Abstract
Selective ubiquitin-dependent autophagy mediates the disposal of superfluous cellular structures and clears cells of protein aggregates such as polyQ proteins linked to Huntington disease. Crucial selectivity factors of this pathway are ubiquitin-Atg8 receptors such as human SQSTM1/p62, which recognize ubiquitinated cargoes and deliver them to phagophores for degradation. Contrasting previous beliefs, we recently showed that ubiquitin-dependent autophagy is not restricted to higher eukaryotes but also exists in yeast with Cue5, harboring a ubiquitin-binding CUE domain, being a ubiquitin-Atg8 receptor. Notably, the human CUE domain protein TOLLIP is functionally similar to yeast Cue5, indicating that Cue5/TOLLIP (CUET) proteins represent a new and conserved class of autophagy receptors. Remarkably, both Cue5 in yeast and TOLLIP in human cells mediate efficient clearance of aggregation-prone polyQ proteins derived from human HTT/huntingtin. Indeed, TOLLIP is potentially more potent in polyQ clearance than SQSTM1/p62 in HeLa cells, suggesting that TOLLIP, also implicated in innate immunity, may be significant for human health and disease.
Protein misfolding and aggregation are potentially toxic for cells, but organisms counteract via various protein quality control pathways. Whereas molecular chaperones prevent protein misfolding or aggregation or even mediate protein repair, proteolysis is used if misfolded or aggregated proteins persist. While soluble proteins are largely degraded by the proteasome, protein assemblies and aggregates are typically eliminated by autophagy. Remarkably, both proteolysis pathways use ubiquitin conjugation (ubiquitination) to earmark proteins (or aggregates) for degradation, and each pathway utilizes dedicated receptors that recognize these ubiquitin marks. For example, Rad23 binds soluble ubiquitinated (typically modified by a polyubiquitin chain) proteins via a ubiquitin-binding UBA domain, whereas another domain of the Rad23 protein interacts with the proteasome. By contrast, the human autophagy receptor SQSTM1/p62, although it recognizes ubiquitinated cargo via a UBA domain as well, promotes autophagy instead, because it binds Atg8/LC3, a ubiquitin-related protein conjugated to lipids of phagophore and autophagosomal membranes. How the alternative receptors discriminate between ubiquitinated proteins destined for different degradation pathways is currently not known, but the type and degree of ubiquitination may be a crucial factor.
Studies using budding yeast Saccharomyces cerevisiae as a model organism were elementary for our current knowledge of the mechanisms of the ubiquitin proteasome system (UPS) and of autophagy. Indeed, pioneering biochemical and genetic screens conducted in yeast identified the core components of the ubiquitination and autophagy machinery and of the autophagy-related cytoplasm-to-vacuole targeting pathway that targets specific enzymes to the vacuole, the yeast equivalent of the lysosome. Surprisingly, however, so far no evidence for ubiquitin-dependent selective autophagy and ubiquitin-Atg8 receptors was found in lower eukaryotes such as yeast, leading to the common concept that this pathway is restricted to higher eukaryotes.
However, revisiting this issue by using a combination of biochemistry and genetics, we recently discovered that ubiquitin-dependent selective autophagy exists in S. cerevisiae as well. A critical paradigm-shifting result was the finding that starvation-induced decay of cellular ubiquitin-protein conjugates is largely blocked when autophagy is impaired genetically (e.g., by using mutants lacking Atg8). Importantly, by searching in an Atg8-pulldown fraction for proteins that possess annotated ubiquitin-binding domains, we also unearthed the previously uncharacterized protein Cue5 as a yeast ubiquitin-Atg8 receptor. Interestingly, different from the human receptors SQSTM1/p62 and NBR1, ubiquitin recognition by Cue5 is mediated through a CUE domain (rather than a UBA domain), which is also present in the ubiquitin-binding proteins Vps9 and Cue1 involved in endocytosis and ER-associated degradation/ERAD, respectively.
By isolating and quantitatively comparing (using a SILAC-based mass spectrometry protocol) ubiquitin-protein conjugates from wild-type cells and mutants lacking Cue5 (cue5Δ) we also identified several Cue5-dependent autophagy substrates. Among them are aggregation-prone proteins but also enzymes (Ubc4, Ubc5) involved in ubiquitination of abnormal proteins. Indeed, a significant fraction of the identified Cue5-dependent autophagy substrates receive their ubiquitin mark by these enzymes in collaboration with the ubiquitin ligase Rsp5. This finding is significant as precisely these yeast enzymes also mediate ubiquitination of abnormal proteins destined for proteasomal degradation. Hence, this observation suggests that the choice of which pathway is used—the UPS or ubiquitin-dependent autophagy—may happen after substrate ubiquitination.
