Abstract
Tissue microarrays (TMAs) have become an invaluable tool in cancer research to evaluate expression and subcellular localization of proteins in cells and tissues. As the catalogs of candidate biomarkers and therapeutic targets become more extensive, there is a need to characterize and validate these targets and biomarkers in cell lines as a primary biological system in research laboratories. Thus, cell microarrays (CMAs) are useful as a high-throughput screening tool. Here, we constructed a CMA containing 32 publicly available immortalized breast cell lines with the goal of creating a method to rapidly screen for antigens of interest in breast cancer research in a relatively easy, rapid and cost-effective manner. As proof of concept, we performed immunocytochemical staining of the HER2 receptor, as the status of this protein is relevant to breast cancer and has previously been reported for these cell lines. We observed a complete concordance of our staining with the published status of HER2 in these cell lines. In addition, we examined the expression of CD44, epithelial markers EpCAM and E-cadherin and tyrosine phosphoproteins. The labeling of these proteins correlates with the known biology of the cell lines. Our results demonstrate the utility of our method to screen for potential biomarkers and therapeutic targets in breast cancer and we suggest that CMAs be used as a general approach in breast cancer research.
Abbreviations
| CMA | = | cell microarray |
| TMA | = | tissue microarray |
| IHC | = | immunohistochemical staining |
| ICC | = | immunocytochemical staining |
| TNBC | = | triple negative breast cancer |
| ER | = | estrogen receptor |
| PR | = | progesterone receptor |
Introduction
Immunohistochemistry (IHC) has become an established tool to evaluate the expression and subcellular localization of proteins and other molecules in tissues.Citation1 It has widespread utility in cancer where it is used as a method to confirm the identity of tissue types, to classify tumors, and to evaluate the presence of specific molecules for therapeutic or prognostic purposes. For instance, in breast cancer, the expression of estrogen receptor (ER), progesterone receptor (PR) and HER2 receptor is evaluated using IHC to classify tumors for determining the appropriate therapy for patients in conjunction with other clinical parameters.Citation2 Expression of ER is an indication for hormone therapy regimens such as tamoxifen, whereas HER2 positivity could serve as an indication for the use of HER2-targeted therapy such as trastuzumab.Citation3,4 In research laboratories, the use of TMAs permits a large number of tissue samples to be screened for the expression of proteins rapidly and with relative ease.Citation5 TMAs of tumor tissues are frequently used to look for the expression of proteins that might correlate with the etiology or pathogenesis of tumors in order to identify biomarkers or therapeutic targets.
In the biomedical research setting, cell lines are still the primary mode of investigation to study biological systems often leading to subsequent studies in animal models and primary tissues. Thus, there is a need for a rapid screening method to choose the appropriate cell lines to be used for a particular study and also to profile the expression of proteins in these cell lines. Our group has previously developed a cell microarray (CMA) of a panel of pancreatic cancer cell lines to evaluate the protein expression and subcellular localization of potential biomarkers in a comprehensive fashion.Citation6 In the current study, we developed a panel of 32 publicly available breast cell lines that broadly represent all major subtypes of breast cancer. As proof of principle, we carried out immunocytochemical (ICC) staining of HER2 receptor as the expression of this receptor in these cell lines has already been reported in the published literature. We found a complete concordance between our staining results and the reported status of HER2 levels. We next applied our method to include a few other molecules that are relevant to breast cancer including CD44, EpCAM, E-cadherin and tyrosine phosphoproteins. We observed distinct staining patterns of each of these molecules in different cell lines, which might correlate with their specific phenotypes. For example, we found that positive staining for CD44 was clustered in basal breast cancer lines while lack of detectable CD44 expression was mostly observed in luminal cell lines. The cell lines also exhibit the same membranous staining pattern for both EpCAM and E-cadherin confirming their roles as adhesion proteins. Overall, our results indicate that CMAs provide a useful high-throughput platform to screen cell lines for studying antigens of interest.
