Abstract
Both in epithelial development as well as in epithelial cancers, the p53 family member p63 plays a crucial role acting as a master transcriptional regulator. P63 steady state protein levels are regulated by the E3 ubiquitin ligase Itch, via a physical interaction between the PPxY consensus sequence (PY motif) of p63 and one of the 4 WW domains of Itch; this substrate recognition process leads to protein-ubiquitylation and p63 proteasomal degradation. The interaction of the WW domains, a highly compact protein-protein binding module, with the short proline-rich sequences is therefore a crucial regulatory event that may offer innovative potential therapeutic opportunity. Previous molecular studies on the Itch-p63 recognition have been performed in vitro using the Itch-WW2 domain and the peptide interacting fragment of p63 (pep63), which includes the PY motif. Itch-WW2-pep63 interaction is also stabilized in vitro by the conformational constriction of the S-S cyclization in the p63 peptide. The PY motif of p63, as also for other proteins, is characterized by the nearby presence of a (T/S)P motif, which is a potential recognition site of the WW domain of the IV group present in the prolyl-isomerase Pin1. In this study, we demonstrate, by in silico and spectroscopical studies using both the linear pep63 and its cyclic form, that the threonine phosphorylation of the (T/S)PPPxY motif may represent a crucial regulatory event of the Itch-mediated p63 ubiquitylation, increasing the Itch-WW domains-p63 recognition event and stabilizing in vivo the Itch-WW-p63 complex. Moreover, our studies confirm that the subsequently trans/cis proline isomerization of (T/S)P motif by the Pin1 prolyl-isomerase, could modulate the E3-ligase interaction, and that the (T/S)pPtransPPxY motif represent the best conformer for the ItchWW-(T/S)PPPxY motif recognition.
Abbreviations
| CXCR4 | = | chemokine receptor |
| HECT | = | Homologous E6-AP Carboxyl Terminus |
| RHS | = | Rapp-Hodgkin syndrome |
| TFE | = | 2, 2, 2-trifluoroethanol |
| RP-HPLC | = | reverse phase high performance chromatography |
| IPTG | = | isopropyl-β-D-thiogalactoside |
| pep63 | = | p63(534–551) peptide |
| Ppep63 | = | phosphorylated pep63 |
| cPpep63 | = | cyclic phosphorylated pep63 |
| TRAF6 | = | TNF receptor-associated factor 6 |
| TNF | = | tumor necrosis factor |
Introduction
Protein ubiquitylation is a post-translational modification leading to either target protein degradation through the proteasome or activation of the target protein via conformational changes, which depends on the ubiquitin linkage.Citation1-3 Ubiquitin E3 ligases, for their ability to accept the charged ubiquitin from the E2 and directly transfers it to a protein substrate, play an essential role in the protein ubiquitylation and, recently, their relevance has been recognized in the pathogenesis of a significant number of human diseases.Citation4,5 Itch is a crucial member of HECT (Homologous to E6-AP carboxyl terminus) ubiquitin E3 ligases Citation6 that impairs TNF-induced NF-κB activation by facilitating A20-mediated ubiquitylation and degradation of the adaptor protein RIP, in the TNF receptor complex in T cells Citation7 and macrophages.Citation8 Accordingly, patients with Itch mutations have autoimmune inflammatory cell infiltration in various tissues.Citation9 Itch is also required for negative regulation of TNF- and lipopolysaccharide (LPS)-mediated TNF receptor-associated factor 6 (TRAF6) ubiquitylation induced RING finger protein 11Citation10 and represent a negative regulator of osteoclastogenesis by promoting de-ubiquitylation of TRAF6.Citation11 Moreover, like other HECTs, Itch is also deregulated in cancer development and represents a potential target for anticancer treatment.Citation3,12 Since Itch is able to regulate chemosensitivity,Citation13 we attempted to develop specific inhibitors. Indeed, a recent high throughput screening for Itch inhibitors has identified compounds that regulate chemo-sensitivity by regulating autophagy.Citation14 Relatively few studies have been performed to identify E3 inhibitors, due to the intrinsic difficulties of the reaction involved, that is, the complexity of the protein components involved in the reaction makes the identification of specific E3 ubiquitin ligase inhibitors challenging in comparison to the identification of inhibitors of other classes of enzymes such as kinases or proteases.Citation15 Much of the research in the quest for E3 inhibitors has been finalized for the p53-HDM2 pathway since p53 is an extremely powerful transcription factor crucial in DNA damage response.Citation16-21 Here, the cellular defense to DNA damage is based on sensors and effectors that activates the cell death pathway,Citation22-24 via p53 Citation25-29 or its family members.Citation30-37 Like for our recent Itch inhibitor screening,Citation14 we believe that in a near future innovative E3 inhibitors will be developed. Here, however, we adopted a different approach to inhibit their function, based on a deeper understanding of the interaction of Itch with its substrate.
The substrate recognition and specificity of the HECTs is depending on specific protein-protein interaction domains, that account for their classifications in (i) HERC-RCC1-like domains (RLDs)-HECTs, (ii) SI(ngle)-HECTs or (iii) C2-WW-HECTs that have a tryptophan-tryptophan (WW) domain. Itch, indeed, recognizes its substrates via a specific WW domain. In the particular, Itch is able to regulate the ubiquitin-mediated proteasomal degradation of several proteins important in cancer, such as: c-Jun, JunB, Notch1, PLC-γ1, Notch1, Thioredoxin, tBid, Smad2, TIEG1, c-Flip-L, ErbB4, RASSF5, LATS1, LAPTM5, CXCR4, TRPV4, A20, itch, p73 and p63, reported in refs. 3,6,14,38. We have previously investigated the molecular mechanism of Itch recognition for the p63 transcription factor Citation39 in particular, because this powerful transcription factor is crucial for the development of epithelia,Citation31,40-45 it is involved in cancer,Citation46-59 and when it is mutated it causes severe genetic diseases.Citation60-62 While Itch is basically ubiquitous, the expression of the p53 family members, that is p63 and p73,Citation63-68 is strictly tissue- and cell-specific; moreover several disorders known as ectodermal dysplasias Citation12,69-75 as well as cancer show a deregulated expression levels of p63 and p73.Citation39,76 Physiologically, p63 and p73 protein levels are normally kept under strict control through Itch-mediated ubiquitylation, and indeed both p63 and p73 have a canonical PY motif (absent in p53 which is regulated by a different E3 ligase) located in their C-terminal which accounts for the binding to Itch through its WW domains. Both Itch and WWP1 target both isoforms of p63 for ubiquitin-mediated proteasomal degradation.Citation3
The WW domains, comprising circa 35 amino acids, are not highly stable and slightly bended 3-stranded β-sheets with a high aromatic content, including several conserved tryptophan residues.Citation77-79 WW domains, widely present in all phyla, are present as single or multiple copies in several proteins with different functions, including regulation of transcription, RNA processing, ubiquitin ligation, protein trafficking and receptor signaling.Citation80-82 The WW domains physically interact with short proline-rich sequences and recently, it has been shown that this WW/PY interaction may also be regulated by tyrosine phosphorylation. According to their ligand binding preferences, the WW domains are in fact grouped into 4 classes: group I binds polypeptides with the core consensus PPxY (PY motif), where ‘x’ can be any residue; group II recognizes the sequence PPLP (PL motif); group III selects for proline-rich sequences with arginine residues (PR motif); and group IV interacts with phospho(serine/threonine)-proline containing peptides (p(S/T)P motif), reviewed in refs 83–86. In the group I WW domains, the first conserved W residue, in concert with an exposed tyrosine residue, forms a concave hydrophobic binding surface for the first 2 proline residues in the PPxY motif of the ligand that packs against the W and Y residue forming a hydrophobic buckle that likely maintains the stable folded structure of the domain. Itch, a group I WW domain protein, and p63 interact with each other through the Itch-WW2 domain and the PPPY sequence (residues 540–543), which is the only PPxY motif in the p63 protein. Although the interaction among the WW2 domain of Itch and the PPxY motif of p63 is crucial for p63 degradation,Citation87,88 more detailed analysis of this interaction is lacking. This could be of relevance for its potential druggability. Previous molecular studies on the Itch-p63 recognition performed in vitro using the WW2 domain and the fragment pep63, which includes the PY motif, show that Itch-WW2-pep63 interaction is stabilized in vitro by the conformational constriction due to S-S cyclization of p63 peptide. The PY motif of p63, similarly to that in p73, is characterized by the close presence of (T/S)P motif, which is a potential recognition site of the WW domain of the IV group present in the prolyl-isomerase, Pin1,Citation89-92 a crucial regulator of p53 function.Citation93 In this study we have demonstrated that the phosphorylation of the threonine residue and proline cis/trans isomerization by prolyl-isomerase, Pin1, of the (T/S)P motif may represent molecular regulatory events of p63 ubiquitylation Itch-mediated, leading to an increase of the stabilization of the WW-p63 complex.
