusion transcripts are recurrently detected in NPC. The presence of this fusion gene was also detected in head and neck cancer, esophageal squamous cell carcinoma (ESCC), and lung cancer. Furthermore, we found certain new isoforms of the FGFR3-TACC3 fusion transcripts, such as a gene fusion between exon 18 of FGFR3 and exon 6 or exon 14 of TACC3 and agene fusion between exon 19 of FGFR3 and exon 11 of TACC3. In addition, we showed that the FGFR3-TACC3 fusion gene promotes cell proliferation, colony formation, and transforming ability in vitro, whereas the FGFR3-TACC3 K508M mutant or treatment with the FGFR inhibitor PD173074 abrogates these effects, suggesting that FGFR3-TACC3 most likely exerts its effects through activation of FGFR kinase activity. This activation likely leads to the development of NPC. Additionally, FGFR3-TACC3 could trigger activation of the ERK and Akt signaling pathways, whereas FGFR3-TACC3 K508M mutant could not, suggesting that these 2 signaling pathways might be involved in the function of FGFR3-TACC3. Taken together, our data demonstrated the oncogenic role of FGFR3-TACC3 in vitro, indicating that FGFR3-TACC3 may be useful as a diagnostic marker and therapeutic target in cancers.
Abbreviations
| FGFR3 | = | fibroblast growth factor receptor 3 |
| TACC3 | = | transforming acidic coiled-coil-containing protein 3 |
| NPC | = | nasopharyngeal carcinoma |
| LTBR | = | lymphotoxin β receptor |
| CCND1 | = | cyclin D1 |
| RT-PCR | = | reverse transcription-PCR |
| FBS | = | fetal bovine serum |
| MTT | = | 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide |
| SDS | = | sodium dodecyl sulfate |
| DTT | = | DL-dithiothreitol |
| DMSO | = | dimethyl sulfoxide |
| PBS | = | phosphate-buffered saline |
| PI | = | propidium iodide |
Introduction
Nasopharyngeal carcinoma (NPC) presents regional differences in incidence, such as high incidences in North Africa, Southeast Asia, and southern China.Citation1,2 Although serum anti-Epstein-Barr virus (EBV) antibodies and plasma EBV DNA are useful tools for diagnosis and prognosis prediction in NPC, more than 60% of NPC patients at diagnosis are in late stagesCitation3 or even present with metastasis.Citation4,6 Therefore, there is still an urgent need to identify diagnostic biomarkers and therapeutic targets in NPC.
A number of chromosomal abnormalities involving specific oncogenes and tumor suppressor genes, including amplification of NPC-associated oncogenes such as lymphotoxin β receptor (LTBR) and cyclin D1 (CCND1)Citation7 and deletion or inactivation of tumor suppressor genes such as p16,Citation8 cytochrome b5 reductase2 (CYB5R2)Citation9 and FEZ family zinc finger 2 (FEZF2),Citation10 have been detected in NPC cases.Citation5 This finding suggests that genomic instability might play an important role in NPC pathogenesis.Citation11 Chromosome aberrations, and especially translocations and relevant gene fusions, play a crucial role in tumor progression. The fusion protein produced by a fusion gene can be oncogenic, as shown in aggressive leukemia with mixed lineage leukemia(MLL) rearrangements.Citation12 Small-molecule inhibitors of the menin-MLL fusion protein can apparently reverse the tumorigenic activity of this fusion protein,Citation12 effectively extending the survival time of cancer patients, which suggests that fusion genes provide effective tools for cancer diagnosis and therapy. A variety of fusion genes have been reported in blood and solid tumors,Citation13 such as the TMPRSS2-ERG fusion gene in prostate cancer,Citation14,15 EML4-ALK fusions in non-small-cell lung cancer,Citation16 and UBR5-ZNF423 in NPC,Citation17 since the Philadelphia chromosome was first reported in chronic myeloid leukemia in 1960.Citation18 However, fusion genes remain poorly characterized in head and neck malignancies, and especially NPC.
Although fibroblast growth factor receptor 3-transforming acidic coiled-coil-containing protein 3 (FGFR3-TACC3) fusion has been reported by Singh,Citation19 Parker,Citation20 Wu,Citation21 and Majewski,Citation22 showing oncogenic potency in glioblastoma multiforme, bladder cancer, and head and neck cancer, herein, we are the first to report that FGFR3-TACC3 fusion transcripts are recurrently detected in NPC and esophageal squamous cell carcinoma (ESCC). In addition, we found that the FGFR3-TACC3 fusion gene promotes cell proliferation, colony formation, and transforming ability, indicating that FGFR3-TACC3 plays an important role in the development of NPC.