Results
Construction of breast cancer cell microarrays
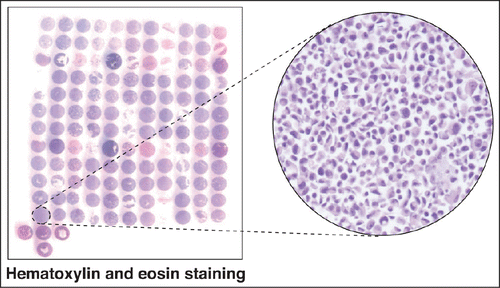
We collected 32 publicly available immortalized breast cell lines that represent major breast cancer molecular subtypes based on gene expression arrays-based classification by Neve et al and Kao et al.Citation7,8 These include 9 luminal cell lines, 11 basal-A cell lines and 12 basal-B cell lines (). All of the basal-B cell lines are also triple negative cell lines indicating that they lack the ER, PR and HER2 receptors. Seven of the 11 basal-A cell lines are triple negative while the remaining 4 exhibit amplification of HER2. The cell lines along with their ER, PR and HER2 receptor status and the source of origin are summarized in . The majority of these lines were immortalized from primary breast tumors or metastases. However, 3 cell lines - MCF10A, MCF12A and HBL100 - were derived from normal tissues or benign breast tumors, are non-tumorigenic in animals and are generally considered to represent benign breast epithelial cells in laboratory studies. Our CMA was constructed by spotting each of the cell lines in duplicate. We also included 2 normal breast tissues on the CMA slides. The panel of cell lines chosen for the CMA represents all major cell lines that are commonly used in breast cancer research. shows a hematoxylin and eosin stained CMA slide along with a magnified view of one of the cell lines showing the intact cellular architecture.
Table 1. List of the breast cell lines used in this study and the immunocytochemical staining scores of the indicated proteins
HER2 receptor
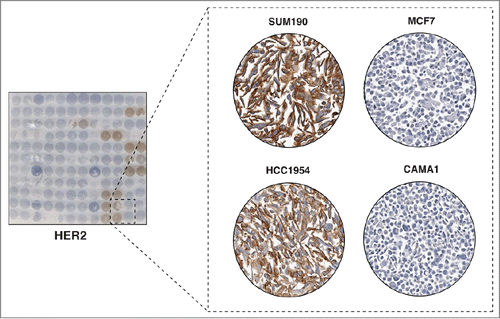
We performed immunocytochemical staining for HER2, one of the 3 molecules that are used clinically to classify breast cancer into molecular subtypes. As the expression status of HER2 on these cell lines is already known, our staining should provide a proof-of-concept of the utility of our method for expression screening. HER2 is a transmembrane receptor which belongs to the epidermal growth factor receptor (EGFR) family. HER2 is overexpressed in 20%-30% of breast cancer cases and the survival of the cells in these tumors relies on the activation of HER2 signaling pathways.Citation9 Patients with hormone receptor negative cancer (i.e. those that lack ER and PR) but with amplification of HER2 are associated with poor prognosis as this subtype has been shown to be aggressive.Citation10,11 When the humanized anti-HER2 antibody, trastuzumab, became available in 1998, the survival of women with this subtype improved.Citation12 In our immunocytochemical staining, we observed strong membrane staining of HER2, which is in agreement with the transmembrane localization of this protein (). More importantly, 7 cell lines in our panel that were reported to overexpress HER2 stained positive for HER2 expression ().Citation7,8 For example, SUM190 and HCC1954 were reported to have HER2 amplification and we observed strong membrane staining of HER2 in these cell lines (). In contrast, no immunocytochemical staining was observed in cell lines reported to be HER2 negative. For example, MCF7 and CAMA1 cell lines, which are both HER2 negative luminal breast cancer cell lines, stained negative for HER2 expression (). These results support the practicality and utility of our CMA as a rapid screening method for protein expression and localization.
