Abstract
Bispecific antibodies have emerged in recent years as a promising field of research for therapies in oncology, inflammable diseases, and infectious diseases. Their capability of dual target recognition allows for novel therapeutic hypothesis to be tested, where traditional mono-specific antibodies would lack the needed mode of target engagement. Among extremely diverse architectures of bispecific antibodies, knobs-into-holes (KIHs) technology, which involves engineering CH3 domains to create either a “knob” or a “hole” in each heavy chain to promote heterodimerization, has been widely applied. Here, we describe the use of a cell-free expression system (Xpress CF) to produce KIH bispecific antibodies in multiple scaffolds, including 2-armed heterodimeric scFv-KIH and one-armed asymmetric BiTE-KIH with tandem scFv. Efficient KIH production can be achieved by manipulating the plasmid ratio between knob and hole, and further improved by addition of prefabricated knob or hole. These studies demonstrate the versatility of Xpress CF in KIH production and provide valuable insights into KIH construct design for better assembly and expression titer.
Abbreviations
| IgG | = | immunoglobulin G |
| Fab | = | antigen-binding fragment |
| Fc | = | fragment crystallizable |
| HC | = | immunoglobulin heavy chain |
| LC | = | immunoglobulin light chain |
| scFv | = | single-chain fragment variable |
| BiTE | = | bispecific T-cell engager |
| PK | = | pharmacokinetics |
| FcR | = | Fc receptor |
| KIH | = | knob-into-hole |
| EpCAM | = | epithelial cell adhesion molecule |
| HER2 | = | human epidermal growth factor receptor 2 |
| LC-MS | = | liquid chromatography-mass spectrometry |
| CHO | = | Chinese hamster ovary |
| FACS | = | fluorescence-activated cell sorting |
| ELISA | = | enzyme-linked immunosorbent assay |
Introduction
With progress in the understanding of complex diseases such as cancer, inflammatory diseases and infectious diseases, more efficacious therapeutics can be designed and developed. One of the desired therapeutic properties for new therapeutics is multispecificity that will: 1) recruit effector cells to enhance killing; 2) prevent crosstalk between parallel/redundant signaling pathways to overcome resistance; 3) differentiate diseased cells from normal cells for improved safety and efficacy; and 4) potentially provoke synergetic effects.Citation1-Citation4 Bispecific antibodies, which comprise a single molecule capable of recognizing 2 targets, have recently drawn attention as an emerging group of therapeutics to fulfill such unmet medical needs. Such characteristics are not only appreciated for the aforementioned functions, but also greatly favored by pharmaceutical companies from a cost-of-goods perspective.
The first approved bispecific antibody, catumaxomab (Removab®), comprises a T-cell engaging arm (anti-CD3) and a tumor-targeting arm (anti-epithelial cell adhesion molecule) in a canonical IgG format.Citation5,Citation6 It is produced from a mouse/rat quadroma (hybrid hybridoma) cell, in theory resulting in 12.5% of the total products possessing the desired dual specificity, though preferential pairing between intra-species HC and LC would most likely give rise to a higher amount of bispecific IgG with correct assembly.Citation3 Such a prodigal process can be largely improved by expression and purification of individual antibodies followed by chemical coupling of 2 IgGs (IgG2) or Fab fragments (F(ab’)2).Citation7 This chemical method has also been utilized to conjugate a pharmacophore peptide heterodimer to the catalytic center of a scaffold IgG, resulting in the CovX-Body.Citation8
