Abstract
Ralstonia solanacearum is the causal agent of bacterial wilt disease. To better understand the molecular mechanisms involved in interaction between Nicotiana benthamiana and R. solanacearum, we focused on Hsp90, RAR1 and SGT1. Appearances of wilt symptom were significantly suppressed in Hsp90, RAR1 and SGT1-silenced plants compared with control plants. In RAR1-silenced plants, population of R. solanacearum increased in a similar manner to control plants. In contrast, multiplication of R. solanacearum was significantly suppressed in Hsp90 and SGT1-silenced plants. In addition, expression of PR genes were increased in Hsp90 and SGT1-silenced plants challenged with R. solanacearum. Therefore, RAR1 might be required for disease development or suppression of disease tolerance. These results also suggested that Hsp90 and/or SGT1 might play an important role in suppression of plant defenses leading to disease susceptibility and disease development.
Abbreviations
| HR | = | hypersensitive response |
| Hsp | = | heat shock protein |
| PCR | = | polymerase chain reaction |
| RT-PCR | = | reverse transcription-polymerase chain reaction |
| qRT-PCR | = | quantitative real time polymerase chain reaction |
| RAR1 | = | required for Mla12 resistance 1 |
| SGT1 | = | suppressor-of-G2-allele-of-skp1 |
| TTSS | = | type III secretion system |
| VIGS | = | virus-induced gene silencing |
Introduction
Plants have evolved sophisticated defense mechanisms that are activated in response to pathogen attacks. In most cases, plants resist infection through active defense mechanisms. The front line of induced defense is triggered by pathogen-associated molecular patterns (PAMPs), also known as PAMP-triggered immunity (PTI). PAMPs are generally conserved compounds, like chitin in fungi and flagellins in bacteria, and PAMP-triggered immunity is induced by all invading pathogens. In contrast, some adapted pathogens suppress PTI by evolving effector molecules. The second line of plant defense is activated via recognition of pathogen effectors by Resistance gene products, followed by triggering of effector-triggered immunity.Citation1–Citation3
Ralstonia solanacearum is a devastating, soil-borne pathogen with a global distribution and wide host range.Citation4 It causes bacterial wilt in several economically important solanaceous crops. In the tomato, resistance to R. solanacearum is controlled by several loci.Citation5,6 In contrast, resistance is monogenic and is conferred by the RRS1-R gene that encodes a R protein in Arabidopsis thaliana, and this resistance is dependent upon salicylic acid and the NDR1 signaling pathway.Citation7 PopP2, type III effector of R. solanacearum was identified and shown to interact with the RRS1-R.Citation8 Recent studies showed that ethylene-, salicylic acid- and MAP kinase-related defense signaling pathways are involved in the resistance of tomato to R. solanacearum.Citation7 We previously reported that cellular components, such as asparagine-rich protein, S-glycoprotein-like protein and translationally controlled tumor protein, have important role in regulation of plant defenses against R. solanacearum.Citation10-Citation12 In addition, our previous report showed that molecular chaperon, Nbshsp17, has a crucial role in regulation of plant defenses, suggesting involvement of molecular chaperons and co-chaperons in plant and R. solanacearum interaction.Citation13
In this study, we focused on Hsp90, RAR1 and SGT1. Although, these chaperons and co-chaperons have been well-known to have important role in regulation of plant immune responses, the role of Hsp90, RAR1 and SGT1 on plant and R. solanacearum interaction is largely unknown. Therefore, we carried out functional analysis of Hsp90, RAR1 and SGT1 using N. benthamiana, and discussed a possible mechanism by which Hsp90, RAR1 and SGT1 affects bacterial wilt disease.