Figure 2. Immunocytochemical staining of HER2. The left panel shows a CMA slide stained with a HER2 antibody. The panel on the right shows a magnified view of 4 representative cell lines with different levels of HER2 staining. Very strong cell membrane staining was observed in 2 HER2 amplified breast cancer cell lines SUM190 and HCC1954. However, negative staining patterns were observed in 2 HER2 negative luminal breast cancer cell lines, MCF7 and CAMA1.
CD44
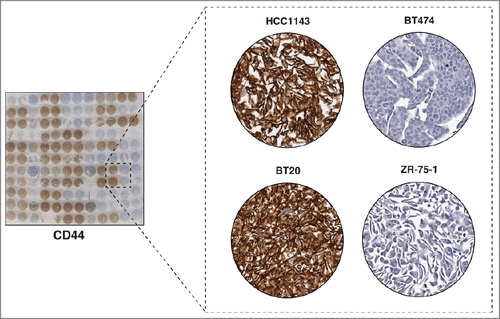
CD44 is a cell surface glycoprotein that is involved in cell-cell and cell-matrix interactions, adhesion and migration. It is encoded by the CD44 gene located on chromosome 11. Alternative splicing and post-translational modifications result in about 20 CD44 isoforms, with proteins ranging from 90 to 250 kDa in size.Citation13 The extracellular domain of CD44 contains binding sites for hyaluronan and other CD44 ligands, whereas the cytoplasmic domain contains binding sites for cytoskeletal adaptor proteins such as ankyrin.Citation14 Expression of CD44, together with lack of expression of CD24, has been used to identify and characterize a population of breast cancer stem cells that possess the ability to self-renew and to possess tumor-initiating properties.Citation15 These cells have also been implicated as having the ability to metastasize and to initiate epithelial-mesenchymal transition.Citation16 We used a pan-CD44 antibody to stain our breast cancer CMA, which revealed exclusive staining on the cell membrane (). More importantly, we observed that 21 out of 22 cell lines in our panel that stained positive for CD44 were from the basal subtype. On the other hand, 9 out of 11 cell lines that were negative for CD44 were of the luminal subtype (), clearly indicating that most basal breast cancers express CD44 while luminal breast cancers do not. This also supports the observation that the basal subtype has a greater stem-cell like phenotype.Citation17 Our results correspond to those of a survey of 240 primary breast tumors, where CD44-positive staining was shown to be enriched in basal breast cancers.Citation16 We demonstrate that CD44-positive staining is also a characteristic of immortalized basal-like breast cancer cell lines implying that the cell lines retain many of the features of the tumors from which they were derived.
Figure 3. Immunocytochemical staining of CD44. The left panel shows a CMA slide stained with a pan-CD44 antibody. The panel on the right shows a magnified view of 4 representative cell lines with different levels of expression of CD44. Very high expression was observed in 2 Basal A breast cancer cell lines HCC1143 and BT20, while the CD44 expression was not detectable in 2 luminal cell lines, BT474 and ZR-75-1.
