Abstract
Supplementation of fish oil rich in omega-3 polyunsaturated fatty acids (n-3 PUFA) during pregnancy has been shown to confer favorable health outcomes in the offspring. In a randomized controlled trial, we have previously shown that n-3 PUFA supplementation in pregnancy was associated with modified immune responses and some markers of immune maturation. However, the molecular mechanisms underlying these heritable effects are unclear. To determine whether the biological effects of maternal n-3 PUFA supplementation are mediated through DNA methylation, we analyzed CD4+ T-cells purified from cryo-banked cord blood samples from a previously conducted clinical trial. Of the 80 mother-infant pairs that completed the initial trial, cord blood samples of 70 neonates were available for genome-wide DNA methylation profiling. Comparison of purified total CD4+ T-cell DNA methylation profiles between the supplement and control groups did not reveal any statistically significant differences in CpG methylation, at the single-CpG or regional level. Effect sizes among top-ranked probes were lower than 5% and did not warrant further validation. Tests for association between methylation levels and key n-3 PUFA parameters, docosahexaenoic acid (DHA), eicosapentaenoic acid (EPA), or total n-3 PUFAs were suggestive of dose-dependent effects, but these did not reach genome-wide significance. Our analysis of the microarray data did not suggest strong modifying effects of in utero n-3 PUFA exposure on CD4+ T-cell methylation profiles, and no probes on the array met our criteria for further validation. Other epigenetic mechanisms may be more relevant mediators of functional effects induced by n-3 PUFA in early life.
Abbreviations
| PUFA | = | polyunsaturated fatty acids |
| n-3 | = | omega 3 |
| DHA | = | docosahexaenoic acid |
| EPA | = | eicosapentaenoic acid |
| CBMC | = | cord blood mononuclear cells |
Introduction
Supplementation of polyunsaturated fatty acids (PUFAs) during pregnancy and during early development has been shown to be potentially beneficial in preventing allergic,Citation1-3 cardiovascular,Citation4,5 and metabolic disordersCitation6 in the offspring. We have previously shown that supplementation with n-3 PUFA DHA (22:6 n-3) and EPA (20:5 n-3) from 20 weeks of gestation until delivery modified inflammatory immune responses in neonatal immune cells.Citation1,Citation7–9 While this dietary intervention affects many biochemical and physiological properties of cells and organs, the exact molecular mechanism by which n-3 PUFAs exert their multi-system benefits is yet to be fully elucidated. Specifically, in the context of heritable effects of maternal exposure, the proposed pathways do not provide a clear molecular link.
There is mounting evidence that dietary factors can change cellular epigenetic marks in association with more favorable clinical outcomes.Citation10 Dietary PUFAs have also been shown to regulate gene expression through epigenetic mechanisms.Citation11–13 Treatment of U937 leukemic macrophage cells with EPA has been shown to increase mRNA expression of CCAAT/enhancer-binding protein delta with concomitant demethylation of specific CpG loci in the gene promoter region,Citation11 whereas treatment of M17 neuroblastoma cells with DHA tended to induce histone modifications that are consistent with active transcription.Citation12 Extending these findings to human cohorts, a recent study reported 27 differentially methylated CpG sites at biologically relevant regions that were associated with n-3 PUFA intake using an “extreme phenotypes” model in a cohort of 185 adults.Citation14 Another study that investigated the association of n-6 PUFA intake and central obesity in 40 young women found that the level of TNF promoter methylation of peripheral white cells was associated with n-6 PUFA intake.Citation15 Of greatest relevance here, emerging data from animal studies support a role for maternal intake of PUFA in pregnancy in the modulation of offspring epigenetic profile.Citation16,17 In a recent study involving pregnant women who had been supplemented with DHA or placebo from the second trimester until delivery, Lee et al. measured the DNA methylation changes in genes associated with Th1, Th2, Th17, and regulatory T-cell development, as well as LINE1 repetitive elements of cord blood mononuclear cells.Citation18 Although there were no significant differences in DNA methylation pattern in genes when comparing the treatment and the placebo groups, overall n-3 supplementation modified effects on methylation in the subgroup of neonates whose mothers smoked during pregnancy. Specifically in this risk group, n-3 supplementation was associated with differences in methylation levels of LINE-1 repetitive element. Collectively, these data suggests maternal n-3 PUFA intake during pregnancy may modify the fetal epigenome, and that this may depend on other environmental exposures.