Results and Discussion
Phosphorylation of (T/S)P motif enhances the recognition of Itch-WW domains with the pep63
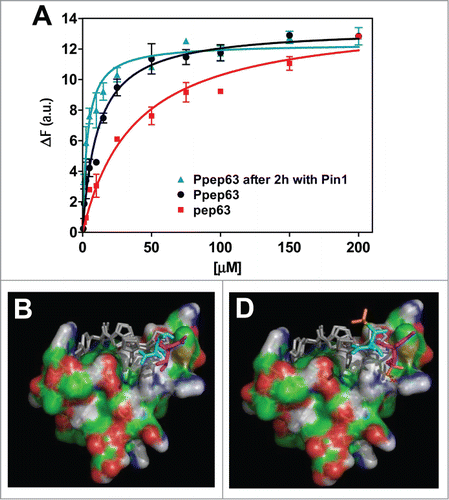
Early events of the physical recognition and interaction of p63 with the E3 ligase Itch, for its proteasomal degradation, seems to occur between the PPxY motif of p63 and WW2 domain of Itch.Citation94-97 In order to understand the biochemical properties of the interaction between these 2 proteins, the interaction between the Itch-WW2 and the synthetic peptide pep63, including the PPxY recognition motif, has been monitored using spectroscopic techniques; the apparent dissociation constant value of the Itch-WW2–pep63 complex has been reported previously.Citation96 The presence of the consensus TP motif, close to the PPxY recognition motif, and also the recent studies on the Pin1/p63 interaction Citation98,99 led us to evaluate the effect of the threonine phosphorylation of the (T/S)PPPxY motif on the binding of the Itch-WW domains to the pep63 peptide. The measured apparent dissociation constants of Itch-WW2 with both pep63 and the phosphorylated Thr form Ppep63 were obtained from the intrinsic fluorescence of the domain upon addition of increasing amounts of the peptides, resulting in a value of 42.09 ± 7.45 μM (for the WW2-pep63), which is in keeping with our previously reported value,Citation96 and 10.97 ± 1.45 μM for the Itch-WW2–Ppep63 (). Molecular docking studies between a structural model of Itch-WW2 domain and p(T/S)PPPxY motif have been also performed in order to evaluate the effects of the threonine phosphorylation on the Itch-WW-PPxY motif binding. shows both the Itch-WW-TPPPPY complexes with and without phosphorylation at the Thr residue. These computerised model simulations are compatible with the above experimental data and suggest that the observed increase in the binding could be due to the electrostatic interaction between the phosphate group of the motif and the Arg residue close to the binding pocket of the WW domain.
The first 2 WW domains of Itch equally bind the Ppep63
Since the relevance of the second Itch-WW domain in the Itch recognition has been highlighted for both p73 and p63 proteins,Citation94,96,100 here we have analyzed the interaction of the Itch-WW1 domain with the phosphorylated form of pep63 by fluorescence spectroscopy, in order to compare the effects of the phosphorylation of (T/S)PPPxY motif on the recognition of the Itch-WW domains. Itch-WW1 domain, consisting of 47 residues, was expressed and purified as described in experimental materials () and the correct folding of the Itch-WW1 domain has been confirmed by CD spectra.Citation101 As shown in , its far-UV CD spectrum with a maximum negative signal at ∼207 nm, was characteristic of a protein with a predominant β sheet secondary structure, in agreement with the conserved 3-stranded β-fold reported for the WW domains.Citation83,84,102 The dissociation constant of the complexes formed by the Itch-WW1 domain and pep63 was determined quantitatively by following the changes of the intrinsic fluorescence of the tryptophan residues in the domain upon addition of increasing amounts of the peptide. The intrinsic fluorescence of Itch-WW1 was quenched by the addition of the Ppep63 and in a concentration-dependent manner, see . The binding profiles of ΔF vs [Ppeptide] show a hyperbolic shape, allowing us to assume the formation of a 1:1 complex, considering the good quality of the fitting obtained using an equation for one-binding site and the Scatchard plot (data not shown). The apparent dissociation constant value, obtained by fitting of the titration binding curve, was 5.1 ± 0.5 μM.
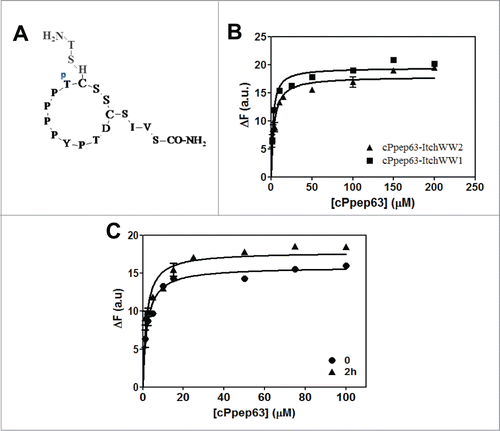
Figure 1. Itch-WW2 domain recognition of the phosphorylated form of pep63. (A) Interactions of Itch-WW2 with pep63 and Ppep63 were experimentally monitored by fluorescence spectroscopy. Intrinsic fluorescence changes of Itch-WW2 (5.0 μM) at the increase of pep63 (▪) and Ppep63 (•) concentration. The quenching of the emission band of Itch-WW2 using an 280 nm λex and 330 nm λem with a slit of 5 nm in 10 mM potassium phosphate buffer, 100 mM NaCl, 0.1 mM EDTA, 5 mM DTT, pH 6.0, at 37°C. Molecular Docking of the (B) TPPPPY or of the (C) TpPPPPY peptide with the WW2 domain model. The phosphate group is shown in orange. The 3D structures were obtained by molecular replacement as described in the materials and methods.
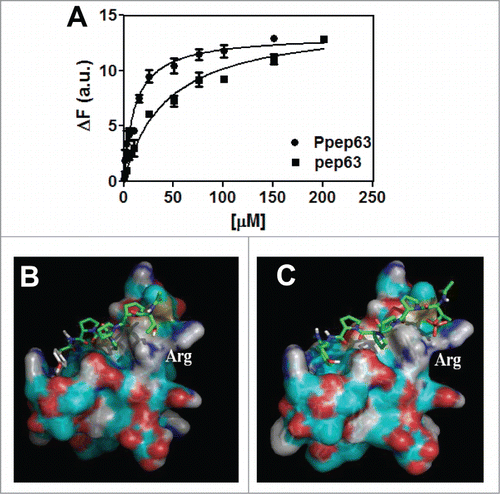
Figure 2. Production and characterization of the recombinant Itch-WW1 domain. (A) Expression of the recombinant GST-Itch-WW1 (29.7 kDa), SDS-PAGE 15% gel acrylamide before (lane 2) and after induction with 1 mM IPTG (lane 1); (B) purified Itch-WW1 before (lane 1) and after (lane 2) cleavage from GST (5.7 kDa); (C) Far-UV CD spectrum (50 μM) and (D) intrinsic fluorescence spectrum of the Itch-WW1 domain (5 μM), in 10 mM phosphate buffer, pH 7.0, with 0.15 M NaCl.
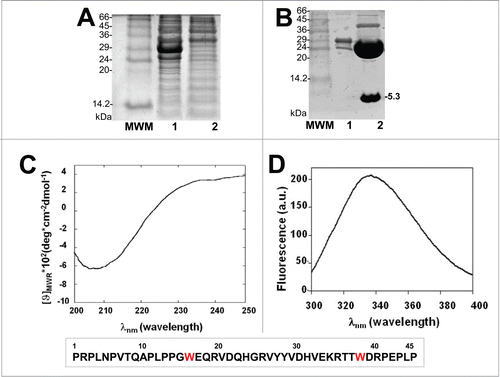
Figure 3. Interaction of Ppep63 peptide with Itch-WW1 domain. (A) Intrinsic fluorescence changes of Itch-WW1 (5 μM) at the increase of P-pep63 concentration. The quenching of the emission band of Itch-WW1 using an 280 nm λex and 330 nm λem in 10 mM potassium phosphate buffer, 100 mM NaCl, 0.1 mM EDTA, 5 mM DTT, pH 6.0, at 37°C; (B) Far-UV spectra of 50 μM ItchWW1 (blue), of 50 μM Ppep63 (red), of the mixture Itch-WW1/Ppep63 in a molar ratio 1:1 (black) and of the arithmetic sum of the spectra of the single components (green). All CD spectra were acquired in 10 mM potassium phosphate buffer, 100 mM NaCl, 0.1 mM EDTA, 5 mM DTT, pH 6.0, at 37°C, using a 0.1 cm path length quartz cell.
The interaction and the effects on the secondary structures were also evaluated by CD spectroscopy; the data show conformational changes induced by the binding of Ppep63 to the protein. In , the CD spectra of the Itch-WW1 domain are shown in the presence of equimolar concentration of Ppep63 and, indeed, the arithmetic sum of Itch-WW1 domain and Ppep63 spectra does not account for the CD spectrum generated by the Itch-WW1-Ppep63 complex. Also in this case, as previously obtained for the interaction of the Itch-WW2-pep63,Citation96 these changes in the dichroic spectrum strongly indicates that relevant changes in the conformation of the molecules are induced upon binding. These changes, in fact, could be due to a different local conformation of Ppep63 induced by the Itch-WW binding. Notably, the partial (or unstable) fold of the WW domains in the absence of the ligands, similar to what has been described for WWP1 with the Nogo-A peptide,Citation103-106 undergoes a significant conformational transitions to become quite structured upon binding to the target peptides. Although, the Itch-WW1 domain shows a stable fold, as also observed in the case of Itch-WW2,Citation96 it is not possible to fully exclude that small conformational arrangements of the more flexible regions of the domain also occur after the binding to the peptide.