EpCAM
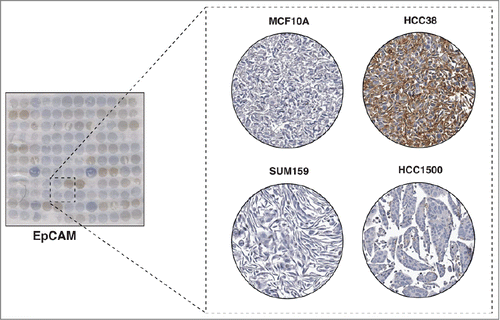
The epithelial cell adhesion molecule, EpCAM, has been shown to be overexpressed in breast cancer.Citation18 Also known as CD326, EpCAM is a transmembrane glycoprotein which is involved in homotypic calcium-independent cell-cell adhesion. It contains a long extracellular domain of 289 amino acids and a short 26 amino acid intracellular domain.Citation19 EpCAM was first identified as a tumor-associated antigen and its expression is frequently correlated with aggressive behavior of cancer cells. In breast cancer, high EpCAM expression is found in larger tumors that are less differentiated with nodal metastases and poor overall survival.Citation20 EpCAM has been shown to be expressed only in epithelial tumors and is not observed in mesenchymal or ectodermal tumors.Citation21 In a study by Martowicz et al. investigating the effects of EpCAM expression on the growth and invasion of breast cancer cell lines, the authors showed that those cell lines with epithelial phenotype require EpCAM to promote growth and invasion whereas the cell lines with mesenchymal phenotype are independent of EpCAM to promote these processes.Citation22 Our staining revealed localization of staining of EpCAM to the plasma membrane (). Variable expression of EpCAM was observed in the breast cancer cell lines represented on the CMA (). The staining pattern of EpCAM in the cell lines is in agreement with Martowicz et al. who showed that MCF7, SKBR3 and T47D have positive EpCAM expression while MDA-MB-231, HS578T and MCF10A do not express EpCAM based on western blot analysis ( and ).Citation22 Notably, the majority of EpCAM-negative cell lines were from the basal subtype and the majority of the luminal cell lines stained positive for EpCAM, which correlates with the epithelial and mesenchymal phenotype that these 2 subtypes exhibit. This suggests that our CMA method could be utilized to make inferences about the roles of proteins by correlating the staining properties with the known biology of the cell lines.
Figure 4. Immunocytochemical staining of EpCAM. The left panel shows a CMA slide stained with a monoclonal antibody against EpCAM. The panel on the right shows a magnified view of 4 representative cell lines showing different levels of expression of EpCAM. Two breast cancer cell lines, SUM159 and HCC1500 and one non-tumorigenic breast epithelial cell line, MCF10A showed very low staining of EpCAM, and HCC38 showed strong staining of EpCAM.
E-cadherin
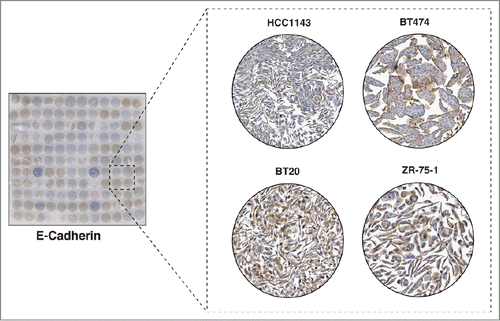
E-Cadherin is another transmembrane protein involved in epithelial cell-cell adhesion and migration. E-cadherin forms the adherens junction, which binds cells to neighboring cells by homotypic binding through the extracellular domain.Citation23 Its intracellular domain interacts with β-catenin, α-catenin, γ-catenin and p120catenin, and this complex links to the actin cytoskeleton. E-cadherin is thought to mediate signal transduction through the interactions with these molecules.Citation24 E-cadherin is relevant to cancer as it is a marker of epithelial cells that is lost when cells undergo the epithelial-mesenchymal transition (EMT) process, which is a hallmark of the invasion and metastasis process.Citation25 Our ICC staining revealed exclusive membrane staining of the protein with different intensity from cell line to cell line (). Notably, we observed a correlation between the patterns of staining of E-cadherin with EpCAM. Cell lines that have low or negative staining of E-cadherin tend to have low or negative staining of EpCAM as well (). This staining pattern is in accordance with the role of both proteins as epithelial markers whose expression is lost when epithelial cells undergo EMT. Furthermore, our results suggest that examining staining patterns for proteins on our CMA could help reveal the roles of those proteins by correlating staining patterns of new proteins with those of known roles in breast cancer biology.