The majority of bispecific antibodies are generated by genetic engineering. As described in a recent comprehensive review by Kontermann,Citation4 more than 45 different bispecific formats have been developed, ranging in molecular mass from 15 kDa (VHH)Citation9 to 150 kDa (natural IgG), to 350 kDa (Dock-and-Lock(DNL)-Fab4-IgG).Citation10 There are 3 broad categories of bispecific antibody design. The first category makes use of a conventional IgGCitation11-Citation13 or FabCitation14 as a platform, and introduces a second functional entity as a single variable domain (sVD), single-chain variable fragment (scFv) or single-chain Fab to the N- or C-terminus of the light chain (LC) or heavy chain (HC) via a flexible peptide linker, leading to tetravalent or hexavalent bispecific antibodies. Among the 3 formats, scFv is most popular because its compact structure of tandem linked VL and VH domains makes the molecule highly amenable to gene engineering, yet it still possesses complete antigen-binding activity.Citation15,Citation16 Alternative formats include DVD-IgTM (dual variable domain immunoglobulin, tandem linkage of the second VH and VL to the N-termini of HC and LC, respectively),Citation17 Tandemab (tandem linkage of 2 VH-CH1 in combination of common LC),Citation18 DNL (natural association of 2 antibodies or antibody fragments anchored with DDD (dimerization and docking domain) from PKA (protein kinase A) and AD (anchoring domain) from A-kinase anchor protein (AKAP), respectively),Citation10,Citation19 LUZ-Y (leucine zipper tethered at the C-termini of HC and later proteolytically removed),Citation20 2-in-1-IgG (same LC and HC capable of dual recognition),Citation21 and mAbCitation2 (engineered loops in CH3 domain of IgG to obtain second specificity).Citation22 Except for the last 3 models described above, bispecific antibodies in this category potentially have higher risk of immunogenicity due to their non-natural IgG structures.
Bispecific antibodies in the second category are characterized by simple structures with variable domains or scFvs as the building blocks. For example, the smallest bispecific antibody, VHH, is made of 2 VH domains in tandem.Citation9 Two formats are widely utilized to generate small bispecific antibodies. One is the Diabody (Db), which is a heterodimer composed of 2 polypeptide chains encoding VLA-VHB and VHA-VLB in the order of VH-VL or VL-VH with a linker of 5 amino acids.Citation23 Its derivatives include, but are not limited to, dsDb (interchain disulfide bond between VL and VH of the same antibody),Citation24 DART (dual-affinity re-targeting, interchain disulfide bond between 2 VL),Citation25 scDb (single chain Diabody),Citation26 and tandAbs (Diabody dimer via flexible linkers in between).Citation27 The other is the BiTE® (bispecific T-cell engager), which is composed of 2 scFvs connected in tandem by an adjustable linker.Citation28 Because of their unique structural features, the more compact Diabody is often employed as a diagnostic tool or drug delivery vehicle,Citation29 while BiTEs are generally developed as anti-cancer therapeutics to co-localize T cells and tumor cells to form immunologic synapses and therefore trigger tumor cell cytotoxicity.Citation6,Citation28 One of the caveats of this category, especially for the BiTE, is also its size (50 kDa), which necessitates continuous intravenous administration to overcome the quick renal serum clearance.Citation28 As such, different fusion techniques have been explored to extend serum half-life. The candidates for fusion include Fc, CH3 (e.g., MinibodyCitation30), albumin, albumin binding peptide,Citation31 and cytokines.Citation19
In the third category, natural IgG-like structures are favored because of their desirable pharmacokinetic (PK) properties, low immunogenicity risk, and retained stability and Fc receptor affinity.Citation3 The dual specificities are conferred by each Fab, requiring a classical IgG assembly but with 4 distinctive polypeptide chains (2 different LCs and 2 different HCs). Random chain associations, namely LC mispairing and unwanted HC homodimerization, initially hindered the development of such bispecific antibodies, but these drawbacks have been progressively overcome by judicious genetic engineering. The application of a common light chain,Citation32,Citation33 orthogonal Fab interface (complementary mutations introduced at LC and HC interface in one Fab and no change to the other Fab),Citation34 or CrossMab (one Fab with switched VH or CH1 domain(s) with the partner VL or CL domain(s) and the other