Results
Discovery of a novel transforming FGFR3-TACC3 fusion gene in NPC
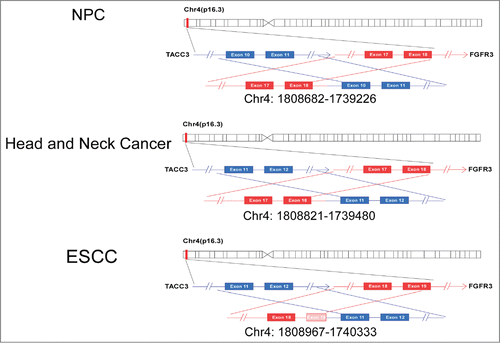
Because an increasing number of fusion genes have been discovered in different types of tumors, we aimed to investigate whether any fusion genes are present in primary NPC patients. We collected 12 tumor samples and 2 non-tumor samples from healthy donors to perform RNA-seq. RNA-seq reads that spanned exon-exon junctions and connected 2 distinct genes indicated fusion gene candidates, with the fusion gene FGFR3-TACC3 being notable (). As shown in , the FGFR3-TACC3 fusion gene consists of FGFR3 and TACC3, which are both located on chromosome 4 (Chr4) and which are separated by 48 kb. The fusion point is between exon 19 of FGFR3 and exon 6 of TACC3 ().
Figure 1. RNA-seq to identify FGFR3-TACC3 fusion genes in NPC. Circos plot of FGFR3-TACC3 fusion genes, as detected by RNA-seq. Chromosome ideograms are shown in the outer layer. The expression level of the genes is shown in the middle layer using a heat map. The FGFR3-TACC3 fusion genes are shown in the central layer. (A) Detection of FGFR3-TACC3 fusion genes using transcriptome sequencing. The reads are aligned across the junction of the predicted fusion transcripts. (B) The genomic fusion point is between exon 19 of FGFR3 and intron 5 of TACC3. (C) Structure of the predicted fusion protein.
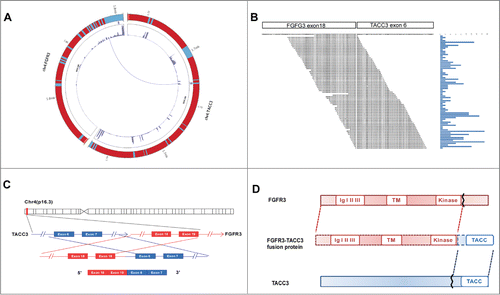
It is known that certain fusion genes promote tumor development through the encoded fusion proteinCitation23 or regulate gene expressionCitation24 to promote tumor development. The predicted fusion protein was deduced based on the sequence of the fusion gene FGFR3-TACC3, as shown in . This protein contains an extracellular domain with 3 Ig-like loops, a highly hydrophobic transmembrane domain (22 amino acids), and an intracellular domain with tyrosine kinase activity from FGFR3 as well as the C-terminus of TACC3. Although the existence of the fusion gene FGFR3-TACC3 has been reported in glioblastoma,Citation19,20 urothelial bladder carcinoma,Citation25 and head and neck cancerCitation21 and lung cancer,Citation22 this is the first demonstration of its presence in NPC and ESCC.
FGFR3-TACC3 fusion gene isoforms are present in different solid tumors
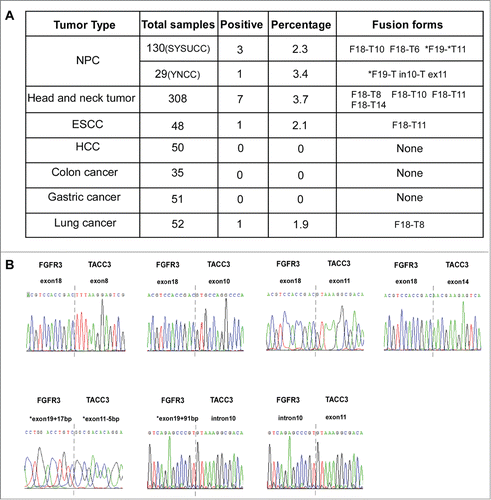
To examine the frequency of the fusion gene FGFR3-TACC3 in NPC, we amplified FGFR3-TACC3 by quantitative reverse transcription-PCR (qRT-PCR) with specific primers for exon 17 of FGFR3 and the 3′-UTR of TACC3 and found the existence of FGFR3-TACC3 in 4 of 159 NPC patients (2.5%). Furthermore, we attempted to determine whether this fusion gene is also present in other tumors. A large number of different types of tumor samples were collected for the detection of FGFR3-TACC3 by RT-PCR, and the amplified PCR products were subjected to Sanger sequencing (). We found the FGFR3-TACC3 fusion gene in other head and neck cancers (7 of 308, 3.7%), ESCC (1 of 48, 2.1%), and lung cancer (1 of 52, 1.9%)(). To our surprise, we found 7 different forms of FGFR3-TACC3 fusion genes, including 1) a fusion between exon 18 of FGFR3 and exon 6 of TACC3 in NPC, 2) a fusion between exon 18 of FGFR3 and exon 8 of TACC3 in head and neck cancer and lung cancer, 3) a fusion between exon 18 of FGFR3 and exon 10 of TACC3 in head and neck cancer and NPC, 4) a fusion between exon 18 of FGFR3 and exon 11 of TACC3 in head and neck cancer and ESCC, 5) a fusion between exon 18 of FGFR3 and exon 14 of TACC3 in head and neck cancer, 6) a fusion between exon 19 of FGFR3 and exon 11 of TACC3 in NPCand 7) a fusion between exon 19 of FGFR3 and part of intron 10 and exon 11 of TACC3 in NPC. In contrast, the FGFR3-TACC3 fusion gene was not detected in hepatocellular carcinoma (0 of 50), colon cancer (0 of 35), or gastric cancer (0 of 51).