Figure 5. Immunocytochemical staining of E-Cadherin. The left panel shows a CMA slide stained with a monoclonal antibody against E-Cadherin. The panel on the right shows a magnified view of 4 representative cell lines showing different levels of expression of E-Cadherin. The Basal A breast cancer cell line, HCC1143 cell line showed low expression of E-Cadherin. However, 2 luminal cell lines, BT474, ZR-75-1 and one basal cell line BT20 showed medium to high expression level of E-Cadherin.
Tyrosine phosphorylated proteins
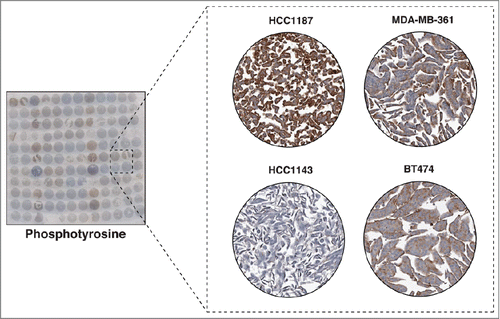
Phosphorylation of proteins is one of the major mechanisms by which a cell conveys its signals, either from the extracellular milieu into the nucleus, between cellular components, or from within a cell to the outside.Citation26 The majority of phosphorylation events in cells occur on serine and threonine residues while a much smaller fraction occurs on tyrosine residues.Citation27 Even though tyrosine phosphorylation accounts for a minority of the total cellular phosphorylation, it regulates important cellular processes including cell proliferation, cell growth, cell survival and metabolism.Citation28 This is why tyrosine kinases are frequently deregulated in cancer. More than half of the tyrosine kinases identified in the human genome have been implicated in cancer through gain-of-function mutations, fusion to partner proteins, gene amplification and overexpression.Citation29 These kinases have become attractive therapeutic targets to treat cancer, starting with the successful introduction of imatinib mesylate (Gleevec) as the first small molecule inhibitor to treat leukemia.Citation30 Use of tyrosine kinase inhibitors for treating breast cancer, especially those with hormone-receptor negative, has been less successful. This could potentially be attributed to differential tyrosine kinase activation between these tumors. Mass spectrometry-based phosphotyrosine profiling of breast cancer cell lines has revealed that protein tyrosine phosphorylation levels are vastly different across these cell lines.Citation31 We used a pan anti-phosphotyrosine antibody to stain the tyrosine-phosphorylated proteins on breast CMAs and observed distinct patterns of staining across the cell lines (). Of the 32 breast cancer cell lines, low staining was observed in 14, moderate staining in 12 and strong staining in 6 cell lines (). Overall, these results indicate that tyrosine phosphorylation levels of proteins in the cell lines greatly vary from each other suggesting differential activation of tyrosine kinases.
Figure 6. Immunocytochemical staining of tyrosine phosphorylated proteins. The left panel shows a CMA slide stained with an anti-phosphotyrosine monoclonal antibody. The panel on the right shows a magnified view of 4 representative cell lines showing different levels of tyrosine phosphorylation. SUM159, BT474 and HCC1500 showed relative high tyrosine phosphorylation levels, while SUM149 demonstrated low tyrosine phosphorylation.
Discussion
In this study, we have developed a cell microarray from a panel of widely used breast cell lines that represent the major subtypes of breast cancer. We show how this cell microarray could be used as a platform to study the expression of candidate biomarkers and therapeutic targets in breast cancer. In comparison with conventional protein gel blot analysis, CMA-based ICC staining offers a high-throughput approach for biomarker screens in a large panel of breast cancer cell lines. Results derived from the ICC staining can not only represent protein expression levels but provide information about the subcellular localization of proteins. As demonstrated, clear membranous staining patterns of HER2, EpCAM, E-cadherin and CD44 were observed in our study. We also showed that this platform could help in making inferences regarding functions of proteins by correlating expression patterns with known biology of the cell lines or the tumors from which they were derived. For example, our CD44 staining clustered all of the basal cell lines together, whereas E-cadherin and EpCAM have similar staining patterns on the cell lines. In addition to proteins, this CMA could also be used to screen the presence of other molecules, such as carbohydrates, lipids, mutant proteins, splice variants and proteins with other post-translational modifications. In summary, our results indicate that CMA is a useful platform for screening of cell lines for study of molecules of interest.