Pro cis/trans isomerization of TP motif modulates the Itch-WW-pep63 recognition
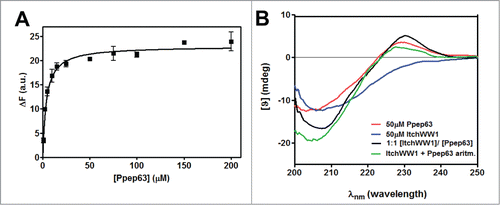
Pin1 isomerase catalyzes interconversion of proline isomers to restore cis-trans equilibrium at a biologically relevant timescale.Citation107 Regulation of the proline “conformational switch” due to peptidil prolyl-isomerases rules over the duration and amplitude of a variety of cellular processes.Citation107 As reported above, p(T/S)P motif is a characteristic recognition binding motif of the WW domain of Pin1 proline isomerase and recent data have shown a catalytic action of Pin1 on p63.Citation99 Therefore we have investigated the effect of the cis/trans isomerization of the first proline on the binding of the Itch-WW–p(T/S)PPPxY motif. Firstly, the binding between Itch-WW2 and Ppep63, following a previous incubation of the Ppep63 with Pin1 aimed at creating a proline cis/trans isomerization, has been analyzed by fluorescence spectroscopy (). The Itch-WW-Ppep63 binding was analyzed by measuring the variation of the intrinsic fluorescence of the Itch-WW2 at different concentrations of Ppep63 in solution, without or with 2 h of pre-incubation of the peptide with Pin1 at 37°C. The results shown in indicate that the pre-incubation of the Ppep63 with Pin1 increases the interaction of the peptide with Itch-WW2, decreasing the apparent Kd from 10.09 ± 0.9 to 4.4 ± 0.5 μM.
Figure 4. Proline cis/trans isomerization of (T/S)PPPxY recognition motif with the WW2-Itch domain. Intrinsic fluorescence changes (A) of Itch-WW2 (5 μM) at the increase of pep63 (—) and Ppep63 concentrations, after 0 (—) and 2 h (—) incubation of the Ppep63 in the presence of Pin1 enzyme. The quenching of the emission band of Itch-WW2 using an 280 nm λex and 330 nm λem with a slit of 5 nm in 10 mM potassium phosphate buffer, 100 mM NaCl, 0.1 mM EDTA, 5 mM DTT, pH 6.0, at 37°C. Molecular Docking of the (B) TPcisPPPY (cyan) and TPtransPPPY (magenta) peptide and (C) TpPcisPPPY (cyan) and TpPtransPPPY (magenta) with the WW2-Itch domain. In orange is shown the phosphate group.
The molecular docking between the model of the Itch-WW2 domain structure and p(T/S)PPPxY motif was also performed using both the proline (cis/trans) isomers of the motif (). As shown in the , the proline trans isomerization of the TPPPPY peptide, in both phosphorylated and not phosphorylated peptide cases, leads to an increase of the Itch-WW-TPPPPY binding with a decrease of the apparent Kd constants. The presence of the Arg residue close to the binding pocket of the WW domain can increase the molecular recognition and stabilize the complex. In the molecular docking, the proline trans isomerization draws up the phosphate group of the TpPPPPY to 2.8 Å from the side chain of the Arg residue of the WW domain. This event can increase the binding leading to a decrease of the calculated apparent Kd from 177.04 to 39.54 μM, according to the experimental results. Altogether, the in vitro and in silico data suggest that the (T/S)pPtransPPxY phosphorylated conformation is the most probable form of the Itch recognition site in p63 protein.
Effect of the phosphorylation and the cis/trans isomerization in the Itch-WW-cyclic pep63 binding
The cyclic form of the Ppep63 (cPpep63) was obtained and characterized as described in previous work ().Citation97 The intrinsic fluorescence changes of both WW1 and WW2 domains of the Itch protein were investigated by addition of the phosphorylated form of the cyclic pep63, in order to evaluate the effects of the phosphorylation on the molecular recognition and stabilization of the ItchWW-cPpep63 complex. shows the ΔFs of the maximal values of the intrinsic fluorescence of both Itch-WW1 and -WW2 at increased concentrations of the cPpep63. The observed apparent dissociation constants for Itch-WW1- and WW2-cPpep63 complexes were 2.1 ± 0.4 and 3.5 ± 0.6 μM respectively. In agreement with the results obtained for the linear form of pep63, the Thr phosphorylation increases the formation of the complex of the cyclic peptide with Itch-WW2, leading the Kd from 13.5 ± 0.8 Citation97 to 3.5 ± 0.6 μM. Moreover, the cyclization and the phosphorylation of the pep63 lead to an increase of about 12 folds in the binding of Itch-WW2 to pep63, with a decrease of the apparent Kd from 43.0 ± 1 to 3.5 ± 0.6 μM. The effect of the cis/trans proline isomerization on the Itch-WW2-cPpep63 binding was also investigated by pre-incubation of cPpep63 with Pin1 before interaction with the WW domain (). After 2 h of pre-incubation of cPpep63 with Pin1 not significant changes were observed in the apparent dissociation constant. This result can be explained considering that the cyclic form of the pep63, as described for many other cyclic peptides containing proline residues, could preferentially assume, for the steric effects, the trans configuration in the proline residue of the TP motif. Moreover, our preliminary NMR studies indicated that the linear form of Ppep63 is characterized by the presence of about 20% of cis proline isomers. These results are in agreement, with other studies that showed the trans form is more favored than the cis form in peptides with Ser(P)-Pro motifs, with populations of cis isomer of 12–20% depending on the adjacent residues.Citation108,109 Thus, altogether these results may be explained with an increase of the trans proline isomers of Ppep63 catalyzed by the Pin1 isomerase during the pre-incubation. The Arg-373 residue of Itch E3-ligase is involved in the interaction with pep63 Citation97 and as previously observed it goes from fast to slow exchange regime when interacts with pep63 and pep63 cyclic form, respectively. This evidence seems to be in agreement with a trans proline conformer of the cyclic form and explain the absence of Kd variation between cPpep63 and both the WW domains after pre-incubation with Pin1.
Figure 5. Interactions of Itch-WW domains with cyclic P-pep63 monitored by fluorescence. (A) Schematic representation of the cyclic form of Ppep63. (B) Intrinsic fluorescence changes of 5 μM Itch-WW1 (▪) or Itch-WW2 (▴) at the increase of cyclic Ppep63 concentration and (C) of 5 μM Itch-WW1 at the increase of cyclic Ppep63 concentration after 0 (•) and 2 h (▴) of incubation of the cyclic peptide with Pin1 enzyme. The quenching of the emission band of Itch-WW domains using an 280 nm λex and 330 nm λem with a slit of 5 nm in 10 mM potassium phosphate buffer, 100 mM NaCl, 0.1 mM EDTA, 5 mM DTT, pH 6.0, at 37°C.
Therefore, in keeping with very recent studies,Citation99 the Thr phosphorylation and the action of Pin1 on p63 directly regulates the physical recognition and interaction of Itch, similar to what has been described in the case of WWP1,Citation98,99,110 affecting the steady-state levels of the p63 protein. This could be also explained with a proline trans to cis isomerization of the p(T/S)P motif by Pin1.
Conclusions
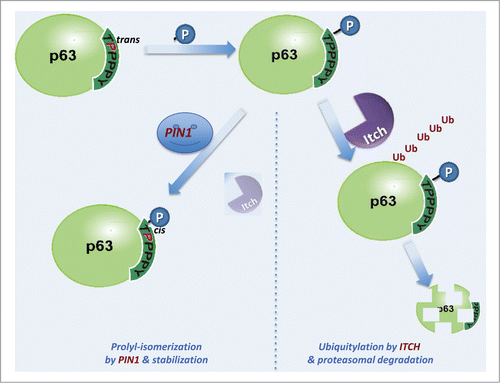
The presence of the conserved Arg (the Arg-373 in the Itch-WW2) (, http://www.ebi.ac.uk/Tools/msa/clustalw2/), close to the binding pocket of WW domains (), seems to be a relevant determinant for the regulation of the WW interaction and phosphorylation. Clearly, the data shown in and indicate a different interactivity of the recognition domain which is depending on the phosphorylation status. Based on these result, we propose that the phosphorylation of p63 in the (T/S)PPPxY recognition motif dictates the interactivity with the WW domain of Itch, and therefore with the proteasomal degradation of p63 itself, while the prolyl isomerization by Pin1, affecting the interaction with the WW domain, diverts the pathway toward a stabilization of the p63 steady state levels. shows a schematic representation of the functionality of the (T/S)PPPxY recognition motif with the WW domain. Indeed, in this schematic potential diagram of the events occurring during the protein-protein interactions, (i) the Thr phosphorylation status of p63 seems to promote the interaction with both proteins containing WW domains, Itch and Pin1, and (ii) the catalytic action of Pin1 may result in a differential availability of p63 to the degradative interaction with Itch. The modulation of the p63-Itch binding by phosphorylation could be a mechanism more widespread and regulate also the interaction of Itch-E3 ligase with other proteins, such as observed in the interaction of Itch with the chemokine receptor CXCR4.Citation111 This last one occurs through WW-Itch domains and a phosphorylated serine residue of a recognition sequence that does not contain proline residues.Citation111
Figure 6. Multiple sequence alignment of the WW domains of Itch and WWP1 E3-ligases. Alignment of the WW1, WW2 and WW3 domains of Itch and WWP1 E3-Ligases involved in the ubiquitylation of p63. The Arg residue corresponding to the Arg-373 in ItchWW2 is shown in blue, and it is conserved in all sequences. The conserved Trp residues, which are characteristics of the WW domains, are in red.