Results
Plant molecular chaperons and co-chaperons such as Hsp70, Hsp90, RAR1 and SGT1, play a critical role in plant-pathogen interaction. Then, we created Hsp70-, Hsp90-, RAR1- and SGT1-silenced plants (), and observed the effect of silencing of these genes on development of bacterial wilt disease. As shown in , the control plants showed wilt symptom from 6 day after inoculation and completely died 12 d after inoculation with R. solanacearum. In Hsp70-silenced plant, wilt symptom was slightly accelerated 6–8 d after inoculation (Fig. S1). Therefore, Hsp70 might play a role in plant defenses similar to previous report.Citation14 In contrast, we could observe significant delay of wilt symptom in Hsp90-, RAR1- and SGT1-silenced plants compared to those in the control plants. RAR1-silenced plants showed suppression of wilt symptom 6–16 d after inoculation. Significant suppression of wilt symptom was also observed 6–19 d after inoculation in Hsp90-silenced plants. Most drastic suppression of wilt symptom was observed in SGT1-silenced plants, and wilt symptom was suppressed during the experiment period up to 20 d after inoculation. Generally, it has been reported that Hsp90, RAR1 and SGT1 have a crucial role in induction of plant defenses. In contrast, our present results suggested that Hsp90, RAR1 and SGT1 might have roles in suppression of defense responses and/or wilt symptom.
Figure 1. Creation of gene-silenced plants. Total RNA was isolated from Control (PVX), Hsp90-(PVX:Hsp90), RAR1-(PVX:RAR1) and SGT1-(PVX:SGT1) silenced plants. Expression values of Hsp70, Hsp90, RAR1 and SGT1 were estimated by qRT-PCR, and expressed as [Qty] after normalization with actin. Values represent the means and SD from triplicate experiments. Asterisks denote values significantly different from empty PVX controls (*; P < 0.05).
![Figure 1. Creation of gene-silenced plants. Total RNA was isolated from Control (PVX), Hsp90-(PVX:Hsp90), RAR1-(PVX:RAR1) and SGT1-(PVX:SGT1) silenced plants. Expression values of Hsp70, Hsp90, RAR1 and SGT1 were estimated by qRT-PCR, and expressed as [Qty] after normalization with actin. Values represent the means and SD from triplicate experiments. Asterisks denote values significantly different from empty PVX controls (*; P < 0.05).](/cms/asset/b3aa16bc-2989-49ca-ace0-916ba385e501/kpsb_a_970410_f0001_b.gif)
Figure 2. Effect of Hsp90, RAR1 and SGT1-silencing on bacterial wilt disease by inoculation with R. solanacearum. Control (PVX), Hsp90- (PVX:Hsp90), RAR1- (PVX:RAR1) and SGT1- (PVX:SGT1) silenced plants were infiltrated with R. solanacearum. (A) Disease development of bacterial wilt was rated daily on a 0–4 disease index in control (open squares) or silenced (solid squares) plants. Asterisks denote values significantly different from those ofcontrol plants (*; P < 0.05, t-test). (B) Characteristic symptoms in control and silenced plants. Photograph was taken 12 d after inoculation with R. solanacearum.
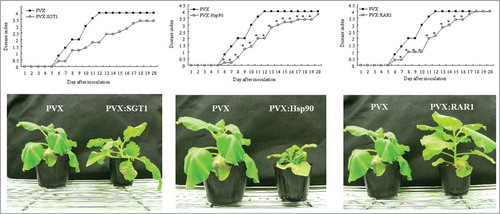
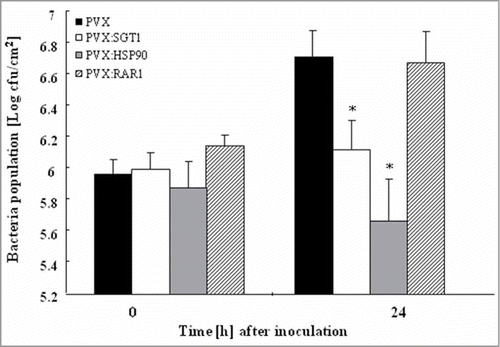
Since suppression of wilt symptom was observed in Hsp90, RAR1 and SGT1-silenced plants, this raised the possibility that disease resistance to R. solanacearum may increase in these silenced plants. To address whether silencing of Hsp90, RAR1 and SGT1 would affect disease resistance, we estimated multiplication of R. solanacearum in Hsp90, RAR1 and SGT1-silenced and control plants. As shown in , growth of R. solanacearum was scarcely affected by RAR1-silencing. In contrast, growth of R. solanacearum was significantly reduced in Hsp90 and SGT1-silenced plants 24h after inoculation in comparison with control plants. Intriguingly, population of type III secretion system (TTSS)-deficient mutant of R. solanacearum increased in control plants as well as Hsp90 and SGT1-silenced plants (Fig. S2).