To determine whether exposure to n-3 PUFA intake during pregnancy alters the fetal epigenome, we compared whole genome DNA methylation profiles from neonatal CD4+ T-cells derived from previously carried out randomized controlled trial of maternal supplementation of fish oil during pregnancy. CD4+ T-cells were chosen as they play a key role in immune responses and have been shown to be susceptible to in utero perturbation from environmental exposures.Citation19,20 We hypothesized that maternal n-3 PUFA supplementation may modulate epigenetic programming of CD4+ T-cells, and that this may modulate later health outcomes.
Results
Characteristics of the cohort
Of the total 83 mother-infant pairs who completed the initial randomized controlled trial, 70 had cord blood available for this epigenome-wide DNA methylation analysis. shows the characteristics of the 70 mother-infant pairs; 36 in the fish oil group and 34 in the control group. There were no significant differences in the key maternal and neonatal parameters between treatment and control groups.
Table 1. Population characteristics of the mothers and infants in the cohort (n = 70)
Maternal supplementation with n-3 PUFA modifies fetal red cell parameters
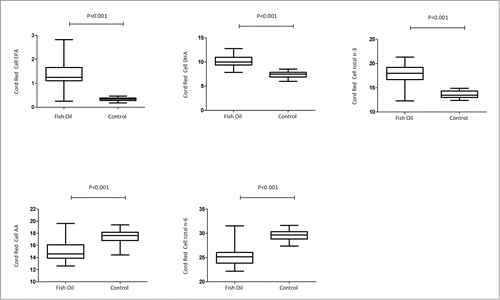
To determine effectiveness of the maternal intervention, fatty acid parameters in cord red blood cells were compared between treatment and intervention group. There was clear evidence of significantly higher total n-3 (fish oil = 17.8; control 13.6; P < 0.001) and lower total n-6 (fish oil = 25.2; control 29.6; P < 0.001) fatty acid levels in treatment group. Key n-3 PUFAs EPA and DHA were also higher in the treatment group, while arachidonic acid, a key n-6 PUFA, was lower, than in the control group (). These readouts provided robust evidence that the maternal supplementation modified fetal lipid profiles.
Figure 1. Cord red blood cell fatty acid measurements in fish oil and control groups. Boxplots represent median and range. Statistical analysis by Man-Whitney U-test. EPA: eicosapentaenoic acid; DHA: docosahexaenoic acid; AA: arachidonic acid; total n-3 PUFAs (Sum 20:5, 22:5, 22:3); total n-6 PUFAs (Sum 18:2, 20:3, 20:4, 22:3, 22:4).
Identification of CpG sites that co-associate with n-3 red cell fatty acid levels
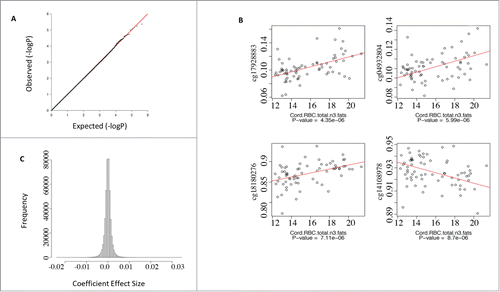
To identify CpG sites associated with n-3 supplementation in the neonatal CD4+ T-cell epigenome, we regressed all probes on neonatal total n-3 red cell fatty acid levels as a continuous variable in the presence of latent covariates (See Materials and Methods). We did not identify any CpG sites that were significantly associated with total n-3 levels that survived multiple testing (). We therefore ranked all CpG sites on the array by unadjusted P-values and examined the top-ranked CpG sites for evidence of an effect using scatterplots. The top 10 ranked CpG were examined by plotting methylation versus total n-3 fatty acid level. A dose-response relationship was apparent among the CpG sites ranked at the top of the list and these relationships were bi-directional at the individual CpG level, with some CpG positively associated with n-3 fatty acid levels and others negatively associated with increasing n-3 fatty acid levels (, 4 CpG shown for convenience). Individually, these CpG sites exhibited a significant association with total n-3 fatty acid levels although the effect sizes were small (less than 5% change in methylation) () and not significant at the genome scale, and, therefore, of unknown biological relevance. We found this to be the case, regardless of whether the data was modeled on cord red cell DHA levels or EPA levels, or whether covariates were included in the model or not. We filtered the data set to include only the top 25% most variable probes on the array and repeated the analysis with the same result (data not shown).