Figure 7. Schematic diagram of the physical interactions between p63 and Itch. Phosphorylation of the TPPPxY recognition motif of the p63 protein facilitates the Itch-p63 recognition promoting the ubiquitylation p63.94,96,97,100,117 This recognition can be modulate by the Pin1 that interacting with the (T/S)pP motif catalyzes the proline isomerization from trans to cis configuration of the recognition motif in p63 and decreases the Itch-p63 interaction, resulting in a stabilization of the protein steady state level. Accordingly, the sequence of events is (i) phosphorylation by a kinase, (ii) recognition by Itch, poly-ubiquitylation and proteasomal degradation (right path), (iii) alternatively, Pin1 recognition and trans-cis prolyl isomerization of the proline, reducing interaction with Itch, hence stabilizing the steady-state protein levels of p63 (left path).
The scheme in can be generalized for other PY interactions with WW domains, as the PY motif of p63, is also present in other proteins including for example RASSF5 where the PPxY motif interact with the WW domain of ItchCitation112 in a manner very similar to p63. All these proteins are characterized by the nearby presence of a (T/S)P motif, which is a potential recognition site of the WW domain of the IV group present in the prolyl-isomerase Pin1. shows that the (T/S)PPPxY recognition motif is present in different crucial cellular proteins, which stability and functional interaction with specific WW domains could affect the fate of the cell. Therefore, this motif could represent a consensus motif for the regulation of E3-ligase- dependent protein-degradation by Ser/Thr phosphorylation and Pin1 trans/cis proline isomerization.
Table 1. Sequences of the Proteins with the (T/S)PPPxY recognition motif
Finally, the results in indicate that a cyclic form of the peptide could specifically affect the PY interactions with the WW domains. According to the data shown, the Itch-WW2-pep63 interaction is stabilized in vitro by the conformational constriction of the S-S cyclization in the p63 peptide, offering a novel potential therapeutic target able to regulate the fate of the cell. Accordingly, cPpep63 peptide could represent a potential model for the design of competitive inhibitors of Itch-protein recognition.
In conclusion, the data presented show, by in silico and spectroscopical studies using both the linear pep63 and its cyclic form, a significant structural difference during the interaction of the different isoforms of the p63 peptide with both the Itch domains, allowing as to predict a differential effect on the steady-state protein levels of p63, regulated by the interaction with the ubiquitin E3 ligase Itch. The threonine phosphorylation of the (T/S)PPPxY motif seems to represent a crucial regulatory event of the Itch-mediated p63 ubiquitylation, increasing the Itch-WW domains-p63 recognition event and stabilizing in vivo the Itch-WW-p63 complex. Furthermore, the identification of the tissue- specific kinases (and/or phosphatases) could clarify the regulation of p63 degradation, also explaining why different E3 ligases act on p63. Finally, our studies confirm that the subsequently tran/cis proline isomerization of (T/S)P motif by the Pin1 prolyl isomerase, could modulate the E3-ligase interaction, and that the (T/S)pPtransPPxY motif represent the best conformer for the Itch-WW-(T/S)PPPxY motif recognition.
Materials and Methods
Peptide synthesis
Synthetic peptides were purchased from Spectra 2000 (Rome, Italy). Analysis of the synthetic peptides by reverse phase high performance chromatography (RP-HPLC) and mass spectrometry revealed a purity >98%. The 2 sequences of the peptides had the following sequences: 18-mer pep63 NH2- TSHCTPPPPYPTDCSIVS-CONH2 (1901.15 m/z) and Ppep63 NH2- TSHCTpPPPPYPTDCSIVS-CONH2 (1981.15 m/z).
Expression and purification of the Itch-WW1 and -WW2 domains
GST-ItchWW1 and WW2 were overexpressed using E.coli BL21 strain in LB medium containing 100 μM /ml ampicillin. Cells were grown at 37°C and the induction of the expression of the proteins were performed by addition of 1 mM IPTG. Cells were grown at 37°C for a further 4 hours, collected by centrifugation and after disrupted by sonication. GST-Itch-WW1 and -WW2 were purified using a GST-Trap FF column (5 ml, GE-Amersham) equilibrated with 100 mM Tris-HCl, pH 7.5, 0.3 M NaCl buffer at 1.0 ml/min and was eluted using 50 mM Tris-HCl, pH 8.0, 10 mM GSH buffer. The Itch-WW1 and -WW2 domains were cleaved from the GST using the PreScission Protease (GE-Amersham) and purified.
Circular dichroism analysis
CD measurements were performed using a Jasco 710 spectropolarimeter (Jasco, Tokyo, Japan) equipped with a thermal controller calibrated with camphor-sulfonic acid.Citation101,113-115 Far-UV CD experiments were carried out to explore the conformation of Itch-WW1 domain and the structural variation after addition of different concentrations of Ppep63. CD spectra were obtained between 200 and 250 nm using a path-length of 0.1 cm and between 600–300 nm using a path-length of 1 cm, a time constant of 1.0 s, a 2 nm bandwidth and a scan rate of 2 nm/min and at 20 or 50 mdeg sensitivity. Each spectrum was averaged over 4 scans and subjected to smoothing following subtraction of the buffer background. The measured ellipticity data were converted to mean molar ellipticity per residue ([θ], deg × cm2 × dmol−1).
Fluorescence interaction studies
The interactions of Itch-WW1 and -WW2 with the different forms of pep63 peptide were monitored by quenching of the emission band of Itch-WW domains excited at 280 nm using an λex and λem slit of 5 nm in 20 mM phosphate buffer, pH 6.0, 25 mM KCl, 2 mM DTT at 37°C. Binding constants were determined by fitting the titration curve of fluorescence changes versus equivalents of peptide provides a titration curve. The ΔF was plotted vs. molar equivalent of peptide and the following equation was used for generating a Scatchard plot from which binding constants were determined ΔF = nK[peptide] ΔFcomplex/(1 + K[peptide])
ΔF was the observed fluorescence changing after addition of the peptide. Fluorescence was measured at the chemical equilibrium at the earliest 2 min after addition of the peptide. The results were plotted using GRAPHPAD PRISM v. 4.0 for Windows (GraphPad Software, San Diego, CA, USA; http://www.graphpad.com).
Before the fluorescence interaction analysis, the pre-incubation of pep63 isoforms with and without Pin1 was performed using 2 mM of peptide, 0.73 μM of Pin1 (specific activity >162 nmoles/min/μg) (PiN3001, ATGen Co Ltd) in 20 mM Tris-HCl buffer, pH 8.0, 5 mM DTT for 2 h at 37°C. The solutions were added at increased concentrations to the WW domain solution in 10 mM phosphate buffer, pH 6.0, buffer, 100 mM NaCl at 37°C. The spectra at time 0 of pre-incubation in the presence of Pin1 were obtained immediately putting the Pin1-peptide solution in ice. The fluorescence of the peptides-Pin1 solutions was subtracted at the respective WW fluorescence's spectra.
Molecular docking procedures
The structure of the Itch-WW2 domain was obtained by homology modeling using pdb file from the protein data bankcode 2 DMv (www.pdb.org). Subsequently, Autodockver 4.2 Citation115 and AutodockVina version 1.1 algorithmsCitation117 were used for the docking of TPPPY, TpPPPY peptides and their trans/cis proline to the Itch-WW domain. The binding of each peptide to the Itch-WW domain was performed by, using the docking procedure for rigid receptor and flexible or semiflexible ligand based on a knowledge guided Protein Docking as already reported by Lu et al.Citation118
All the molecules were prepared for docking using Autodock Tools version1.4.Citation116 The default settings were used for all other parameters. Algorithms validation was conducted by re-docking native ligands to their receptors. The algorithms are considered valid if the re-docking results have a root square mean deviation (RSMD) less than 2 Å from original structure. After that, the binding energies of peptide to each receptor were calculated. Visualization of the binding site after docking analysis was performed by Autodock Tools version 1.4.Citation116,117
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work has been supported by the Medical Research Council, UK; grants from “Alleanza contro il Cancro” (ACC12), AIRC (2011-IG11955), AIRC 5 × mille (MCO #9979), Telethon Grant GGPO9133, Min. Salute (RicOncol 26/07) and IDI-IRCCS (RF08 c.15, RF07 c.57) to GM; from MAE GR:Italia-Albania 2012-2014 to SM.