Figure 3. Growth of Ralstonia solanacearum in Hsp90, RAR1 and SGT1-silenced plants Control (PVX), Hsp90- (PVX:Hsp90), RAR1- (PVX:RAR1) and SGT1- (PVX:SGT1) silenced plants were infiltrated with R. solanacearum (108 CFU/ml). Bacterial population was determined by plating at specified time points. Values are means of 4 replicate experiments with SD. Asterisks denote values significantly different from those of empty PVX controls (*; P < 0.05, t-test).
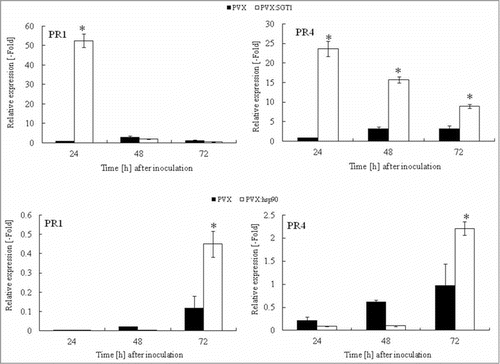
We could observe growth inhibition of R. solanacearum in Hsp90 and SGT1-silenced plants, suggesting the up-regulation of defense responses in Hsp90 and SGT1-silenced plants. To further determine the role of Hsp90 and SGT1 in defense responses against R. solanacearum, we analyzed salicylic acid-dependent PR-1a expression and jasmonic acid-dependent PR-4 expression. In control plants inoculated with R. solanacearum, expression of PR-1a and PR-4 increased at 24-72 hours after inoculation. Expression levels of PR-1a and PR-4 transcript were greatly enhanced in Hsp90- and SGT1-sielnced plants ().
Figure 4. Acceleration of PR genes expression in Hsp90 and SGT1-silenced plants in response to Ralstonia solanacearum infection. Total RNA was isolated from control (PVX) and Hsp90 (PVX:Hsp90) and SGT1 (PVX:SGT1)-silenced plants inoculated with R. solanacearum (108 CFU/ml). Relative expression of PR-1a and PR-4 transcripts were normalized with actin and calculated as relative to the non-treated control. Values represent the means and SD from triplicate experiments. Asterisks denote valuessignificantly different from empty PVX controls (*; P < 0.05).
Discussion
It has been shown that molecular chaperones act as a critical component of plant defense responses. Hsp90 is reportedly required for HR mediated by the resistant genes, Pto and Rx.Citation14 Another Hsp, Hsp70, is required not only for induction of HR in response to INF1 elicitin and Pseudomonas cichorii, but also basal resistance against P. syringae. Citation15,16 The RAR1 gene is reportedly required for N-mediated HR.Citation14 SGT1 is also a critical signaling component required for R gene-mediated HR in several plant species against various plant pathogens, including fungi, bacteria and viruses.Citation17-Citation20 Recent study showed that pepper SGT1 act as a host interactor of AvrBsT during the HR induction.Citation21 Our present result showed that acceleration of bacterial wilt symptom was observed in Hsp70-silenced plant, but bacterial manipulation was not affected (Fig. S1). Therefore, Hsp70 might be required for suppression of bacterial wilt disease and/or induction of disease tolerance.