Figure 2. Different forms of FGFR3-TACC3 fusion transcripts present in a variety of solid tumors. (A) Frequency and different forms of the FGFR3-TACC3 fusion gene identified in different types of cancers. (B) Identification of the fusion breakpoint using Sanger sequencing.
In addition, we examined the fusion point of FGFR3 and TACC3 at the genomic DNA level and found different genomic DNA breakpoints on Chr4 in various samples, such as the breakpoint between 1808682 and 1739226 in an NPC patient, the breakpoint between 1808821 and 1739480 in a head and neck cancer patient, and the breakpoint between 1808967 and 1740333 in an ESCC patient (). Taken together, these data demonstrate the existence and prevalence of FGFR3-TACC3 fusions in tumor samples, showing a relatively higher frequency in head and neck tumors, including NPC.
The FGFR3-TACC3 fusion protein promotes NPC cell proliferation
The endogenous expression of the fusion protein FGFR3-TACC3 was not detected in common head and neck tumor cell lines (data not shown). Thus, to explore the role of the FGFR3-TACC3 fusion gene in cancer, we successfully established stable NPC cell lines (HNE1 and HK1) expressing human wild-type FGFR3 (isoform 3); wild-type TACC3; the fusion protein FGFR3-TACC3 (FGFR3 exon18 and TACC3 exon10); or the FGFR3-TACC3 K508M (Lys508→Met508) mutant, a mutation that abrogates FGFR kinase activity. The pBabe-vector was used as a negative control ().
Figure 4. The FGFR3-TACC3 fusion gene can promote the development of NPC. Representative pictures of the stable expression of wild-type FGFR3(F), wild-typeTACC3(T),FGFR3-TACC3(FT) fusion protein and the FGFR3-TACC3 fusion protein with the K508M mutation (FGFR3-TACC3 K508M) in HNE1 and HK1 NPC cell lines, as detected by western blotting. (A) MTT assay measuring the viability of the stable HNE1 and HK1 cell lines expressing wild-type FGFR3, wild-type TACC3, the FGFR3-TACC3 fusion protein, or FGFR3-TACC3 K508M, or the vector control. (B) MTT assay measuring the viability of the FGFR3-TACC3 fusion protein-expressing stable HNE1 and HK1 cell lines treated with 500 nM or 1000 nM PD173074. DMSO served as a negative control. The error bars denote the SEM. * refers to differences between DMSO and 500 nM PD173074 treatment, whereas # refers to differences between DMSO and 1000 nM PD173074 treatment. *** and ### mean P < 0.001 by 2-way ANOVA. (C) Cell cycle analysis of the FGFR3-TACC3 fusion protein- or vectorcontrol-expressing stable HNE1 and HK1 cell lines. * refers to differences between FT and the pBabe vector, P < 0.05 by unpaired t-test. (D) Colony formation assay of the stable HNE1 and HK1 cell lines expressing the FGFR3-TACC3 fusion protein, the FGFR3-TACC3 fusion protein with the K508M mutation (FGFR3-TACC3 K508M), or the vector control. * refers to differences between FT and the pBabe vector or FT-K, * means P < 0.05, ** means P < 0.01, *** means P < 0.001 by paired t-test. (E) Anchorage-independent growth in soft agar by the stable HNE1 cell lines expressing the FGFR3-TACC3 fusion protein, the FGFR3-TACC3 fusion protein with the K508M mutation (FGFR3-TACC3 K508M), or the vector control. 40X magnification. (F) Representative pictures of phosphorylated Akt (p-Akt), total Akt (t-Akt), phosphorylated ERK (p-ERK), total ERK (t-ERK), FGFR3 and β-actin in stable HNE1 cell lines, as determined by protein gel blotting.