Figure 2. Regression analysis of CpG sites that co-associate with cord red cell total n-3 fatty acid levels. (A) qq plot of P-values from the association test between CpGs and total n-3 fatty acids did not deviate from the null hypothesis. (B) Scatterplots of the top 4 CpGs ranked by P-value for the association with total n-3 fatty acids suggested a dose-response relationship. (C) Histogram of the coefficients from the regression model indicated a small effect size.
Comparison of methylation profiles between treatment and control groups
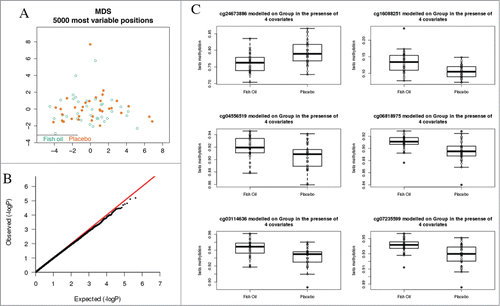
We next compared genome-wide methylation profiles between treatment and control groups using multidimensional scaling analysis. Neither genome-wide methylation profiles, nor the top 5000 most variable probes discriminated the 2 groups (). We compared treatment and control groups for differential methylation in a case control comparison by regressing all probes on treatment group in the presence of covariates (described in Materials and Methods). Genome-wide profiles of methylation were not significantly different (), nor did we detect significant differences when we restricted the analysis to promoter regions or CpG island-associated regions only. We ranked CpG sites by unadjusted P-value and examined the top ranked candidates. There was no clear evidence for a substantial effect of fish oil on the neonatal CD4+ T-cell methylome (). This analysis was repeated for the top 25% most variable probes, which did not show a significant difference between groups (not shown). We also report no evidence against the null hypothesis using a sliding windows analysis to test for regional differences in adjacent CpG sites (not shown).
Figure 3. Comparative analysis of DNA methylation profiles between treatment and intervention groups. (A) MDS scaling analysis did not discriminate treatment groups. Samples are indicated by open circles and projected along axes of the first 2 principal components of the top 5000 most variable CpG sites. (B) qq plot of P-values from the moderated t-test between intervention and control groups did not deviate from the null hypothesis. (C) Boxplots of data from 6 top-ranked CpG sites for the between-group comparison did not indicate substantive differences in methylation levels. Boxplots show median and range.
Discussion
This study builds upon emerging literature suggesting dietary components can potentially modify epigenetic regulation in humans.Citation10 Foundational work by othersCitation11-14 suggests dietary n-3 PUFAs may alter promoter methylation in blood cells.Citation14 Here, we examined this in the context of supplementation of n-3 PUFA during pregnancy. In line with the emerging evidence that maternal dietary factors can influence the neonatal epigenetic profile in modulating immune development,Citation22,23 we investigated a potential role of DNA methylation as an underlying mechanism mediating biological effects of maternal n-3 fish oil supplementation. Our primary hypothesis was that high maternal supplementation of n-3 PUFA modifies neonatal CD4+ T-cell DNA methylation profiles. Despite the clear evidence of a modifying effect of maternal supplementation on red cell lipid profiles, we did not observe substantial changes in the methylome, at least not at baseline in CD4+ T-cells. Analysis of CpG sites that co-vary with total n-3 red cell fatty acid levels suggests there may be small effects at specific CpG sites, but these did not appear to differ in a biologically meaningful way between the treatment and control groups in this study. Our data do not support the idea that n-3 PUFA supplementation in pregnancy alters the developing fetal methylome. While our analysis was restricted to a single purified cell type, we expect that any effects due to n-3 PUFA exposure during in utero development would be likely to propagate throughout the haematopoietic system and be present in other blood cell types, as we have reported before.Citation19 Despite this, it remains possible that other blood cell components, such as erythrocyte populations, may exhibit cell-type specific effect not observed in CD4s. Moreover, investigating the immune response capacity of CD4+T-cells was beyond the scope of the present study and, therefore it remains possible that neonatal fish oil exposure may modify activation-induced effects on the neonatal DNA methylome. Follow-up studies are required to address this, given that our data suggest any effects on the establishment and maintenance of DNA methylation marks in the neonatal epigenome are likely to be subtle. Other epigenetic marks, such as histone modifications, which in general terms are more dynamically responsive to environment than DNA methylation, may be key modulators of n-3 induced effects on gene transcription. These represent further avenues of investigation.