Recent studies also showed that plant molecular chaperons and co-chaperons play important roles not only in disease resistance but also disease development and disease susceptibility. Mutations in 2 Hsp90 genes lead to heightened accumulation of immune receptors, including SNC1, RPS2 and RPS4. Hsp90s, and enhanced disease resistance, suggesting involvement of Hsp90 in the negative regulation of immune receptor accumulation.Citation22 SGT1 has been shown to be involved in cell death that promotes the pathogenesis of Botrytis cinereaCitation23 and Fusarium culmorum.Citation24 SGT1 was also reportedly to be required for coronatine-induced chlorosis and cell death during bacterial speck disease on tomato.Citation25 Development of disease-associated cell death caused by the P. syringae pv. tabaci, require for SGT1.Citation26 Our previous report showed Hsp70 is required for tabtoxinine-β-lactum induced cell death and wildfire disease development by P. syringae pv tabaci.Citation27 Previous report showed SGT1 tightly interacts with RAR1 is required for early stages of Agrobacterium-mediated plant transformation, suggesting that RAR1, along with SGT1 is important for virulence function.Citation28 Recent reports also suggested that the plant chaperon and co-chaperon act as type III effector targets during establishment of disease susceptibility. The major target of HopI1, a virulence type III effector from P. syringae, was reported as plant heat shock protein Hsp70.Citation16 P. syringae effector protein AvrB enhances plant susceptibility by interaction with Hsp90 and/or RAR1.Citation29,30 Salmonella typhimurium type III effector-mediated phenotypes required its catalytic E3 ubiquitin ligase activity and interaction with the conserved host protein SGT1 in both plants and mammals.Citation31
Our present data showed the requirement of plant chaperon and co-chaperon in bacterial wilt. Silencing of RAR1 significantly reduced bacterial wilt symptom, but not affected on bacterial manipulation (). We previously reported that PopA-mutant of R. solanacearum, which grew in intercellular spaces and systemically infected into tobacco plants similarly to wild type R. solanacearum, did not cause wilt on tobacco plants, suggesting suppression for disease development or induction of disease tolerance.Citation32 Therefore, RAR1 might be required for disease development or suppression of disease tolerance. In contrast, hyper-induction of defense related PR-gene expression and suppression of R. solanacearum growth were observed in Hsp90 and SGT1-silenced plants (). Intriguingly, silencing of Hsp90 and SGT1 did not affected on multiplication of TTSS-mutant of R. solanacearum (Fig. S2). These results suggested that Hsp90 and SGT1 might be type III effector targets for during the pathogenesis of R. solanacearum, and play an important role in suppression of plant defenses leading to disease susceptibility.
In conclusion we found out that in addition to positive regulation of plant immune responses by SGT1, RAR1 and Hsp90, they are also involved in the negative regulation of immune responses. Further research about plant intracellular proteins related to SGT1, RAR1 and Hsp90, and cognate bacterial effectors of Hsp90 and SGT1 will be required to clarify molecular mechanisms of plants-R. solanacearum interactions.
Materials and Methods
Plant materials, bacterial isolates and chemicals
Nicotiana benthamiana was grown in a growth room.Citation13 Ralstonia solanacearum strain OE1–1 and the corresponding hrpY (encoding Hrp pilus)-mutant of R. solanacearum strain OE1–1 was cultured in PY medium containing 20 μg/mL rifampicin or 20 μg/mL kanamycin, respectively.Citation12 Bacterial suspension were infiltrated into N. benthamiana leaves as described previously.Citation13
Bacterial Population and Disease Index
The population of R. solanacearum and TTSS-deficient mutant of R. solanacearum was determined by plating on Hara-Ono plates. Plants inoculated with R. solanacearum were coded and inspected daily for wilting symptoms for 20 d For each plant, a disease index on a scale of 0 to 4 was calculated as described elsewhere.Citation13
Isolation of RNA and cDNA synthesis
Total RNA was isolated from N. benthamiana leaves with RNAiso (Takara), and RNA samples were treated with DNase I (RNase-free; Takara) to degrade contaminating genomic DNA as described previously.Citation13 cDNA (cDNA) was synthesized by PrimeScript RT reagent Kit (Takara).
Vector constructs and seedling infection for virus-induced gene silencing
Construction of virus vectors for VIGS experiments were described previously.Citation27 The VIGS experiment was carried out with Agrobacterium tumefaciens strain GV3101Citation33 and inoculated into N. benthamiana leaves as described previously.Citation3 We observed characteristic dwarf phenotypes of SGT1-, Hsp70- and Hsp90-silenced plants as shown previously.Citation27
Quantitative real time PCR
Quantitative real time PCR was carried out according to the method of Maimbo et al.Citation13 Reverse transcription was carried out with 1 μg total RNA using the PrimeScript RT reagent Kit (Takara). Quantitative reverse-transcription PCR (qRT-PCR) was carried out with a 20 μL reaction mixture containing 1 μL of the cDNA stock and 0.4 μL of the respective primers (10 pM; Supplemental Table 1), using the SYBR GreenER qPCR Reagent System (Invitrogen, Tokyo Japan), with an Applied Biosystems 7300 real time PCR system. Standard deviations and differences between expression ratios of non-treated controls and other samples were tested for statistical significance using the t-test.