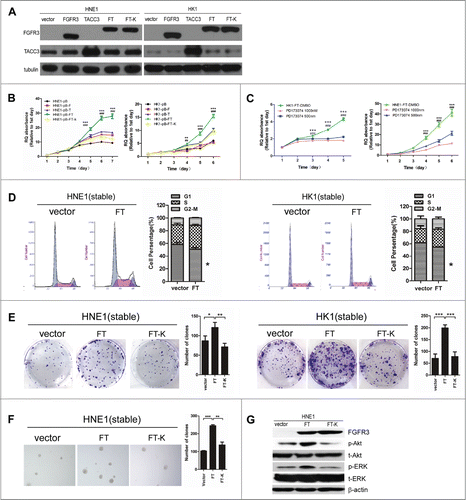
We performed an MTT assay to investigate whether the FGFR3-TACC3 fusion protein promotes cell growth. The results showed that the FGFR3-TACC3-expressing HNE1 and HK1 cells grew faster than the stable cell lines overexpressing the empty vector, wild-type FGFR3, or wild-type TACC3 did (), suggesting that FGFR3-TACC3 can promote the proliferation of NPC cells. Moreover, the growth rate of the cells expressing FGFR3-TACC3 K508M was greatly reduced compared with that of the cells expressing FGFR3-TACC3 (), indicating that the kinase activity of FGFR3 is essential for the ability of the FGFR3-TACC3 fusion protein to enhance cellular proliferation. In addition, we treated stable cell lines with different concentrations of the FGFR kinase-specific inhibitor PD173074 and found that treatment with the FGFR3 kinase inhibitor PD173074 can slow the cell proliferation rate in a dose-dependent manner (). This finding further convinced us that the kinase activity of FGFR3 in the FGFR3-TACC3 fusion protein plays an important role in the growth of NPC cells.
Next, we examined whether FGFR3-TACC3 has any effect on the cell cycle. As shown in , the percentage of cells expressing FGFR3-TACC3 that were in the G1 phase was less than in the control, with statistical significance in both the HNE1 and the HK1 stable cell lines. This finding suggested that the FGFR3-TACC3 fusion protein regulates the G0/G1 phase in the cell cycle, most likely promoting cell cycle progression in NPC cells.
The FGFR3-TACC3 fusion protein enhances the transforming ability of NPC cells
To investigate whether the FGFR3-TACC3 fusion protein can enhance the transforming ability of NPC cells, we used both an anchorage-independent growth assay and a colony formation assay. As shown in , FGFR3-TACC3 fusion protein-expressing HNE1 or HK1 cells formed significantly more colonies than the empty vector-expressing cells did. Because HK1 is a highly differentiated NPC cell line, it is difficult for these cells to form colonies in an anchorage-independent assay. We observed that HNE1 cells overexpressing FGFR3-TACC3 formed more and larger colonies than the control cells did (), suggesting that the FGFR3-TACC3 fusion gene promotes cell transformation in NPC. However, the kinase-dead form of FGFR3-TACC3 K508M reduced the induction of the number of colonies in the colony formation assay and anchorage-independent assay (), indicating that FGFR3 kinase activity is important for the FGFR3-TACC3 fusion gene to enhance the transforming ability of NPC cells.
To investigate the potential downstream pathways of FGFR3-TACC3, we examined both the ERK and the Akt signaling pathways and found that ERK and Akt phosphorylation was induced by FGFR3-TACC3, suggesting that FGFR3-TACC3 triggers activation of the Akt and ERK signaling pathways (). However, the kinase-dead form, FGFR3-TACC3K508M, abrogated this induction, suggesting that the FGFR3 kinase activity in the FGFR3-TACC3 fusion protein plays an essential role in the activation of the downstream signaling pathways ().
Discussion
To date, high-throughput technologies have led to the discovery of a plethora of fusion genes in different solid tumors.Citation26 However, the current study is the first to identify the FGFR3-TACC3 fusion gene, which promotes cell proliferation and transformation in NPC, in a cohort of NPC patients. Several fusion genes, such as UBR5-ZNF423, have been detected by RNA-seq in 6 EBV (+) NPC tumor lines.Citation17 In the present study, we performed RNA-seq on 12 primary NPC patients and found the fusion of FGFR3 and TACC3. These results more accurately reflect chromosomal aberration in NPC patients.
We are also the first to detect FGFR3-TACC3 fusion genes in ESCC and to examine the existence of FGFR3-TACC3 fusion genes in head and neck tumors and lung cancer, as stated earlier. The results indicate that this abnormal fusion is not restricted to a certain cancer type, but rather might commonly exist in cancer, which may account for the development and progression of cancers. However, the FGFR3-TACC3 fusion was not detected in hepatocellular carcinoma, colon cancer, or gastric cancer, most likely due to the limited sample size. Thus, we will further expand the sample size to examine the FGFR3-TACC3 fusion in a future study.
Furthermore, we found certain new forms of FGFR3-TACC3 fusion transcripts, such as the fusion between exon 18 of FGFR3 and exon 6 or exon 14 of TACC3 and the fusion between exon 19 of FGFR3 and exon 6 or intron 10 and exon 11 of TACC3, which are different from those already reported.Citation19-22,Citation25 In previous studies, the FGFR3 fusion mainly involved exons 18 and 19;Citation20 the kinase domain of FGFR3is located in exon 17.Citation27 Therefore, we designed a primer specific for exon 17 of FGFR3 to amplify FGFR3-TACC3, which might have resulted in certain fusion forms of FGFR3-TACC3 being undetected. In the future, we will attempt to detect FGFR3-TACC3 fusion genes with primers specific for different exons of FGFR3.