To date, only a few animal studies have reported the potential effects of maternal PUFA intake on the fetal epigenome.Citation16,17 One such study reported differing levels of FADS2 promoter methylation in the liver tissue from offspring exposed to linoleic acid during gestation.Citation17 This was replicated in a rat model in which dietary fatty acid exposure during gestation was associated with differential methylation of 4 CpG dinucleotides within the FADS2 promoter.Citation16 In humans, the evidence for these effects on the neonatal epigenome are lacking, and certainly our own study did not reproduce these findings in animal models. Similarly, Lee and coworkers analyzed a comparatively larger birth cohort (n = 261) and reported that maternal intervention significantly modulated the LINE-1 methylation, which is a surrogate marker of global methylation.Citation18 However, the difference in the methylation level of LINE-1 sequences between supplemented and control group was about 1%, in line with our own findings in non-repeat regions of the genome and, therefore, of unknown biological significance. This is a critical issue in DNA methylation studies, as the finite distribution of methylation measurements is readily amenable to achieving statistical significance in situations where the variance distribution across cases and controls is small. However, it is unclear whether these small effects bear any biological relevance. DNA methylation levels mitigate functional effects in specific genomic contexts and, therefore, it is imperative that functionality is established through examination of effect sizes (with >10% widely considered to be biologically meaningful) and genomic context (location of methylation site relative to gene promoter regions, splice sites etc.) Further research is therefore warranted to uncover the molecular effects of dietary fatty acid on neonatal immune development. Based on our observations, we anticipate large cohorts or animal models will be required to address this using genome-wide association tests and, in this respect, our study informs future EWAS designs with similar goals.
The strength of this study is the design in which a priori evidence for a modification effect on neonatal red blood cell lipid profiles was a driver of our primary investigation. Moreover, the analysis of purified CD4+ T-cells is more homogenous than whole blood or mononuclear cells; therefore, it is subjected to less unwanted variation due to cell composition and is thus expected to provide good sensitivity.Citation24 In summary, we conducted a hypothesis-free epigenome-wide search to identify genomic regions in neonatal immune cells that is modulated by maternal n-3 PUFA supplementation. Our data argues against substantial modifying effects on the neonatal CD4+ T-cell methylome. This suggests that the well-described biological effects of n-3 PUFA are more likely to be mediated by other epigenetic or posttranscriptional effects that modulate cellular function.
Materials and Methods
Study population
We used samples of the well-established study cohort, which was a double-blind randomized trial of pregnant mothers who were atopic and had clinical allergy (either allergic rhinitis or asthma) but were otherwise healthy.Citation1 They were randomized to receive either 3.7 g of fish oil (with 56.0% as DHA and 27.7% as EPA) or placebo in capsules daily from 20 weeks of gestation until delivery. A total of 83 women and their healthy full-term babies completed the trial. All study procedures were carried out in full accordance with institutional ethic guidelines. Full details of this cohort have been published elsewhere.Citation1
Data and blood sample collections
Maternal fasting blood samples were collected at 20 weeks of gestation to determine the maternal baseline fatty acids levels and at 37 weeks to calculate the increment in fatty acids levels during the study period. Women also completed a validated semi-quantitative food frequency questionnaire.
Cord blood samples were collected at birth. Cord blood mononuclear cells (CBMC) were harvested from blood samples within 12 hours of collection according to standard protocols.Citation25 Of the completed participants, a total of 70 cord bloods were available for the present study. Maternal antenatal and socio-demographic factors and infant clinical outcomes at 1 y were also available.
Erythrocyte fatty acid composition analysis
Both maternal and cord blood samples were collected and processed according to standard protocols to determine the fatty acid composition in red cells.Citation26 Fatty acids were expressed as a percentage of the weight of the total fatty acids measured. The total sum of n-3 PUFAs (20:5n-3, 22:5n-3, and 22:6n-3) and n-6 PUFAs (18:2n-6, 20:3n-6, 20:4n-6, 22:3n-6, and 22:4n-6), as well as the ratio of n3 to n6 fatty acids, were also expressed.