Control (PVX) and Hsp70 (PVX:Hsp70)-silenced plants were infiltrated with R. solanacearum (10Citation8 CFU/ml). (A) Total RNA was isolated from Control (PVX) and Hsp70-(PVX:Hsp70) silenced plants. Expression values of Hsp70 was estimated by qRT-PCR, and expressed as [Qty] after normalization with actin. Values represent the means and SD from triplicate experiments. Asterisks denote values significantly different from empty PVX controls (*; P < 0.05). (B) Disease development of bacterial wilt was rated daily on a 0–4 disease index in control or silenced plants. Asterisks denote values significantly different from those of control plants (*; P < 0.05, t-test). (C) Characteristic symptoms in control and silenced plants. Photograph was taken 8 d after inoculation with R. solanacearum. (D) Bacterial population was determined by plating at specified time points. Values are means of 4 replicate experiments with SD. Asterisks denote values significantly different from those of empty PVX controls (*; P < 0.05, t-test).
Control, Hsp90 (PVX:Hsp90)- and SGT1 (PVX:SGT1)-silenced plant were inoculated with type III secretion system-deficient R. solanacearum (108 CFU/ml). Bacterial population was determined by plating at specified time points. Values are means of 4 replicate experiments with SD. Asterisks denote values significantly different from those of empty PVX controls.
Table S1 List of primers used for qRT-PCR in this study.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Table_S1.tiff
Download TIFF Image (88.2 KB)Figure_S2.tiff
Download TIFF Image (32.2 KB)Figure_S1.tiff
Download TIFF Image (612.6 KB)Acknowledgments
The authors thank Dr. D. Baulcombe for the pPVX201 vector.
Supplemental Materials
Supplemental data for this article can be accessed on the publisher's website.
Funding
This work was supported by Grants in Aid for Scientific Research to AK (16780031 and 18780029) and to YH (15028214 and 16380037) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
References
- Bittel P, Robatzek S. Microbe-associated molecular patterns (MAMPs) probe plant immunity. Curr Opin Plant Biol 2007; 10:335-341; PMID:17652011; http://dx.doi.org/10.1016/j.pbi.2007.04.021
- Boller T, He SY. Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 2009; 324:742-4; PMID:19423812; http://dx.doi.org/10.1126/science.1171647
- Jones JD, Dangl JL. The plant immune system. Nature 2006; 444:323-9; PMID:17108957; http://dx.doi.org/10.1038/nature05286
- Hayward HC. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Ann Rev Phytopathol 1991; 29:65-87; http://dx.doi.org/10.1146/annurev.py.29.090191.000433
- Thoquet P, Olivier J, Sperisen C, Rogowsky P, Laterrot H, Grimsley, N. Quantitative trait loci determining resistance to bacterial wilt in tomato cultivar Hawaii 7996. Mol Plant-Microbe Interact 1996; 9:826-36; http://dx.doi.org/10.1094/MPMI-9-0826
- Thoquet P, Olivier J, Sperisen C, Rogowsky P, Prior P, Anaïs G, Mangin B, Bazin B, Nazer R, Grimsley N. Polygenic resistance of tomato plants to bacterial wilt in the French West Indies. Mol Plant-Microbe Interact 1996; 9:837-42; http://dx.doi.org/10.1094/MPMI-9-0837
- Deslandes L, Olivier J, Theulieres F, Hirsch J, Feng DX, Bittner-Eddy P, Beynon J, Marco Y. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc Natl Acd Sci USA 2002; 99:24048.
- Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acd Sci USA 2003; 100:8024-9; http://dx.doi.org/10.1073/pnas.1230660100
- Chen YY, Lin YM, Chao TC, Wang JF, Liu AC, Ho FI, Cheng CP. Virus-induced gene silencing reveals the involvement of ethylene-, salicylic acid- and mitogen-activated protein kinase-related defense pathways in the resistance of tomato to bacterial wilt. Physiol Plant 2009 136:32435; PMID:19470092; http://dx.doi.org/10.1111/j.1399-3054.2009.01226.x
- Komori D, Nishihara M, Takahashi A, Gupta M, Yoshioka H, Mizumoto H, Ohnishi K, Hikichi Y, Kiba A. Isolation and characterization of an asparagine-rich protein that regulates hypersensitive cell death-mediated resistance in Nicotiana plants. Plant Biotechnology 2012; 29:292-300; http://dx.doi.org/10.5511/plantbiotechnology.12.0213b
- Gupta M, Yoshioka H, Ohnishi K, Mizumoto H, Hikichi Y, Kiba A. A translationally controlled tumor protein negatively regulates the hypersensitive response in Nicotiana benthamiana. Plant Cell Physiol 2013; 54:1403-14; PMID:23788648; http://dx.doi.org/10.1093/pcp/pct090
- Maimbo M, Ohnishi K, Hikichi Y, Yoshioka H, Kiba A. S glycoprotein-like protein regulates defense responses in Nicotiana plants against Ralstonia solanacearum. Plant Physiol 2010; 152:2023-35; PMID:20118275; http://dx.doi.org/10.1104/pp.109.148189
- Maimbo M, Ohnishi K, Hikichi Y, Yoshioka H, Kiba A. Induction of a small heat shock protein and its functional roles in Nicotiana Plants in the defense response against Ralstonia solanacearum. Plant Physiol 2007; 145:1588-99; PMID:17965181; http://dx.doi.org/10.1104/pp.107.105353
- Liu Y, Schiff M, Marathe R, Dinesh-Kummar S. Tobacco Rar1, EDS1 and NPR1NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J 2002; 30:415-29; PMID:12028572; http://dx.doi.org/10.1046/j.1365-313X.2002.01297.x
- Kanzaki H, Saitoh H, Ito A, Fujisawa S, Kamoun S, Katou S, Yoshioka H., Terauchi R. Cytosolic HSP90 and HSP70 are essential components of Inf1-mediated hypersensitive response and non-host resistance to Pseudomonas cichorii in Nicotiana benthamiana. Mol Plant Pathol 2003; 4:383-91; PMID:20569398; http://dx.doi.org/10.1046/j.1364-3703.2003.00186.x
- Jelenska J, van Hal JA, Greenberg JT. Pseudomonas syringae hijacks plant stress chaperone machinery for virulence. PNAS 2010; 107:13177-82; PMID:20615948
- Austin MJ, Muskett P, Kahn K, Feys BJ, Jones JDG, Parker JE. Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 2002; 295 2077-80; PMID:11847308; http://dx.doi.org/10.1126/science.1067747
- Azevedo C, Sadanandom A, Kitagawa, K, Freialdenhoven A, Shirasu K, Schulze-Lefert P. The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 2002; 295:2073-76; PMID:11847307; http://dx.doi.org/10.1126/science.1067554
- Tör M, Gordon P, Cuzick A, Eulgem T, Sinapidou E, Mert-Türk F, Can C, Dangl JL, Holub EB. Arabidopsis SGT1b Is Required for Defense Signaling Conferred by Several Downy Mildew Resistance Genes. Plant Cell 2002; 14:993-1003; PMID:12034892; http://dx.doi.org/10.1105/tpc.001123
- Liu Y, Nakayama N, Schiff M, Litt A, Irish VF, and Dinesh-Kumar SP. Virus induced gene silencing of a DEFICIENS ortholog in Nicotiana benthamiana. Plant Mol Biol 2004; 54:701-11; PMID:15356389; http://dx.doi.org/10.1023/B:PLAN.0000040899.53378.83