The FGFR3-TACC3 fusion was discovered in glioblastoma multiforme, in which it displayed oncogenic activity due to encoding of a fusion protein with constitutive kinase activity. This activity induced a loss of mitotic fidelity and aneuploidy, and thus contributed to tumorigenesis.Citation19 FGFR3 itself and its related mutations also serve as prognostic factors or targets in urothelial cell carcinoma.Citation28,29 Overexpression of FGFR3-TACC3 was found to induce cell proliferation in breast cancer, whereas an inhibitor of FGFR3 abolished the increased cell growth of FGFR3-TACC3-expressing cells.Citation21 These findings suggest that FGFR3 kinase activity plays an important role in the function of FGFR3-TACC3. In our study, we showed that overexpression of the FGFR3-TACC3 fusion protein induced a significant increase in cell proliferation and anchorage-independent cell growth in vitro compared with controls (). Amino acid 508 (Lys) is regarded as the key site of the functional kinase activity of FGFR3,Citation10 and the introduction of FGFR3-TACC3 K508M had no effect on cell proliferation, colony formation, or transformation ability (). Taken together, our data suggest that the kinase activity of FGFR3 is essential for the oncogenic function of the FGFR3-TACC3 fusion protein in NPC and that amino acid 508 of FGFR3 plays an important role in the activation of FGFR3.
The ERK signaling pathway has been previously reported as one of the downstream signaling pathways of FGFR3-TACC321. The Akt pathway has been shown to play an important role in cell growth. Therefore, we examined both the ERK and the Akt signaling pathways and found that both pathways are activated by FGFR3-TACC3 ().
In addition, FGFR3-TACC3 has been shown to be tumorigenic via deletion of the 3′-UTR of FGFR3, resulting in escape of miR-99a regulation.Citation11 A low miR-99a level has been demonstrated in head and neck cancer,Citation30 suggesting that the dysregulation of miR-99a in cancer might lead to the activation of FGFR3, further contributing to tumorigenesis. Nonetheless, miR-99a might not be distributed evenly in cancers. Further investigation of the expression of miR-99a and the correlation between miR-99a, FGFR3, and FGFR3-TACC3is needed, as is further exploration of the underlying mechanisms by which the FGFR3-TACC3 fusion gene functions during NPC development.
In conclusion, herein, we are the first to report FGFR3-TACC3 fusion in NPC and ESCC. We found that this fusion enhanced cell proliferation and transformation in NPC, suggesting that FGFR3-TACC3 may be useful as a diagnostic marker and therapeutic target for personalized medicine for cancers. Thus, it is urgent to identify potential inhibitors or small molecules against the FGFR3-TACC3 fusion gene to prevent the oncogenic function of this fusion gene in the development of NPC or other tumors.
Materials and Methods
Tumor samples and sequencing
Tumor samples, including 130 NPC samples, 308 head and neck tumor samples, 48 ESCC samples, 50 hepatocellular carcinoma samples, 35 colon cancer samples, 51 gastric cancer samples, and 52 lung cancer samples, were acquired from Sun Yat-Sen University Cancer Center's Tumor Tissue Bank. Another 29 NPC samples were acquired from the Tumor Tissue Bank of Yunnan Cancer Center in China. The Institute Research Medical Ethics Committee of Sun Yat-Sen University Cancer Center and Yunnan Cancer Center granted IRB approval for this study. We obtained informed consent from all of the participants involved in the study. Tissues were obtained from surgery and snap-frozen and then immediately immersed in RNA later reagent (Ambion, TX, USA) overnight at 4ºC. These tissues were stored at −80°C prior to RNA extraction.
Paired-end libraries (n = 14) including NPEC2, C666-1, and 12 NPC tissues were prepared according to the protocol provided by Illumina (San Diego, CA, USA) with the mRNA-seq Sample Prep Kit (Illumina). Images generated by sequencers were converted into nucleotide sequences by a base-calling pipeline. Raw reads were saved in fast format. Removing the dirty raw reads was necessary before analyzing the data. The reads were aligned to the human genome (hg18 version). During alignment, certain reads could only be aligned when they were divided into 2 segments, known as 2-segment alignment. Next, according to information on 2-segment alignments, we used a Perl script to retrieve fusion sites. Fusion was detected if the fusion sites were located within known exon boundaries from 2 different genes and if there was at least one paired-end read to support it.
RNA extraction and reverse transcription
Each tissue was transferred to a liquid nitrogen-precooled mortar and ground into a powder using a pestle, and the E.Z.N.A. DNA/RNA Isolation Kit (Omega Bio-tek, USA) was used to extract RNA according to the manufacturer's instructions. The RNA was eluted with 50μl nuclease-free water. Approximately 2μg total RNA was reverse transcribed using the Thermo Script RT-PCR System (Invitrogen, CA, USA), with oligo (dT) as the primer.