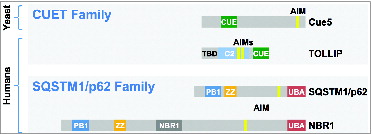
Given that yeast Cue5 uses a CUE rather than a UBA domain for ubiquitin binding, we argued that humans might have CUE domain autophagy receptors as well, in addition to the well-studied UBA domain autophagy receptors SQSTM1/p62 and NBR1. Indeed, we found that the human CUE domain protein TOLLIP (toll interacting protein), implicated previously in ubiquitin binding, endocytosis, and innate immunity, also binds Atg8/LC3 and functions as a ubiquitin-Atg8 receptor. Cue5 and TOLLIP are in fact distantly related (termed CUET proteins), but their domain organizations are different : the most notable difference being the additional presence in TOLLIP of a phospholipid-binding C2 domain and a Tom1-binding/TBD domain, which may, however, be linked to the other roles TOLLIP seems to play.
Figure 1. Scheme of ubiquitin-Atg8 receptors. S. cerevisiae Cue5 and human TOLLIP (CUET family) proteins possess CUE domains for ubiquitin recognition, whereas human SQSTM1/p62 and NBR1 (SQSTM1/p62 family) recognize ubiquitin via UBA domains. The relative positions of Atg8 (LC3) binding sites (yellow) and additional domains are shown (see text for TOLLIP).
Selective autophagy in humans is suggested to clear cells of protein aggregates such as polyQ proteins linked to Huntington disease. Notably, when we expressed in yeast aggregation-prone versions of a polyQ model protein derived from exon 1 of the human protein HTT, we observe enhanced cytotoxicity if Cue5 is absent (cue5Δ). Similar to other Cue5-dependent autophagy substrates the polyQ protein is ubiquitinated via the yeast ubiquitin ligase Rsp5 and accumulates in aggregates when Cue5 is absent. Remarkably, overexpression of human TOLLIP in HeLa cells very efficiently removes co-expressed aggregation-prone polyQ from cells, and is seemingly more efficient than overexpressed SQSTM1/p62. Correspondingly, cytotoxicity in HeLa cells caused by overexpression of polyQ proteins is more strongly enhanced upon TOLLIP depletion (by RNAi) than upon SQSTM1/p62 depletion. Double depletion of TOLLIP and SQSTM1/p62 enhances polyQ cytotoxicity even further, suggesting that the different autophagy receptors might act cooperatively. Indeed, immunoprecipitation of TOLLIP co-isolates both SQSTM1/p62 and NBR1, indicating that ubiquitinated aggregates may attract different ubiquitin-Atg8 autophagy receptors concurrently.
We think that the identification of a new class of ubiquitin-Atg8 receptors (CUET proteins) opens new avenues for studies of autophagy, protein aggregation, and potentially human neurodegenerative diseases caused by protein aggregation. Remarkably, yeast Cue5 can partially substitute for TOLLIP in human cells and human TOLLIP can reduce polyQ cytotoxicity of yeast cells lacking Cue5. This finding gives strong support to the concept that the genetically tractable S. cerevisiae is highly suitable for basic studies related to polyQ diseases. Furthermore, the observation that abnormal proteins eliminated by the UPS and by ubiquitin-dependent selective autophagy frequently receive their destruction marks by the same ubiquitination machinery also supports the concept of hierarchically organized protein quality control pathways. Similarly, the finding that TOLLIP, in addition to its suggested roles in innate immunity and ubiquitin-dependent endocytosis, additionally mediates autophagy through Atg8/LC3 binding, exemplifies the complexity of cellular networks. Thus, important future directions are to find out how pathway decisions are made, and judging from previous studies and our work, we propose that critical choices are made at the level of ubiquitin-binding receptors.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
Work in Jentsch laboratory is supported by Max Planck Society, Deutsche Forschungsgemeinschaft, Center for Integrated Protein Science Munich, ERC Advanced Grant, and the Louis-Jeantet Foundation.