- Kim NH, Kim DS, Chung EH, Hwang BK. Pepper suppressor of the G2 allele of skp1 interacts with the receptor-like cytoplasmic kinase1 and type III effector AvrBsT and promotes the hypersensitive cell death response in a phosphorylation-dependent manner. Plant Physiol 2014; 165:76-91; PMID:24686111; http://dx.doi.org/10.1104/pp.114.238840
- Huang S, Monaghan J, Zhong X, Lin L, Sun T, Dong OX, Li X. HSP90s are required for NLR immune receptor accumulation in Arabidopsis. Plant J 2014; 79:427-39; http://dx.doi.org/10.1111tpj.12573
- EI Oirdi M, Bouarab K. SGT1 positively regulates the process of plant cell death during both compatible and incompatible plant-pathogen interactions. New Phytol 2007; 175:131-9; PMID:17547673; http://dx.doi.org/10.1111/j.1469-8137.2007.02086.x
- Cuzick A, Maguire K, Hammond-Kosack KE. Lack of the plant signalling component SGT1b enhances disease resistance to Fusarium culmorum in Arabidopsis buds and flowers. New Phytologist 2009; 181:901-12; PMID:19140951; http://dx.doi.org/10.1111/j.1469-8137.2008.02712.x
- Uppalapati SR, Ishiga Y, Ryu CM, Ishiga T, Wang K, Noe LD, Parker JE, Mysore KS. SGT1 contributes to coronatine signaling and Pseudomonas syringae pv. tomato disease symptom development in tomato and Arabidopsis. New Phytologist 2011; 189:83-93; PMID:20854394; http://dx.doi.org/10.1111/j.1469-8137.2010.03470.x
- Wang K, Uppalapati SR, Zhu X, Dihesh-Kumar SP, Mysore KS. SGT1 positively regulates the process of plant cell death during both compatible and incompatible plant–pathogen interactions. Mol Plant Pathol 2010; 11:597-611; PMID:20695999
- Ito M, Yamamoto Y, Kim CS, Ohnishi K, Hikichi Y, Kiba A. Heat shock protein 70 is required for tabtoxinine-β-lactam-induced cell death in Nicotiana benthamiana. J Plant Physiol 2014; 171:173-8; PMID:24331433; http://dx.doi.org/10.1016/j.jplph.2013.10.012
- Anand A, Mysore KS. The role of RAR1 in Agrobacterium-mediated plant transformation. Plant Signal Behav. 2013; 9:e26784; http://dx.doi.org/10.4161/psb.26784
- Cui H, Wang Y, Xue L, Chu J, Yan C, Fu J, Chen M, Innes RW, Zhou JM. Pseudomonas syringae effector protein AvrB perturbs Arabidopsis hormone signaling by activating MAP kinase 4. Cell Host & Microbe 2010; 7:164-75; PMID:20159621; http://dx.doi.org/10.1016/j.chom.2010.01.009
- Shang Y, Li X, Cui H, He P, Thilmony R, Chintamanani S, Zwiesler-Vollick J, Gopalan S, Tang X, Zhou JM. RAR1, a central player in plant immunity, is targeted by Pseudomonas syringae effector AvrB. PNAS 2006; 103:19200-05; PMID:17148606; http://dx.doi.org/10.1073/pnas.0607279103
- Bhavsar AP, Brown NF, Stoepel J, Wiermer M, Martin DDO, Hsu KJ, Imami K, Ross CJ, Hayden MR, Foster LJ, et al. The Salmonella type III effector SspH2 specifically exploits the NLR co-chaperone activity of SGT1 to subvert immunity. Plos Pathogen 2013; 9:e1003518; http://dx.doi.org/10.1371/journal.ppat.1003518
- Kanda A, Ohnishi K, Kiba A, Hikichi Y. Implication of C-terminal mutation of PopA of Ralstonia solanacearum strain OE1-1 in suppression of bacterial wilt. Plant Pathology 2009; 58:159-69; http://dx.doi.org/10.1111/j.1365-3059.2008.01938.x
- Baulcombe DC, Chapman S, Cruz SS. Jellyfish green fluorescent protein as a reporter for virus infections. Plant J 1995; 7:1045-53; PMID:7599646; http://dx.doi.org/10.1046/j.1365-313X.1995.07061045.x