Isolation of CD4+ T-cells
CD4+ T-cells were isolated from CBMC using a 2-stage positive isolation strategy with magnetic DYNAL beads (Invitrogen). CBMC were incubated with CD8+ magnetic beads as per the recommended protocol (Invitrogen) and the CD8− fraction was then incubated with CD4+ magnetic beads as per the recommended protocol. Routine purity tests were conducted by flow cytometry using antibodies CD19-FITC, CD3-PE, CD8-PerCP, CD11c-PE Cy7, CD4-APC and CD14-APC Cy7 (BD Biosciences) and appropriate concentration matched isotype controls. CD4+ cell purities ranged from 91–96% pure. CD4+ T-cells were lysed with RLT buffer containing 2-mercaptoethanol (Qiagen Allprep kit) and stored at -80 ºC until genomic DNA was extracted for methylation analysis.
Nucleic acid extraction
Genomic DNA was purified from CD4+ T- cells using Qiagen Allprep kits according to manufacturer's instructions.
DNA methylation analysis
DNA samples were submitted to the NXT-Dx service facility (Belgium) for bisulphite conversion and DNA methylation profiling using the Illumina Human Methylation 450K platform. Samples were randomized across arrays according to treatment group, age and maternal allergy status. Raw data (iDAT files) were processed using the MinfiCitation27 package from the bioconductor project (http://www.bioconductor.org) in the R statistical environment (http://cran.r-project.org/). The minfi package was used evaluate control probes on the array and both sample-dependent and assay-dependent control probes indicated high quality data. Arrays were preprocessed using the stratified quantile normalization method. Technical bias attributable to different probe chemistries between Type 1 and Type II probes were adjusted in this procedure. Probes with a detection P-value call >0.01 in 1 or more samples were removed. Probes on the X and Y-chromosomes were removed to eliminate gender bias. Probes previously demonstrated to potentially cross-hybridize non-specifically in the genome were removed.Citation28 Probes containing a polymorphic SNP at the single-base extension site with a minor allele frequency of <0.05 were removed. The log2 ratio for methylated probe intensity to unmethylated probe intensity, the M value, was subsequently derived and used for statistical inference. Beta values were derived from intensities as defined by the ratio of methylated to unmethylated probes given by B = M/(U/M*100) and were used to complement M-value, as a measure of effect size. Cluster analysis was used in conjunction with the chip-wide medians of the methylated and unmethylated channels to identify any outlying samples, all samples passed QC. The final data set included 424 803 probes.
Data analysis
Following pre-processing a surrogate variable analysis was conducted on normalized M-values to estimate any residual variation not associated with the primary outcomes of interest using the method of Storey and Leek.Citation29 This method estimates unwanted variation empirically from the observed data and latent variables constructed from this analysis were included in the regression model as covariates to negate their effects. To identify CpG sites associated with treatment group or total n-3 fatty acid levels, all probes were regressed on these independent variables and the empirical Bayes method of Smyth et al.Citation30 was used to compare treatment groups. For continuous variables, we used the CpGassoc statistical packageCitation31 and included surrogate variables as the covariate matrix. QQ-plots of P-value distributions and scatterplots were used to visualize data. Adjustment for multiple testing was performed using the Benjamini Hochberg methodCitation32 with adjusted P-values <0.1 considered significant. Our criteria for identifying significant hits for further validation studies included a P-value <0.05 after correction for multiple testing, and an effect size greater than 5%.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was funded by the National Health and Medical Research Council of Australia. SP is supported by an NHMRC practitioner fellowship, DM is supported by NHMRC early carrier fellowship and RS is funded by an NHMRC Senior Research Fellowship.