Materials and Methods
Cell lines
A total of 32 publicly available cell lines derived from breast tissues and tumors were used to construct the breast cancer cell microarray. BT20, BT474, BT549, CAMA1, HBL100, HCC1143, HCC1187, HCC1500, HCC1599, HCC1806, HCC1937, HCC1954, HCC38, HCC70, HS578T, MCF10A, MCF12A, MCF7, MDA-MB-157, MDA-MB-231, MDA-MB-361, MDA-MB-435, MDA-MB-468, SKBR3, T47D, ZR-75-1 and ZR-75-30 were purchased from ATCC and cell lines SUM 1315M02, SUM 225CWN, SUM149, SUM159 and SUM190 were a gift from Dr Stephen Ethier (Medical University of South Carolina). All cell lines were cultured in their recommended media and culture conditions and were grown until ∼70% confluency in tissue culture dishes before the media was removed and replaced with serum-free media for overnight incubation. The cells were washed 3 times with phosphate buffered saline and fixed with 10% formalin in phosphate buffered saline. The fixed cells were then detached, centrifuged at 500 g and added to 2% agarose. The cells were fixed for 2 days before paraffin embedded blocks were constructed to generate the cell microarray slides.
Immunocytochemical staining
Immunocytochemical staining was essentially performed as previously described.Citation6 Briefly, the CMA slides were heated in 60°C for 2 h followed by deparaffinization in xylene for 10 min. The rehydration process was carried out by keeping the slides in 100% ethanol for 2 min, then transferring to 95%, 70%, 50%, distilled water and phosphate buffered saline, each with 2 min immersion. Antigen retrieval was carried out by putting the slides in preheated target retrieval buffer (Dako C# S1699) in boiling steamer for 20 min. Peroxidase blocker was then applied for 20 min to discard any cellular peroxidase activity and the slides were then blocked with 5% goat serum for 20 minutes. Primary antibodies for pY100 (Cell Signaling Technology), HER2 (BD Biosciences), CD44 (Cell Signaling Technology), E-cadherin (Millipore), and EpCAM (Cell Signaling Technology) were diluted in antibody diluent before adding on to the slides for overnight incubation in 4°C. Slides were then incubated with HRP-conjugated secondary antibody (Dako, mouse, rabbit) for 30 min. DAB substrate was then added for 2-5 minutes until brown staining is observed. Slides were then counterstained in Mayer's hematoxylin (Dako) by dipping 3 times before quenching with ammonium hydroxide solution. The dehydration process was performed by 2 minute, step-wise immersion in water, 50% ethanol, 70% ethanol, 95% ethanol, and 100% ethanol. The slides were then transferred into xylene twice, each for 10 min before mounting with the mounting reagent. Cell lines were examined on a Nikon DS-Fil microscope equipped with a microscope operated using NIS-Elements F package and were scanned using an Aperio ScanScope CS. The staining was assessed by an experienced pathologist (EG) with scores ranging from 0 to 3.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the Tissue Microarray Core at Johns Hopkins University for their assistance.
Funding
This study was supported by a Career Catalyst Award from the Susan G. Komen for the Cure to XW, NCI's Clinical Proteomic Tumor Analysis Consortium initiative (U24CA160036 to AP), the NIH SPORE (Specialized Programs of Research Excellence) in Gastrointestinal Cancer Grant (CA62924 to AP), an NIH roadmap grant for Technology Centers of Networks and Pathways (U54GM103520 to AP), a DOD Era of Hope Scholar Award (BC051652 to AP), and The Avon Foundation Breast Cancer Research Foundation (AP). We thank Majlis Amanah Rakyat (MARA) of Government of Malaysia for the funding of MSZ. We thank the Department of Biotechnology of the Government of India for research support to the Institute of Bioinformatics, Bangalore, India. SR is a recipient of Senior Research Fellowship from University Grants Commission.