References
- Schulman BA, Chen ZJ. Protein ubiquitination: CHIPping away the symmetry. Mol Cell 2005; 20(5):653-5; PMID:16337587; http://dx.doi.org/10.1016/j.molcel.2005.11.019
- Chen ZJ. Ubiquitin signalling in the NF-kappaB pathway. Nat Cell Biol 2005; 7(8):758-65; http://dx.doi.org/10.1038/ncb0805-758
- Bernassola F, Karin M, Ciechanover A, Melino G. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell 2008; 14(1):10-21; PMID:18598940; http://dx.doi.org/10.1016/j.ccr.2008.06.001
- Vinuesa CG, Cook MC, Angelucci C, Athanasopoulos V, Rui L, Hill KM, Yu D, Domaschenz H, Whittle B, Lambe T, et al. A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature 2005; 435(7041):452-8; PMID:15917799; http://dx.doi.org/10.1038/nature03555
- Wiesner S, Ogunjimi AA, Wang HR, Rotin D, Sicheri F, Wrana JL, Forman-Kay JD. Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell 2007; 130(4):651-62; PMID:17719543; http://dx.doi.org/10.1016/j.cell.2007.06.050
- Melino G, Gallagher E, Aqeilan RI, Knight R, Peschiaroli A, Rossi M, Scialpi F, Malatesta M, Zocchi L, Browne G, et al. Itch: a HECT-type E3 ligase regulating immunity, skin and cancer. Cell Death Differ 2008; 15(7):1103-12; PMID:18552861; http://dx.doi.org/10.1038/cdd.2008.60
- Shembade N, Harhaj NS, Parvatiyar K, Copeland NG, Jenkins NA, Matesic LE, Harhaj EW. The E3 ligase Itch negatively regulates inflammatory signaling pathways by controlling the function of the ubiquitin-editing enzyme A20. Nat Immunol 2008; 9(3):254-62; http://dx.doi.org/10.1038/ni1563
- Tao M, Scacheri PC, Marinis JM, Harhaj EW, Matesic LE, Abbott DW. ITCH K63-ubiquitinates the NOD2 binding protein, RIP2, to influence inflammatory signaling pathways. Curr Biol 2009; 19(15):1255-63; PMID:19592251; http://dx.doi.org/10.1016/j.cub.2009.06.038
- Lohr NJ, Molleston JP, Strauss KA, Torres-Martinez W, Sherman EA, Squires RH, Rider NL, Chikwava KR, Cummings OW, Morton DH, et al. Human ITCH E3 ubiquitin ligase deficiency causes syndromic multisystem autoimmune disease. Am J Hum Genet 2010; 86(3):447-53; PMID:20170897; http://dx.doi.org/10.1016/j.ajhg.2010.01.028
- Shembade N, Parvatiyar K, Harhaj NS, Harhaj EW. The ubiquitin-editing enzyme A20 requires RNF11 to downregulate NF-kappaB signalling. EMBO J 2009; 28(5):513-22; PMID:19131965; http://dx.doi.org/10.1038/emboj.2008.285
- Zhang H, Wu C, Matesic LE, Li X, Wang Z, Boyce BF, Xing L. Ubiquitin E3 ligase Itch negatively regulates osteoclast formation by promoting deubiquitination of tumor necrosis factor (TNF) receptor-associated factor 6. J Biol Chem 2013; 288(31):22359-68; PMID:23782702; http://dx.doi.org/10.1074/jbc.M112.442459
- Amiel J, Bougeard G, Francannet C, Raclin V, Munnich A, Lyonnet S, Frebourg T. TP63 gene mutation in ADULT syndrome. Eur J Hum Genet 2001; 9(8):642-5; PMID:11528512; http://dx.doi.org/10.1038/sj.ejhg.5200676
- Hansen TM, Rossi M, Roperch JP, Ansell K, Simpson K, Taylor D, Mathon N, Knight RA, Melino G. Itch inhibition regulates chemosensitivity in vitro. Biochem Biophys Res Commun 2007; 361(1):33-6; PMID:17640619; http://dx.doi.org/10.1016/j.bbrc.2007.06.104
- Rossi M, Rotblat B, Ansell K, Amelio I, Caraglia M, Misso G, Bernassola F, Cavasotto CN, Knight RA, Ciechanover A, et al. High throughput screening for inhibitors of the HECT ubiquitin E3 ligase ITCH identifies antidepressant drugs as regulators of autophagy. Cell Death Dis 2014; 5:e1203; http://dx.doi.org/10.1038/cddis.2014.113
- Eldridge AG, O'Brien T. Therapeutic strategies within the ubiquitin proteasome system. Cell Death Differ 2010; 17(1):4-13; PMID:19557013; http://dx.doi.org/10.1038/cdd.2009.82
- Manzl C, Fava LL, Krumschnabel G, Peintner L, Tanzer MC, Soratroi C, Bock FJ, Schuler F, Luef B, Geley S, et al. Death of p53-defective cells triggered by forced mitotic entry in the presence of DNA damage is not uniquely dependent on Caspase-2 or the PIDDosome. Cell Death Dis 2013; 4:e942; http://dx.doi.org/10.1038/cddis.2013.470
- Louwen F, Yuan J. Battle of the eternal rivals: restoring functional p53 and inhibiting Polo-like kinase 1 as cancer therapy. Oncotarget 2013; 4(7):958-71; PMID:23948487
- Neise D, Sohn D, Stefanski A, Goto H, Inagaki M, Wesselborg S, Budach W, Stühler K, Jänicke RU. The p90 ribosomal S6 kinase (RSK) inhibitor BI-D1870 prevents gamma irradiation-induced apoptosis and mediates senescence via RSK- and p53-independent accumulation of p21WAF1/CIP1. Cell Death Dis 2013; 4:e859; http://dx.doi.org/10.1038/cddis.2013.386
- Sayan BS, Sayan AE, Knight RA, Melino G, Cohen GM. p53 is cleaved by caspases generating fragments localizing to mitochondria. J Biol Chem 2006; 281(19):13566-73; PMID:16531411; http://dx.doi.org/10.1074/jbc.M512467200
- Melino G, Knight RA, Nicotera P. How many ways to die? How many different models of cell death? Cell Death Differ 2005; 12 Suppl 2:1457-62; PMID:16247490; http://dx.doi.org/10.1038/sj.cdd.4401781
- Chillemi G, Davidovich P, D'Abramo M, Mametnabiev T, Garabadzhiu AV, Desideri A, Melino G. Molecular dynamics of the full-length p53 monomer. Cell Cycle 2013; 12(18):3098-108; PMID:23974096; http://dx.doi.org/10.4161/cc.26162
- Evangelou K, Bartkova J, Kotsinas A, Pateras IS, Liontos M, Velimezi G, Kosar M, Liloglou T, Trougakos IP, Dyrskjot L, et al. The DNA damage checkpoint precedes activation of ARF in response to escalating oncogenic stress during tumorigenesis. Cell Death Differ 2013; 20(11):1485-97; PMID:23852374; http://dx.doi.org/10.1038/cdd.2013.76
- Salah Z, Cohen S, Itzhaki E, Aqeilan RI. NEDD4 E3 ligase inhibits the activity of the Hippo pathway by targeting LATS1 for degradation. Cell Cycle 2013; 12(24):3817-23; PMID:24107629; http://dx.doi.org/10.4161/cc.26672
- Zambetti GP. Expanding the reach of the p53 tumor suppressor network. Cell Death Differ 2014; 21(4):505-6; PMID:24608846; http://dx.doi.org/10.1038/cdd.2014.13
- Selvarajah J, Nathawat K, Moumen A, Ashcroft M, Carroll VA. Chemotherapy-mediated p53-dependent DNA damage response in clear cell renal cell carcinoma: role of the mTORC1/2 and hypoxia-inducible factor pathways. Cell Death Dis 2013; 4:e865; http://dx.doi.org/10.1038/cddis.2013.395
- Roh JL, Kim EH, Park HB, Park JY. The Hsp90 inhibitor 17-(allylamino)-17-demethoxygeldanamycin increases cisplatin antitumor activity by inducing p53-mediated apoptosis in head and neck cancer. Cell Death Dis 2013; 4:e956; http://dx.doi.org/10.1038/cddis.2013.488
- Bartoletti-Stella A, Mariani E, Kurelac I, Maresca A, Caratozzolo MF, Iommarini L, Carelli V, Eusebi LH, Guido A, Cenacchi G, et al. Gamma rays induce a p53-independent mitochondrial biogenesis that is counter-regulated by HIF1α. Cell Death Dis 2013; 4:e663; http://dx.doi.org/10.1038/cddis.2013.187
- Montero J, Dutta C, van Bodegom D, Weinstock D, Letai A. p53 regulates a non-apoptotic death induced by ROS. Cell Death Differ 2013; 20(11):1465-74; PMID:23703322; http://dx.doi.org/10.1038/cdd.2013.52
- Yu Y, Huang H, Li J, Zhang J, Gao J, Lu B, Huang C. GADD45β mediates p53 protein degradation via Src/PP2A/MDM2 pathway upon arsenite treatment. Cell Death Dis 2013; 4:e637; http://dx.doi.org/10.1038/cddis.2013.162