References
- Dunstan JA, Mori TA, Barden A, Beilin LJ, Taylor AL, Holt PG, Prescott SL. Fish oil supplementation in pregnancy modifies neonatal allergen-specific immune responses and clinical outcomes in infants at high risk of atopy: a randomized, controlled trial. J Allergy Clin Immunol 2003 Dec; 112(6):1178-84; PMID:14657879; http://dx.doi.org/10.1016/j.jaci.2003.09.009
- Palmer DJ, Sullivan T, Gold MS, Prescott SL, Heddle R, Gibson RA, Makrides M. Effect of n-3 long chain polyunsaturated fatty acid supplementation in pregnancy on infants’ allergies in first year of life: randomised controlled trial. BMJ 2012; 344:e184; PMID:22294737; http://dx.doi.org/10.1136/bmj.e184
- Klemens CM, Berman DR, Mozurkewich EL. The effect of perinatal omega-3 fatty acid supplementation on inflammatory markers and allergic diseases: a systematic review. BJOG 2011 Jul; 118(8):916-25; PMID:21658192; http://dx.doi.org/10.1111/j.1471-0528.2010.02846.x
- Skilton MR, Ayer JG, Harmer JA, Webb K, Leeder SR, Marks GB, Celermajer DS. Impaired fetal growth and arterial wall thickening: a randomized trial of omega-3 supplementation. Pediatrics 2012 Mar; 129(3):e698-703; PMID:22351892; http://dx.doi.org/10.1542/peds.2011-2472
- Forsyth JS, Willatts P, Agostoni C, Bissenden J, Casaer P, Boehm G. Long chain polyunsaturated fatty acid supplementation in infant formula and blood pressure in later childhood: follow up of a randomised controlled trial. BMJ 2003 May 3;326(7396):953; PMID:12727766; http://dx.doi.org/10.1136/bmj.326.7396.953
- Innis SM. Metabolic programming of long-term outcomes due to fatty acid nutrition in early life. Matern Child Nutr 2011 Apr; 7 Suppl 2:112-23; PMID:21366871; http://dx.doi.org/10.1111/j.1740-8709.2011.00318.x
- Prescott SL, Barden AE, Mori TA, Dunstan JA. Maternal fish oil supplementation in pregnancy modifies neonatal leukotriene production by cord-blood-derived neutrophils. Clin Sci (Lond) 2007 Nov; 113(10):409-16; PMID:17596121
- Barden AE, Mori TA, Dunstan JA, Taylor AL, Thornton CA, Croft KD, Beilin LJ, Prescott SL. Fish oil supplementation in pregnancy lowers F2-isoprostanes in neonates at high risk of atopy. Free Radic Res 2004 Mar; 38(3):233-9; PMID:15129731; http://dx.doi.org/10.1080/10715760310001656722
- Prescott SL, Irvine J, Dunstan JA, Hii C, Ferrante A. Protein kinase Czeta: a novel protective neonatal T-cell marker that can be upregulated by allergy prevention strategies. J Allergy Clin Immunol 2007 Jul; 120(1):200-6; PMID:17544492; http://dx.doi.org/10.1016/j.jaci.2007.03.045
- McKay JA, Mathers JC. Diet induced epigenetic changes and their implications for health. Acta Physiol (Oxf) 2011 Jun; 202(2):103-18; PMID:21401888
- Ceccarelli V, Racanicchi S, Martelli MP, Nocentini G, Fettucciari K, Riccardi C, Marconi P, Di Nardo P, Grignani F, Binaglia L, et al. Eicosapentaenoic acid demethylates a single CpG that mediates expression of tumor suppressor CCAAT/enhancer-binding protein delta in U937 leukemia cells. J Biol Chem 2011 Aug 5; 286(31):27092-102; PMID:21659508; http://dx.doi.org/10.1074/jbc.M111.253609
- Sadli N, Ackland ML, De Mel D, Sinclair AJ, Suphioglu C. Effects of zinc and DHA on the epigenetic regulation of human neuronal cells. Cell Physiol Biochem 2012; 29(1-2):87-98; PMID:22415078; http://dx.doi.org/10.1159/000337590
- Cho Y, Turner ND, Davidson LA, Chapkin RS, Carroll RJ, Lupton JR. Colon cancer cell apoptosis is induced by combined exposure to the n-3 fatty acid docosahexaenoic acid and butyrate through promoter methylation. Exp Biol Med (Maywood) 2014 Mar; 239(3):302-10; PMID:24495951; http://dx.doi.org/10.1177/1535370213514927
- Aslibekyan S, Wiener HW, Havel PJ, Stanhope KL, O’Brien DM, Hopkins SE, Absher DM, Tiwari HK, Boyer BB. DNA methylation patterns are associated with n-3 fatty acid intake in Yup’ik people. J Nutr 2014 Apr; 144(4):425-30; PMID:24477300; http://dx.doi.org/10.3945/jn.113.187203