References
- Shi S-R, Shi Y, Taylor CR. Antigen retrieval immunohistochemistry review and future prospects in research and diagnosis over two decades. J Histochem Cytochem 2011; 59:13-32; PMID:21339172; http://dx.doi.org/10.1369/jhc.2010.957191
- Zhao L, Yang X, Khan A, Kandil D. Diagnostic role of immunohistochemistry in the evaluation of breast pathology specimens. Arch Pathol Lab Med 2013; 138:16-24; http://dx.doi.org/10.5858/arpa.2012-0440-RA
- Early Breast Cancer Trialists' Collaborative Group. Tamoxifen for early breast cancer: an overview of the randomised trials. The Lancet 1998; 351:1451-67; http://dx.doi.org/10.1016/S0140-6736(97)11423-4.
- Tremolada L, Magni F, Valsecchi C, Sarto C, Mocarelli P, Perego R, Cordani N, Favini P, Galli Kienle M, Sanchez J-C, et al. Characterization of heat shock protein 27 phosphorylation sites in renal cell carcinoma. PROTEOMICS 2005; 5:788-95; PMID:15682460; http://dx.doi.org/10.1002/pmic.200401134
- Kampf C, Andersson A-C, Wester K, Björling E, Uhlen M, Ponten F. Antibody-based tissue profiling as a tool for. Clin Proteomics 2004; 1:285-99; http://dx.doi.org/10.1385/CP:1:3-4:285
- Kim M-S, Kuppireddy SV, Sakamuri S, Singal M, Getnet D, Harsha HC, Goel R, Balakrishnan L, Jacob HKC, Kashyap MK, et al. Rapid characterization of candidate biomarkers for pancreatic cancer using cell microarrays (CMAs). J Proteome Res 2012; 11:5556-63; PMID:22985314; http://dx.doi.org/10.1021/pr300483r
- Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe J-P, Tong F, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 2006; 10:515-27; PMID:17157791; http://dx.doi.org/10.1016/j.ccr.2006.10.008
- Kao J, Salari K, Bocanegra M, Choi Y-L, Girard L, Gandhi J, Kwei KA, Hernandez-Boussard T, Wang P, Gazdar AF, et al. Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PLoS ONE 2009; 4:e6146; PMID:19582160; http://dx.doi.org/10.1371/journal.pone.0006146
- Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987; 235:177-82; PMID: 3798106; http://dx.doi.org/10.1126/science.3798106
- Sjögren S, Inganäs M, Lindgren A, Holmberg L, Bergh J. Prognostic and predictive value of c-erbB-2 overexpression in primary breast cancer, alone and in combination with other prognostic markers. J Clin Oncol 1998; 16:462-9; PMID:9469329
- Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science 1989; 244:707-12; PMID: 2470152; http://dx.doi.org/10.1126/science.2470152
- Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med 2001; 344:783-92; PMID:11248153; http://dx.doi.org/10.1056/NEJM200103153441101
- Fox SB, Fawcett J, Jackson DG, Collins I, Gatter KC, Harris AL, Gearing A, Simmons DL. Normal human tissues, in addition to some tumors, express multiple different CD44 isoforms. Cancer Res 1994; 54:4539-6; PMID:7519124
- Louderbough JMV, Schroeder JA. Understanding the dual nature of CD44 in breast cancer progression. Mol Cancer Res 2011; 9:1573-86; PMID:21970856; http://dx.doi.org/10.1158/1541-7786.MCR-11-0156
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci 2003; 100:3983-8; PMID:12629218; http://dx.doi.org/10.1073/pnas.0530291100