- Candi E, Dinsdale D, Rufini A, Salomoni P, Knight RA, Mueller M, Krammer PH, Melino G. TAp63 and DeltaNp63 in cancer and epidermal development. Cell Cycle 2007; 6(3):274-85; PMID:17264681; http://dx.doi.org/10.4161/cc.6.3.3797
- Candi E, Rufini A, Terrinoni A, Giamboi-Miraglia A, Lena AM, Mantovani R, Knight R, Melino G. DeltaNp63 regulates thymic development through enhanced expression of FgfR2 and Jag2. Proc Natl Acad Sci USA 2007; 104(29):11999-2004; http://dx.doi.org/10.1073/pnas.0703458104
- Rufini A, Niklison-Chirou MV, Inoue S, Tomasini R, Harris IS, Marino A, Federici M, Dinsdale D, Knight RA, Melino G, et al. TAp73 depletion accelerates aging through metabolic dysregulation. Genes Dev1 2012; 26(18):2009-14; PMID:22987635; http://dx.doi.org/10.1101/gad.197640.112
- Martynova E, Pozzi S, Basile V, Dolfini D, Zambelli F, Imbriano C, Pavesi G, Mantovani R. Gain-of-function p53 mutants have widespread genomic locations partially overlapping with p63. Oncotarget 2012; 3(2):132-43; PMID:22361592
- Neilsen PM, Noll JE, Suetani RJ, Schulz RB, Al-Ejeh F, Evdokiou A, Lane DP, Callen DF. Mutant p53 uses p63 as a molecular chaperone to alter gene expression and induce a pro-invasive secretome. Oncotarget 2011; 2(12):1203-17; PMID:22203497
- Tomasini R, Secq V, Pouyet L, Thakur AK, Wilhelm M, Nigri J, Vasseur S, Berthezene P, Calvo E, Melino G, et al. TAp73 is required for macrophage-mediated innate immunity and the resolution of inflammatory responses. Cell Death Differ 2013; 20(2):293-301; PMID:22976836; http://dx.doi.org/10.1038/cdd.2012.123
- Kadakia MP, Caron de Fromentel C, Sabapathy K. The 5th International p63/p73 Workshop: much more than just tumour suppression. Cell Death Differ 2012; 19(3):549-50; PMID:22240899; http://dx.doi.org/10.1038/cdd.2011.204
- Alexandrova EM, Petrenko O, Nemajerova A, Romano RA, Sinha S, Moll UM. ΔNp63 regulates select routes of reprogramming via multiple mechanisms. Cell Death Differ 2013; 20(12):1698-708; PMID:24013722; http://dx.doi.org/10.1038/cdd.2013.122
- Scialpi F, Malatesta M, Peschiaroli A, Rossi M, Melino G, Bernassola F. Itch self-polyubiquitylation occurs through lysine-63 linkages. Biochem Pharmacol 2008; 76(11):1515-21; PMID:18718449; http://dx.doi.org/10.1016/j.bcp.2008.07.028
- Melino G. p63 is a suppressor of tumorigenesis and metastasis interacting with mutant p53. Cell Death Differ 2011; 18(9):1487-99; PMID:21760596; http://dx.doi.org/10.1038/cdd.2011.81
- Yallowitz AR, Alexandrova EM, Talos F, Xu S, Marchenko ND, Moll UM. p63 is a prosurvival factor in the adult mammary gland during post-lactational involution, affecting PI-MECs and ErbB2 tumorigenesis. Cell Death Differ 2014; 21(4):645-54; PMID:24440910; http://dx.doi.org/10.1038/cdd.2013.199
- Chari NS, Romano RA, Koster MI, Jaks V, Roop D, Flores ER, Teglund S, Sinha S, Gruber W, Aberger F, et al. Interaction between the TP63 and SHH pathways is an important determinant of epidermal homeostasis. Cell Death Differ 2013; 20(8):1080-8; PMID:23686138; http://dx.doi.org/10.1038/cdd.2013.41
- Senoo M, Pinto F, Crum CP, McKeon F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell 2007; 129(3):523-36; PMID:17482546; http://dx.doi.org/10.1016/j.cell.2007.02.045
- Paris M, Rouleau M, Pucéat M, Aberdam D. Regulation of skin aging and heart development by TAp63. Cell Death Differ 2012; 19(2):186-93; PMID:22158419; http://dx.doi.org/10.1038/cdd.2011.181
- Giacobbe A, Bongiorno-Borbone L, Bernassola F, Terrinoni A, Markert EK, Levine AJ, Feng Z, Agostini M, Zolla L, Agrò AF, et al. p63 regulates glutaminase 2 expression. Cell Cycle 2013; 12(9):1395-405; PMID:23574722; http://dx.doi.org/10.4161/cc.24478
- Luh LM, Kehrloesser S, Deutsch GB, Gebel J, Coutandin D, Schäfer B, Agostini M, Melino G, Dötsch V. Analysis of the oligomeric state and transactivation potential of TAp73α. Cell Death Differ 2013; 20(8):1008-16; PMID:23538419; http://dx.doi.org/10.1038/cdd.2013.23
- Wu G, Nomoto S, Hoque MO, Dracheva T, Osada M, Lee CC, Dong SM, Guo Z, Benoit N, Cohen Y, et al. DeltaNp63alpha and TAp63alpha regulate transcription of genes with distinct biological functions in cancer and development. Cancer Res 2003; 63(10):2351-7; PMID:12750249
- Wu G, Osada M, Guo Z, Fomenkov A, Begum S, Zhao M, Upadhyay S, Xing M, Wu F, Moon C, et al. DeltaNp63alpha up-regulates the Hsp70 gene in human cancer. Cancer Res 2005; 65(3):758-66; PMID:15705872
- Huang Y, Jeong JS, Okamura J, Sook-Kim M, Zhu H, Guerrero-Preston R, Ratovitski EA. Global tumor protein p53/p63 interactome: making a case for cisplatin chemoresistance. Cell Cycle 2012; 11(12):2367-79; PMID:22672905; http://dx.doi.org/10.4161/cc.20863
- Huang Y, Guerrero-Preston R, Ratovitski EA. Phospho-ΔNp63α-dependent regulation of autophagic signaling through transcription and micro-RNA modulation. Cell Cycle 2012; 11(6):1247-59; PMID:22356768; http://dx.doi.org/10.4161/cc.11.6.19670
- Huang Y, Kesselman D, Kizub D, Guerrero-Preston R, Ratovitski EA. Phospho-ΔNp63α/microRNA feedback regulation in squamous carcinoma cells upon cisplatin exposure. Cell Cycle 2013; 12(4):684-97; PMID:23343772; http://dx.doi.org/10.4161/cc.23598
- Viticchiè G, Lena AM, Latina A, Formosa A, Gregersen LH, Lund AH, Bernardini S, Mauriello A, Miano R, Spagnoli LG, Knight RA, Candi E, Melino G. MiR-203 controls proliferation, migration and invasive potential of prostate cancer cell lines. Cell Cycle 2011 Apr 1; 10(7):1121-31; http://dx.doi.org/10.4161/cc.10.7.15180
- Celardo I, Antonov A, Amelio I, Annicchiarico-Petruzzelli M, Melino G. p63 transcriptionally regulates the expression of matrix metallopeptidase 13. Oncotarget 2014; 5(5):1279-89; PMID:24658133
- Celardo I, Grespi F, Antonov A, Bernassola F, Garabadgiu AV, Melino G, Amelio I. Caspase-1 is a novel target of p63 in tumor suppression. Cell Death Dis 2013; 4:e645; http://dx.doi.org/10.1038/cddis.2013.175
- Marcel V, Petit I, Murray-Zmijewski F, Goullet de Rugy T, Fernandes K, Meuray V, Diot A, Lane DP, Aberdam D, Bourdon JC. Diverse p63 and p73 isoforms regulate Δ133p53 expression through modulation of the internal TP53 promoter activity. Cell Death Differ 2012; 19(5):816-26; PMID:22075982; http://dx.doi.org/10.1038/cdd.2011.152
- He Z, Liu H, Agostini M, Yousefi S, Perren A, Tschan MP, Mak TW, Melino G, Simon HU. p73 regulates autophagy and hepatocellular lipid metabolism through a transcriptional activation of the ATG5 gene. Cell Death Differ 2013; 20(10):1415-24; PMID:23912709; http://dx.doi.org/10.1038/cdd.2013.104
- Amelio I, Grespi F, Annicchiarico-Petruzzelli M, Melino G. p63 the guardian of human reproduction. Cell Cycle 2012; 11(24):4545-51; PMID:23165243; ttp://dx.doi.org/10.4161/cc.22819
- Hutt K, Kerr JB, Scott CL, Findlay JK, Strasser A. How to best preserve oocytes in female cancer patients exposed to DNA damage inducing therapeutics. Cell Death Differ 2013; 20(8):967-8; PMID:23832146; http://dx.doi.org/10.1038/cdd.2013.54
- Mattiske S, Ho K, Noll JE, Neilsen PM, Callen DF, Suetani RJ. TAp63 regulates oncogenic miR-155 to mediate migration and tumour growth. Oncotarget 2013; 4(11):1894-903; PMID:24177167
- Shekhar MP, Kato I, Nangia-Makker P, Tait L. Comedo-DCIS is a precursor lesion for basal-like breast carcinoma: identification of a novel p63/Her2/neu expressing subgroup. Oncotarget 2013; 4(2):231-41; PMID:23548208