- Hermsdorff HH, Mansego ML, Campion J, Milagro FI, Zulet MA, Martinez JA. TNF-alpha promoter methylation in peripheral white blood cells: relationship with circulating TNFalpha, truncal fat and n-6 PUFA intake in young women. Cytokine 2013 Oct; 64(1):265-71; PMID:23796695; http://dx.doi.org/10.1016/j.cyto.2013.05.028
- Hoile SP, Irvine NA, Kelsall CJ, Sibbons C, Feunteun A, Collister A, Torrens C, Calder PC, Hanson MA, Lillycrop KA, et al. Maternal fat intake in rats alters 20:4n-6 and 22:6n-3 status and the epigenetic regulation of Fads2 in offspring liver. J Nutr Biochem 2013 Jul; 24(7):1213-20; PMID:23107313; http://dx.doi.org/10.1016/j.jnutbio.2012.09.005
- Niculescu MD, Lupu DS, Craciunescu CN. Perinatal manipulation of alpha-linolenic acid intake induces epigenetic changes in maternal and offspring livers. FASEB J 2013 Jan; 27(1):350-8; http://dx.doi.org/10.1096/fj.12-210724
- Lee HS, Barraza-Villarreal A, Hernandez-Vargas H, Sly PD, Biessy C, Ramakrishnan U, Romieu I, Herceg Z. Modulation of DNA methylation states and infant immune system by dietary supplementation with omega-3 PUFA during pregnancy in an intervention study. Am J Clin Nutr 2013 Aug; 98(2):480-7; http://dx.doi.org/10.3945/ajcn.112.052241
- Amarasekera M, Martino D, Ashley S, Harb H, Kesper D, Strickland D, Saffery R, Prescott SL. Genome-wide DNA methylation profiling identifies a folate-sensitive region of differential methylation upstream of ZFP57-imprinting regulator in humans. FASEB J 2014 Jun 2.
- Martino D, Joo JE, Sexton-Oates A, Dang T, Allen K, Saffery R, Prescott S. Epigenome-wide association study reveals longitudinally stable DNA methylation differences in CD4+ T cells from children with IgE-mediated food allergy. Epigenetics 2014 Apr 24; 9(7); http://dx.doi.org/10.4161/epi.28945
- Prescott SL. Early-life environmental determinants of allergic diseases and the wider pandemic of inflammatory noncommunicable diseases. J Allergy Clin Immunol 2013 Jan; 131(1):23-30; PMID:23265694; http://dx.doi.org/10.1016/j.jaci.2012.11.019
- Martino DJ, Prescott SL. Silent mysteries: epigenetic paradigms could hold the key to conquering the epidemic of allergy and immune disease. Allergy 2010 Jan; 65(1):7-15; http://dx.doi.org/10.1111/j.1398-9995.2009.02186.x
- Amarasekera M, Prescott SL, Palmer DJ. Nutrition in early life, immune-programming and allergies: the role of epigenetics. Asian Pac J Allergy Immunol. 2013 Sep; 31(3 175-82.
- Adalsteinsson BT, Gudnason H, Aspelund T, Harris TB, Launer LJ, Eiriksdottir G, Smith AV, Gudnason V. Heterogeneity in white blood cells has potential to confound DNA methylation measurements. PLoS One 2012; 7(10):e46705; http://dx.doi.org/10.1371/journal.pone.0046705
- Martino DJ, Tulic MK, Gordon L, Hodder M, Richman T, Metcalfe J, Prescott SL, Saffery R. Evidence for age-related and individual-specific changes in DNA methylation profile of mononuclear cells during early immune development in humans. Epigenetics 2011 Sep 1; 6(9).
- Dunstan JA, Roper J, Mitoulas L, Hartmann PE, Simmer K, Prescott SL. The effect of supplementation with fish oil during pregnancy on breast milk immunoglobulin A, soluble CD14, cytokine levels and fatty acid composition. Clin Exp Allergy 2004 Aug; 34(8):1237-42; PMID:15298564; http://dx.doi.org/10.1111/j.1365-2222.2004.02028.x
- Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014 May 15; 30(10):1363-9; PMID:24478339; http://dx.doi.org/10.1093/bioinformatics/btu049
- Chen YA, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, Gallinger S, Hudson TJ, Weksberg R. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 2013 Feb; 8(2):203-9; PMID:23314698; http://dx.doi.org/10.4161/epi.23470
- Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet 2007 Sep; 3(9):1724-35.
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004; 3:Article3; PMID:16646809
- Barfield RT, Kilaru V, Smith AK, Conneely KN. CpGassoc: an R function for analysis of DNA methylation microarray data. Bioinformatics. 2012 May 1; 28(9):1280-1; PMID:22451269; http://dx.doi.org/10.1093/bioinformatics/bts124
- Benjamini Y HY. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. JSTOR 1995; 57(1):289-300.