Reverse transcription-PCR (RT-PCR)
The occurrence rate of the FGFR3-TACC3 fusion gene in different types of tumor samples was determined by real-time RT-PCR using Light Cycler 480 SYBR Green I Master(Roche, Indianapolis, IN, USA). The real-time PCR and data collection were performed using a Light Cycler 480 II (Roche). Quantitative determination of the RNA levels was performed in triplicate in 3 independent experiments. All of the PCR products were subjected to sequencing. The sequences of the real-time PCR primers were as follows:
FGFR3-sense:5′-CGTGAAGATGCTGAAAGACGATG-3′
TACC3-anti-sense:5′-AAACGCTTGAAGAGGTCGGAG-3′
Plasmid construction and retrovirus generation
cDNAs encoding human wild-type FGFR3 and wild-type TACC3 were amplified from the RNA of the immortalized nasopharyngeal epithelial cell line NP69 by PCR using specific primers (FGFR3:5′-ATGGGCGCCCCTGCCTG-3′, 5′-TCACGTCCGCGAGCCCC-3′; TACC3:5′-ATGAGTCTGCAGGTCTTAAACGACA-3′,5′-TCAGATCTTCTCCATC-TTGGAGATG-3′). The cDNA encoding the fusion gene FGFR3-TACC3 was amplified by PCR using total RNA from an FGFR3-TACC3-positive NPC patient. K508M-mutated FGFR3-TACC3 was generated by single-primer mutagenesis of the relevant sequence (AAG→ATG). The cDNA fragments were cloned into the TA vector pCR2.1 (Invitrogen) and then subcloned into the pBabe vector. The pBabe plasmids and packaging plasmid were transfected into the 293FT cell line using the calcium phosphate precipitation transfection method. The transfected cells were incubated at 37°C for 6 h, and the medium was then replaced with fresh medium. Cell supernatants containing the viral particles were harvested at 18–42 h after transfection and stored at −80°C.
Cell culture
Two well-differentiated NPC cell lines (HNE1 and HK1) were maintained in Roswell Park Memorial Institute (RPMI)1640 medium (Life Technologies, Carlsbad, CA, USA) supplemented with 5% fetal bovine serum (FBS) in a humidified 5% CO2 incubator at 37°C.
Establishment of stable cell lines
The NPC cell lines HNE1 and HK1 were infected with viral particles containing human wild-type FGFR3, wild-type TACC3, the fusion gene FGFR3-TACC3, or K508M-mutated FGFR3-TACC3. The empty vector was used as a negative control. The cells were selected and maintained in puromycin (1 μg/ml).
Western immunoblotting
Cell lysates were prepared in SDS sample buffer (62.5 mM Tris-HCl (pH 6.8), 3% sodium dodecyl sulfate (SDS), 10% glycerol, 50 mM DL-dithiothreitol (DTT), and 0.1% bromophenolblue) with protease inhibitors (Roche, Indianapolis, IN, USA). The proteins were separated by SDS-PAGE and transferred to a polyvinylidene fluoride membrane. The membrane was blocked with 5% bovine serum albumin in PBS-T (25 mM Tris, 0.8% NaCl, 2.68 mM KCl (pH 7.4), and 0.1% Tween 20) and subsequently incubated with an anti-FGFR antibody (Ab) (sc-13121; Santa Cruz, CA, USA), an anti-TACC3 Ab(ab134154; Abcam, Cambridge, MA, USA),an anti-α-tubulin Ab (#3873; Cell Signaling Technology, Danvers, MA, USA), an anti-phosphor-AKT Ab (#13038; Cell Signaling Technology) and a total Akt Ab (#4691; Cell Signaling Technology), an anti-phosphor-ERK Ab (#4390; Cell Signaling Technology)and an anti-total ERK Ab (#9194; Cell Signaling Technology), or anti-β-actin Ab (A5441; Sigma-Aldrich) at 4°C overnight. The blots were then treated with an HRP-conjugated secondary Ab (Pierce, Thermo Fisher Scientific Inc., Rockford, IL, USA).
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium-bromide (MTT) assay
To measure cell viability, MTT assays were conducted. Cells were seeded at 1500 cells/well in a 96-well plate in sextuplicate and allowed to attach overnight. To test the effects of an FGFR inhibitor on the proliferation of the NPC cells, the FGFR inhibitor PD173074 (Selleck Chemicals, TX, USA) was added to the cultures at a final concentration of 1000 nM or 500 nM 24 h post-plating. The viability of the HNE1 or HK1 cells was measured by incubating the cells with 0.5 mg/ml MTT reagent in phosphate-buffered saline (PBS) (Sigma-Aldrich) at 24, 48, 72, 96, or 120 h post-plating. After approximately 4 h of incubation with MTT, the reagent was aspirated, and the cells were lysed with 100% dimethyl sulfoxide (DMSO). The absorbance was measured at 490 nm.
Colony formation assay
After counting, 200 HNE1 cells or 2000 HK1 cells were plated in triplicate in 6-well plates and cultured in complete RPMI 1640 culture medium for 9 d. After most of the colonies had expanded to more than 100 cells, they were washed twice with PBS, fixed in methanol for 15 min, and dyed with crystal violet for 15 min at room temperature. After removing the dye by washing, the plates were photographed. At least 3 independent experiments were performed for each assay.