- Ianakiev P, Kilpatrick MW, Toudjarska I, Basel D, Beighton P, Tsipouras P. Split-hand/split-foot malformation is caused by mutations in the p63 gene on 3q27. Am J Hum Genet 2000; 67(1):59-66; PMID:10839977; http://dx.doi.org/10.1086/302972
- Huang YP, Wu G, Guo Z, Osada M, Fomenkov T, Park HL, Trink B, Sidransky D, Fomenkov A, Ratovitski EA. Altered sumoylation of p63alpha contributes to the split-hand/foot malformation phenotype. Cell Cycle 2004; 3(12):1587-96; PMID:15539951; http://dx.doi.org/10.4161/cc.3.12.1290
- Kantaputra PN, Hamada T, Kumchai T, McGrath JA. Heterozygous mutation in the SAM domain of p63 underlies Rapp-Hodgkin ectodermal dysplasia. J Dent Res 2003; 82(6):433-7; PMID:12766194; http://dx.doi.org/10.1177/154405910308200606
- Levine AJ, Tomasini R, McKeon FD, Mak TW, Melino G. The p53 family: guardians of maternal reproduction. Nat Rev Mol Cell Biol 2011; 12(4):259-65; http://dx.doi.org/10.1038/nrm3086
- Billon N, Terrinoni A, Jolicoeur C, McCarthy A, Richardson WD, Melino G, Raff M. Roles for p53 and p73 during oligodendrocyte development. Development 2004; 131(6):1211-20; PMID:14960496; http://dx.doi.org/10.1242/dev.01035
- Gonzalez-Cano L, Hillje AL, Fuertes-Alvarez S, Marques MM, Blanch A, Ian RW, Irwin MS, Schwamborn JC, Marín MC. Regulatory feedback loop between TP73 and TRIM32. Cell Death Dis 2013; 4:e704; http://dx.doi.org/10.1038/cddis.2013.224
- Masse I, Barbollat-Boutrand L, Molina M, Berthier-Vergnes O, Joly-Tonetti N, Martin MT, Caron de Fromentel C, Kanitakis J, Lamartine J. Functional interplay between p63 and p53 controls RUNX1 function in the transition from proliferation to differentiation in human keratinocytes. Cell Death Dis 2012; 3:e318; http://dx.doi.org/10.1038/cddis.2012.62
- Tomasini R, Mak TW, Melino G. The impact of p53 and p73 on aneuploidy and cancer. Trends Cell Biol 2008; 18(5):244-52; PMID:18406616; http://dx.doi.org/10.1016/j.tcb.2008.03.003
- Burnley P, Rahman M, Wang H, Zhang Z, Sun X, Zhuge Q, Su DM. Role of the p63-FoxN1 regulatory axis in thymic epithelial cell homeostasis during aging. Cell Death Dis 2013; 4:e932; http://dx.doi.org/10.1038/cddis.2013.460
- Celli J, Duijf P, Hamel BC, Bamshad M, Kramer B, Smits AP, Newbury-Ecob R, Hennekam RC, Van Buggenhout G, van Haeringen A, et al. Heterozygous germline mutations in the p53 homolog p63 are the cause of EEC syndrome. Cell 1999; 99(2):143-53; PMID:10535733; http://dx.doi.org/10.1016/S0092-8674(00)81646-3
- Bertola DR, Kim CA, Albano LM, Scheffer H, Meijer R, van Bokhoven H. Molecular evidence that AEC syndrome and Rapp-Hodgkin syndrome are variable expression of a single genetic disorder. Clin Genet 2004; 66(1):79-80; PMID:15200513; http://dx.doi.org/10.1111/j.0009-9163.2004.00278.x
- Bougeard G, Hadj-Rabia S, Faivre L, Sarafan-Vasseur N, Frébourg T. The Rapp-Hodgkin syndrome results from mutations of the TP63 gene. Eur J Hum Genet 2003; 11(9):700-4; PMID:12939657; http://dx.doi.org/10.1038/sj.ejhg.5201004
- McGrath JA, Duijf PH, Doetsch V, Irvine AD, de Waal R, Vanmolkot KR, Wessagowit V, Kelly A, Atherton DJ, Griffiths WA, et al. Hay-Wells syndrome is caused by heterozygous missense mutations in the SAM domain of p63. Hum Mol Genet 2001; 10(3):221-9; PMID:11159940; http://dx.doi.org/10.1093/hmg/10.3.221
- Prontera P, Escande F, Cocchi G, Donti E, Martini A, Sensi A. An intermediate phenotype between Hay-Wells and Rapp-Hodgkin syndromes in a patient with a novel P63 mutation: confirmation of a variable phenotypic spectrum with a common aetiology. Genet Couns 2008; 19(4):397-402; PMID:19239083
- van Bokhoven H, Hamel BC, Bamshad M, Sangiorgi E, Gurrieri F, Duijf PH, Vanmolkot KR, van Beusekom E, van Beersum SE, Celli J, et al. p63 Gene mutations in eec syndrome, limb-mammary syndrome, and isolated split hand-split foot malformation suggest a genotype-phenotype correlation. Am J Hum Genet 2001; 69(3):481-92; PMID:11462173; http://dx.doi.org/10.1086/323123
- van Bokhoven H, McKeon F. Mutations in the p53 homolog p63: allele-specific developmental syndromes in humans. Trends Mol Med 2002; 8(3):133-9; PMID:11879774; http://dx.doi.org/10.1016/S1471-4914(01)02260-2
- Crum CP, McKeon FD. p63 in epithelial survival, germ cell surveillance, and neoplasia. Annu Rev Pathol 2010; 5:349-71; PMID:20078223; http://dx.doi.org/10.1146/annurev-pathol-121808-102117
- Chong PA, Lin H, Wrana JL, Forman-Kay JD. An expanded WW domain recognition motif revealed by the interaction between Smad7 and the E3 ubiquitin ligase Smurf2. J Biol Chem 2006; 281(25):17069-75; PMID:16641086; http://dx.doi.org/10.1074/jbc.M601493200
- Ilsley JL, Sudol M, Winder SJ. The WW domain: linking cell signalling to the membrane cytoskeleton. Cell Signal 2002; 14(3):183-9; PMID:11812645; http://dx.doi.org/10.1016/S0898-6568(01)00236-4
- Ingham RJ, Colwill K, Howard C, Dettwiler S, Lim CS, Yu J, Hersi K, Raaijmakers J, Gish G, Mbamalu G, et al. WW domains provide a platform for the assembly of multiprotein networks. Mol Cell Biol 2005; 25(16):7092-106; PMID:16055720; http://dx.doi.org/10.1128/MCB.25.16.7092-7106.2005
- Lim SK, Orhant-Prioux M, Toy W, Tan KY, Lim YP. Tyrosine phosphorylation of transcriptional coactivator WW-domain binding protein 2 regulates estrogen receptor α function in breast cancer via the Wnt pathway. FASEB J 2011; 25(9):3004-18; PMID:21642474; http://dx.doi.org/10.1096/fj.10-169136
- Chong PA, Lin H, Wrana JL, Forman-Kay JD. Coupling of tandem Smad ubiquitination regulatory factor (Smurf) WW domains modulates target specificity. Proc Natl Acad Sci USA 2010; 107(43):18404-9; http://dx.doi.org/10.1073/pnas.1003023107
- Chan SW, Lim CJ, Huang C, Chong YF, Gunaratne HJ, Hogue KA, Blackstock WP, Harvey KF, Hong W. WW domain-mediated interaction with Wbp2 is important for the oncogenic property of TAZ. Oncogene 2011; 30(5):600-10; PMID:20972459; http://dx.doi.org/10.1038/onc.2010.438
- Macias MJ, Hyvönen M, Baraldi E, Schultz J, Sudol M, Saraste M, Oschkinat H. Structure of the WW domain of a kinase-associated protein complexed with a proline-rich peptide. Nature 1996; 382(6592):646-9; PMID:8757138; http://dx.doi.org/10.1038/382646a0
- Macias MJ, Wiesner S, Sudol M. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett 2002; 513(1):30-7; PMID:11911877; http://dx.doi.org/10.1016/S0014-5793(01)03290-2
- Karplus PA. Hydrophobicity regained. Protein Sci 1997; 6(6):1302-7; PMID:9194190; http://dx.doi.org/10.1002/pro.5560060618
- Sudol M, Hunter T. NeW wrinkles for an old domain. Cell 2000; 103(7):1001-4; PMID:11163176; http://dx.doi.org/10.1016/S0092-8674(00)00203-8
- Oberst A, Malatesta M, Aqeilan RI, Rossi M, Salomoni P, Murillas R, Sharma P, Kuehn MR, Oren M, Croce CM, Bernassola F, et al. The Nedd4-binding partner 1 (N4BP1) protein is an inhibitor of the E3 ligase Itch. Proc Natl Acad Sci USA 2007; 104(27):11280-5; http://dx.doi.org/10.1073/pnas.0701773104
- Bakkers J, Camacho-Carvajal M, Nowak M, Kramer C, Danger B, Hammerschmidt M. Destabilization of DeltaNp63alpha by Nedd4-mediated ubiquitination and Ubc9-mediated sumoylation, and its implications on dorsoventral patterning of the zebrafish embryo. Cell Cycle 2005; 4(6):790-800; PMID:15908775; http://dx.doi.org/10.4161/cc.4.6.1694