- Honeth G, Bendahl P-O, Ringnér M, Saal LH, Gruvberger-Saal SK, Lövgren K, Grabau D, Fernö M, Borg Å, Hegardt C. The CD44+/CD24- phenotype is enriched in basal-like breast tumors. Breast Cancer Res 2008; 10:R53; PMID:18559090
- Yehiely F, Moyano JV, Evans JR, Nielsen TO, Cryns VL. Deconstructing the molecular portrait of basal-like breast cancer. Trends Mol Med 2006; 12:537-44; PMID:17011236; http://dx.doi.org/10.1016/j.molmed.2006.09.004
- Osta WA, Chen Y, Mikhitarian K, Mitas M, Salem M, Hannun YA, Cole DJ, Gillanders WE. EpCAM is overexpressed in breast cancer and is a potential target for breast cancer gene therapy. Cancer Res 2004; 64:5818-24; PMID:15313925; http://dx.doi.org/10.1158/0008-5472.CAN-04-0754
- Baeuerle PA, Gires O. EpCAM (CD326) finding its role in cancer. Br J Cancer 2007; 96:417-23; PMID: 17211480; http://dx.doi.org/10.1038/sj.bjc.6603494
- Spizzo G, Went P, Dirnhofer S, Obrist P, Simon R, Spichtin H, Maurer R, Metzger U, von Castelberg B, Bart R, et al. High Ep-CAM expression is associated with poor prognosis in node-positive breast cancer. Breast Cancer Res Treat 2004; 86:207-13; PMID: 15567937; http://dx.doi.org/10.1023/B:BREA.0000036787.59816.01
- Momburg F, Moldenhauer G, Hämmerling GJ, Möller P. Immunohistochemical study of the expression of a Mr 34,000 human epithelium-specific surface glycoprotein in normal and malignant tissues. Cancer Res 1987; 47:2883-91; PMID:3552208
- Martowicz A, Spizzo G, Gastl G, Untergasser G. Phenotype-dependent effects of EpCAM expression on growth and invasion of human breast cancer cell lines. BMC Cancer 2012; 12:501; PMID:23110550; http://dx.doi.org/10.1186/1471-2407-12-501
- Takeichi M. Morphogenetic roles of classic cadherins. Curr Opin Cell Biol 1995; 7:619-27; PMID:8573335; http://dx.doi.org/10.1016/0955-0674(95)80102-2
- Bryant DM, Stow JL. The ins and outs of E-cadherin trafficking. Trends Cell Biol 2004; 14:427-34; PMID: 15308209; http://dx.doi.org/10.1016/j.tcb.2004.07.007
- Canel M, Serrels A, Frame MC, Brunton VG. E-cadherin–integrin crosstalk in cancer invasion and metastasis. J Cell Sci 2013; 126:393-401; PMID:23525005; http://dx.doi.org/10.1242/jcs.100115
- Graves JD, Krebs EG. Protein phosphorylation and signal transduction. Pharmacol Ther 1999; 82:111-21; PMID:10454190; http://dx.doi.org/10.1016/S0163-7258(98)00056-4
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 2006; 127:635-48; PMID:17081983; http://dx.doi.org/10.1016/j.cell.2006.09.026
- Hunter T. Tyrosine phosphorylation: thirty years and counting. Curr Opin Cell Biol 2009; 21:140-6; PMID: 19269802; http://dx.doi.org/10.1016/j.ceb.2009.01.028
- Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature 2001; 411:355-65; PMID:11357143; http://dx.doi.org/10.1038/35077225
- Krause DS, Van Etten RA. Tyrosine kinases as targets for cancer therapy. N Engl J Med 2005; 353:172-87; PMID: 16014887; http://dx.doi.org/10.1056/NEJMra044389
- Albeck JG, Brugge JS. Uncovering a tumor suppressor for triple-negative breast cancers. Cell 2011; 144:638-40; PMID:21376226; http://dx.doi.org/10.1016/j.cell.2011.02.030