Soft-agar assays
Soft agar was prepared by mixing equivalent volumes of 1% agarose in PBS with 2X fresh medium. This mixture was added to each well of a 6-well plate and kept at 4°C for 15 min. Cells were suspended in 0.333% soft agar at 6000 cells/ml. This cell suspension (1 ml) was added on top of the prepared base agar layer. The plates were incubated for 2 weeks, and the colonies were counted under a microscope.
Cell cycle analysis
Cells were trypsinized and washed with 1X PBS to remove the residual serum and trypsin. Next, 1–2×106 cells were resuspended in 0.5 ml 1X PBS and vortexed gently, and 4.5 ml ice-cold 70% ethanol was added dropwise over a period of 30 sec to 1 min. The cells were then incubated at 4°C overnight, centrifuged, and washed twice with 1X PBS. The cells were resuspended in 0.5 ml propidium iodide (PtdIns) staining solution (450 μl 1X PBS, 50 μl 500 mg/ml PI, and 5 μl RNase), incubated for 30 min at 37°C, and analyzed by flow cytometry.
Statistical analysis
Statistical analysis was performed using SPSS software, version 17.0 (SPSS, Chicago, IL, USA). All of the assays were repeated at least 3 times. The data are presented as the mean ± SEM. The MTT assays were analyzed by 2-way ANOVA, and the rest of the statistical analyses were performed using unpaired t-test analysis. A P-value less than 0.05 was considered as statistically significant in all cases.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This study was supported by grants from the National Natural Science Foundation of China (81025014 and 81230045).The funder had no role in the study design, data collection or analysis, decision to publish, or preparation of the manuscript.
References
- Chan ATC, Teo PML, Johnson PJ. Nasopharyngeal carcinoma. Ann Oncol 2002; 13:1007-15; PMID:12176778; http://dx.doi.org/10.1093/annonc/mdf179
- Dolly P H, Johnson PJ. Introduction: nasopharyngeal cancer. Semin Cancerbiol 2002; 12:419.
- Kwok Wai L, Ka Fai T, Huang ADP. Focus on nasopharyngeal carcinoma. Cancer C 2004; 5:423-28; http://dx.doi.org/10.1016/S1535-6108(04)00119-9
- Razak AR, Siu LL, Liu FF, Ito E, O'Sullivan B, Chan K. Nasopharyngeal carcinoma: the next challenges. Eur J Cancer 2010; 46:1967-78; PMID:20451372; http://dx.doi.org/10.1016/j.ejca.2010.04.004
- Tang LQ, Chen QY, Guo SS, Chen WH, Li CF, Zhang L, Lai XP, He Y, Xu YX, Hu DP, et al. The impact of plasma epstein-barr virus DNA and fibrinogen on nasopharyngeal carcinoma prognosis: an observational study. British J Cancer 2014; 111:1102-11; PMID:25051405; http://dx.doi.org/10.1038/bjc.2014.393
- Sun D, Yang Z, Fu Y, Chen Y, Wang S, Zhang Y, Ma Y, Zhang X. Clinical value of serum Epstein-Barr virus DNA assay in the diagnosis of nasopharyngeal carcinoma. Tumour Biol: The J Int Soc Oncodev Biol Med 2014; PMID:24879626; http://dx.doi.org/10.1007/s132770142148x
- Lo KW, Chung GT, To KF. Deciphering the molecular genetic basis of NPC through molecular, cytogenetic, and epigenetic approaches. Semin Cancer Biol 2012; 22:79-86; PMID:22245473; http://dx.doi.org/10.1016/j.semcancer.2011.12.011
- Lo K-W, Huang DP. Genetic and epigenetic changes in nasopharyngeal carcinoma. Semin Cancer Biol 2002; 12:451-62; PMID:12450731; http://dx.doi.org/10.1016/S1044579X02000883
- Xiao X, Zhao W, Tian F, Zhou X, Zhang J, Huang T, Hou B, Du C, Wang S, Mo Y, et al. Cytochrome b5 reductase 2 is a novel candidate tumor suppressor gene frequently inactivated by promoter hypermethylation in human nasopharyngeal carcinoma. Tumour Biol: The J Int Soc Oncodevel Biol Med 2014; 35:3755-63; PMID:24338690; http://dx.doi.org/10.1007/s13277-013-1497-1
- Shu XS, Li L, Ji M, Cheng Y, Ying J, Fan Y, Zhong L, Liu X, Tsao SW, Chan AT, et al. FEZF2, a novel 3p14 tumor suppressor gene, represses oncogene EZH2 and MDM2 expression and is frequently methylated in nasopharyngeal carcinoma. Carcinogenesis 2013; 34:1984-93; PMID:23677067; http://dx.doi.org/10.1093/carcin/bgt165
- Lo K-W, Huang DP. Genetic and epigenetic changes in nasopharyngeal carcinoma. Semin Cancer Biol 2002; 12:451-62; PMID:12450731; http://dx.doi.org/10.1016/S1044579X02000883
- Grembecka J, He S, Shi A, Purohit T, Muntean AG, Sorenson RJ, Showalter HD, Murai MJ, Belcher AM, Hartley T, et al. Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat Chem Biol 2012; 8:277-84; PMID:22286128; http://dx.doi.org/10.1038/nchembio.773
- Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer 2007; 7:233-45; PMID:17361217; http://dx.doi.org/10.1038/nrc2091
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 2005; 310:644-8; PMID:16254181; http://dx.doi.org/10.1126/science.1117679
- Nam RK, Sugar L, Wang Z, Yang W, Kitching R, Klotz LH, Venkateswaran V, Narod SA, Seth A. Expression of TMPRSS2:ERG gene fusion in prostate cancer cells is an important prognostic factor for cancer progression. Cancer Biol Ther 2007; 6:40-5; PMID:17172822; http://dx.doi.org/10.4161/cbt.6.1.3489
- Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007; 448:561-6; PMID:17625570; http://dx.doi.org/10.1038/nature05945
- Chung GT, Lung RW, Hui AB, Yip KY, Woo JK, Chow C, Tong CY, Lee SD, Yuen JW, Lun SW, et al. Identification of a recurrent transforming UBR5-ZNF423 fusion gene in EBV-associated nasopharyngeal carcinoma. J Pathol 2013; 231:158-67; PMID:23878065; http://dx.doi.org/10.1002/path.4240
- Nowell PC. A minute chromosome in human chronic granulocytic leukemia. Science 1960; 132:1497.