- Nicole Tsang YH, Wu XW, Lim JS, Wee Ong C, Salto-Tellez M, Ito K, Ito Y, Chen LF. Prolyl isomerase Pin1 downregulates tumor suppressor RUNX3 in breast cancer. Oncogene 2013; 32(12):1488-96; PMID:22580604; http://dx.doi.org/10.1038/onc.2012.178
- Shi Z, Woody RW, Kallenbach NR. Is polyproline II a major backbone conformation in unfolded proteins? Adv Protein Chem 2002; 62:163-240; PMID:12418104; http://dx.doi.org/10.1016/S0065-3233(02)62008-X
- Blanch EW, Morozova-Roche LA, Cochran DA, Doig AJ, Hecht L, Barron LD. Is polyproline II helix the killer conformation? A Raman optical activity study of the amyloidogenic prefibrillar intermediate of human lysozyme. J Mol Biol 2000; 301(2):553-63; PMID:10926527; http://dx.doi.org/10.1006/jmbi.2000.3981
- Creamer TP, Campbell MN. Determinants of the polyproline II helix from modeling studies. Adv Protein Chem 2002; 62:263-82; PMID:12418106; http://dx.doi.org/10.1016/S0065-3233(02)62010-8
- Girardini JE, Napoli M, Piazza S, Rustighi A, Marotta C, Radaelli E, Capaci V, Jordan L, Quinlan P, Thompson A, et al. A Pin1/mutant p53 axis promotes aggressiveness in breast cancer. Cancer Cell 2011; 20(1):79-91; PMID:21741598; http://dx.doi.org/10.1016/j.ccr.2011.06.004
- Rossi M, De Laurenzi V, Munarriz E, Green DR, Liu YC, Vousden KH, Cesareni G, Melino G. The ubiquitin-protein ligase Itch regulates p73 stability. EMBO J 2005; 24(4):836-48; PMID:15678106; http://dx.doi.org/10.1038/sj.emboj.7600444
- Cicero DO, Falconi M, Candi E, Mele S, Cadot B, Di Venere A, Rufini S, Melino G, Desideri A. NMR structure of the p63 SAM domain and dynamical properties of G534V and T537P pathological mutants, identified in the AEC syndrome. Cell Biochem Biophys 2006; 44(3):475-89; PMID:16679535; http://dx.doi.org/10.1385/CBB:44:3:475
- Bellomaria A, Barbato G, Melino G, Paci M, Melino S. Recognition of p63 by the E3 ligase ITCH: Effect of an ectodermal dysplasia mutant. Cell Cycle 2010; 9(18):3730-9; PMID:20855944; http://dx.doi.org/10.4161/cc.9.18.12933
- Bellomaria A, Barbato G, Melino G, Paci M, Melino S. Recognition mechanism of p63 by the E3 ligase Itch: novel strategy in the study and inhibition of this interaction. Cell Cycle 2012; 11(19):3638-48; PMID:22935697; http://dx.doi.org/10.4161/cc.21918
- Li Y, Zhou Z, Chen C. WW domain-containing E3 ubiquitin protein ligase 1 targets p63 transcription factor for ubiquitin-mediated proteasomal degradation and regulates apoptosis. Cell Death Differ 2008; 15(12):1941-51; PMID:18806757; http://dx.doi.org/10.1038/cdd.2008.134
- Li C, Chang DL, Yang Z, Qi J, Liu R, He H, Li D, Xiao ZX. Pin1 modulates p63α protein stability in regulation of cell survival, proliferation and tumor formation. Cell Death Dis 2013; 4:e943; http://dx.doi.org/10.1038/cddis.2013.468
- Rossi M, Aqeilan RI, Neale M, Candi E, Salomoni P, Knight RA, Croce CM, Melino G. The E3 ubiquitin ligase Itch controls the protein stability of p63. Proc Natl Acad Sci USA 2006; 103(34):12753-8; http://dx.doi.org/10.1073/pnas.0603449103
- Rabanal F, Ludevid MD, Pons M, Giralt E. CD of proline-rich polypeptides: application to the study of the repetitive domain of maize glutelin-2. Biopolymers 1993; 33(7):1019-28; PMID:8343583; http://dx.doi.org/10.1002/bip.360330704
- Zarrinpar A, Lim WA. Converging on proline: the mechanism of WW domain peptide recognition. Nat Struct Biol 2000; 7(8):611-3; http://dx.doi.org/10.1038/77891
- Chen HI, Einbond A, Kwak SJ, Linn H, Koepf E, Peterson S, Kelly JW, Sudol M. Characterization of the WW domain of human yes-associated protein and its polyproline-containing ligands. J Biol Chem 1997; 272(27):17070-7; PMID:9202023; http://dx.doi.org/10.1074/jbc.272.27.17070
- Koepf EK, Petrassi HM, Ratnaswamy G, Huff ME, Sudol M, Kelly JW. Characterization of the structure and function of W –>F WW domain variants: identification of a natively unfolded protein that folds upon ligand binding. Biochemistry 1999; 38(43):14338-51; PMID:10572009; http://dx.doi.org/10.1021/bi991105l
- Koepf EK, Petrassi HM, Sudol M, Kelly JW. WW: an isolated three-stranded antiparallel beta-sheet domain that unfolds and refolds reversibly; evidence for a structured hydrophobic cluster in urea and GdnHCl and a disordered thermal unfolded state. Protein Sci 1999; 8(4):841-53; PMID:10211830; http://dx.doi.org/10.1110/ps.8.4.841
- Qin H, Pu HX, Li M, Ahmed S, Song J. Identification and structural mechanism for a novel interaction between a ubiquitin ligase WWP1 and Nogo-A, a key inhibitor for central nervous system regeneration. Biochemistry 2008; 47(51):13647-58; PMID:19035836; http://dx.doi.org/10.1021/bi8017976
- Lu KP, Finn G, Lee TH, Nicholson LK. Prolyl cis –trans isomerization as a molecular timer. Nat Chem Biol 2007; 3:619-629; PMID:17876319; http://dx.doi.org/10.1038/nchembio.2007.35
- Schutkowski M, Bernhardt A, Zhou XZ, Shen M, Reimer U, Rahfeld JU, Lu KP, Fischer G. Role of phosphorylation in determining the backbone dynamics of the serine/threonine-proline motif and Pin1 substrate recognition. Biochemistry 1998; 37:5566-75; PMID:9548941; http://dx.doi.org/10.1021/bi973060z
- Werner-Allen JW, Lee CJ, Liu P, Nicely NI, Wang S, Greenleaf AL, et al. cis-Proline-mediated Ser(P)5 dephosphorylation by the RNA polymerase II C-terminal domain phosphatase Ssu72. J Biol Chem 2011; 286:5717-26; PMID:21159777; http://www.jbc.org/content/286/7/5717
- Peschiaroli A, Scialpi F, Bernassola F, El Sherbini el S, Melino G. The E3 ubiquitin ligase WWP1 regulates ΔNp63-dependent transcription through Lys63 linkages. Biochem Biophys Res Commun 2010; 402(2):425-30; PMID:20951678; http://dx.doi.org/10.1016/j.bbrc.2010.10.050
- Bhandari D, Robia SL, Marchese A. The E3 ubiquitin ligase atrophin interacting protein 4 binds directly to the chemokine receptor CXCR4 via a novel WW domain-mediated interaction. Mol Biol Cell 2009; 20:1324-39; PMID:19116316; http://dx.doi.org/10.1091/mbc.E08-03-0308
- Suryaraja R, Anitha M, Anbarasu K, Kumari G, Mahalingam S. The E3 ubiquitin ligase Itch regulates tumor suppressor protein RASSF5/NORE1 stability in an acetylation-dependent manner. Cell Death Dis 2013; 4:e565; http://dx.doi.org/10.1038/cddis.2013.91
- Uversky VN. Natively unfolded proteins: a point where biology waits for physics. Protein Sci 2002; 11(4):739-56; PMID:11910019; http://dx.doi.org/10.1110/ps.4210102
- Woody RW, Sugeta H, Kodama TS. [Circular dichroism of proteins: recent developments in analysis and prediction]. Tanpakushitsu Kakusan Koso 1996; 41(1):56-69; PMID:8584742
- Buck M. Trifluoroethanol and colleagues: cosolvents come of age. Recent studies with peptides and proteins. Q Rev Biophys 1998; 31(3):297-355; PMID:10384688; http://dx.doi.org/10.1017/S003358359800345X
- Morris GM., Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. Autodock4 and AutoDockTools4: automated docking with selective receptor flexiblity. J Comput Chem 2009; 16: 2785-91; http://dx.doi.org/10.1002/jcc.21256
- Trott O, Olson AJ. AutoDockVina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading, J Comput Chem 2010; 31: 455-461
- Lu H, Li H, BanuBte SR, Shamima, Leow W, Liou Y-C. Knowledge-Guided Docking of WW Domain Proteins and Flexible Ligands. Pattern Recognition in Bioinformatics. Comput Sci 2009; 5780: 175-186