- Singh D, Chan JM, Zoppoli P, Niola F, Sullivan R, Castano A, Liu EM, Reichel J, Porrati P, Pellegatta S, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 2012; 337:1231-5; PMID:22837387; http://dx.doi.org/10.1126/science.1220834
- Brittany CP, Matti JA, David EC, Kirsi JG, Yan S, Ping J, Xia L, Joy G, Hong Z, Limei H, et al. The tumorigenic FGFR3-TACC3 gene fusion escapes miR-99a regulation in glioblastoma. J Clin Invest 2013; 123:855-65; PMID:23298836
- Wu YM, Su F, Kalyana-Sundaram S, Khazanov N, Ateeq B, Cao X, Lonigro RJ, Vats P, Wang R, Lin SF, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Disc 2013; 3:636-47; PMID:23558953; http://dx.doi.org/10.1158/2159-8290.CD-13-0050
- Majewski IJ, Mittempergher L, Davidson NM, Bosma A, Willems SM, Horlings HM, de Rink I, Greger L, Hooijer GK, Peters D, et al. Identification of recurrent FGFR3 fusion genes in lung cancer through kinome-centred RNA sequencing. J Pathol 2013; 230:270-6; PMID:23661334; http://dx.doi.org/10.1002/path.4209
- Falini B, Mason DY. Proteins encoded by genes involved in chromosomal alterations in lymphoma and leukemia: clinical value of their detection by immunocytochemistry. Blood 2002; 99:409-26; PMID:11781220; http://dx.doi.org/10.1182/blood.V99.2.409
- Kas K, Voz ML, Roijer E, Astrom AK, Meyen E, Stenman G, Van de Ven WJ. Promoter swapping between the genes for a novel zinc finger protein and beta-catenin in pleiomorphic adenomas with t(3;8)(p21;q12) translocations. Nat Genet 1997; 15:170-4; PMID:9020842; http://dx.doi.org/10.1038/ng0297-170
- Williams SV, Hurst CD, Knowles MA. Oncogenic FGFR3 gene fusions in bladder cancer. Human Mol Genet 2013; 22:795-803; PMID:23175443; http://dx.doi.org/10.1093/hmg/dds486
- Annala MJ, Parker BC, Zhang W, Nykter M. Fusion genes and their discovery using high throughput sequencing. Cancer Lett 2013; 340:192-200; PMID:23376639; http://dx.doi.org/10.1016/j.canlet.2013.01.011
- Bellus GA, McIntosh I, Smith EA, Aylsworth AS, Kaitila I, Horton WA, Greenhaw GA, Hecht JT, Francomano CA. A recurrent mutation in the tyrosine kinase domain of fibroblast growth factor receptor 3 causes hypochondroplasia. Nat Genet 1995; 10:357-9; PMID:7670477; http://dx.doi.org/10.1038/ng0795-357
- Iyer G, Milowsky MI. Fibroblast growth factor receptor-3 in urothelial tumorigenesis. Urol Oncol 2013; 31:303-11; PMID:22285006; http://dx.doi.org/10.1016/j.urolonc.2011.12.001
- Pandith AA, Shah ZA, Siddiqi MA. Oncogenic role of fibroblast growth factor receptor 3 in tumorigenesis of urinary bladder cancer. Urol Oncol 2013; 31:398-406; PMID:20822928; http://dx.doi.org/10.1016/j.urolonc.2010.07.014
- Yen YC, Shiah SG, Chu HC, Hsu YM, Hsiao JR, Chang JY, Hung WC, Liao CT, Cheng AJ, Lu YC, et al. Reciprocal regulation of microRNA-99a and insulin-like growth factor I receptor signaling in oral squamous cell carcinoma cells. Mol Cancer 2014; 13:6; PMID:24410957; http://dx.doi.org/10.1186/1476-4598-13-6