Fab untouched)Citation35 have essentially eliminated the mispairing of LCs. Alternatively, replacing Fab with scFv or BiTE as the binding entity has been applied to overcome LC mispairing, resulting in scFv-knobs-into-holes (KIH)-Fc or BiTE-KIH-Fc.Citation4 Meanwhile, a number of engineering strategies in CH3 domains have also significantly enhanced HC heterodimerization based on stericCitation36-Citation38 or electrostatic complementarity.Citation39,Citation40 Among them, the most successful and widely applied route is knobs-into-holes, in which a “knob” is created by replacing T366 with a bulky residue W on one HC, and the corresponding “hole” is made by triple mutations of T366S, L368A and Y407V on the partner HC.Citation32,Citation33,Citation36,Citation41 KIH-directed HC heterodimer formation renders more than 90% desired product under co-expression conditions, making it feasible for scalable production to satisfy clinical and market needs.Citation36 In addition, a more intense engineering approach has created alternating human IgG and IgA fragments in CH3, resulting in the so-called SEEDbody (Strand-Exchange Engineered Domain) to guide HC heterodimerization.Citation42
Production of KIH bispecific antibodies to high purity has always been challenging due to the inherent complexity of the molecule. For example, the efficient co-expression of KIH relies on approximately equal expression of both knob and hole. Therefore, the selection of such a stable expression clone is a prerequisite for KIH production using E. coli or mammalian cells, and could be a laborious and time-consuming process.Citation3 Although downstream processes such as selective Protein A purification of CH3 mutantsCitation43 and in vitro assembly (separate expression of knob and hole followed by mixture of knob and hole at appropriate redox condition to induce assembly)Citation37,Citation40,Citation44,Citation45 have demonstrated their feasibility in generating high quality KIH molecules, additional obstacles may appear, such as immunogenicity induced by mutations or cost increase. As such, co-expression remains the preferable method for KIH production.
Cell-free synthesis (Xpress CF) is an in vitro transcription/translation system using E. coli based extract.Citation46,Citation47 Without the limitation of cell viability imposed by conventional expression systems, Xpress CF is capable of protein expression at g/L scale within hours, and has been demonstrated to retain high productivity in transition from micro- to manufacturing scale. Its open and flexible nature can tolerate multifarious manipulations in extract, DNA template, and chemical and protein additives,Citation46,Citation48,Citation49 which makes it amenable for high-throughput screening by automation.Citation50 Over the years, its utilization has expanded to include production of IgGs and antibody derivatives,Citation46 antibody-drug conjugates,Citation49 vaccines,Citation51 membrane proteins,Citation52 metalloproteins,Citation53 and viral proteins.Citation54
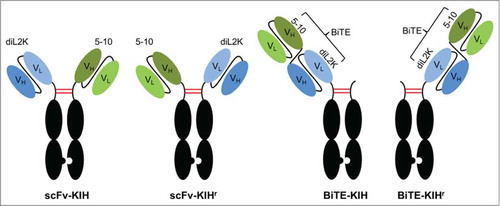
We report bispecific antibody expression, in particular KIH, using Xpress CF in this study. The high manipulability of the systemCitation46,Citation50,Citation55 enables expeditious optimization of knob:hole plasmid ratio to achieve most efficient KIH assembly, as well as an “add-back” approach with prefabricated knob or hole for higher titer. Multiple KIH scaffolds have been investigated, including 2-armed heterodimeric scFv-KIH and one-armed asymmetric BiTE-KIH with tandem scFv (), providing insights to guide rational KIH design for better assembly and titer. Furthermore, Xpress CF has been demonstrated to scale in a linear fashion from small scale expression to pilot scale production,Citation46,Citation47 making it capable of KIH production at a large scale.
Figure 1. KIH scaffolds. Two different scFv (in circles) are anchored on Fc(K) and Fc(H), respectively, defined as scFv-KIH or scFv-KIHr (the reversed arrangement of scFv-KIH). Alternatively, 2 scFv are connected with a flexible linker, resulting in a BiTE. BiTE-KIH is defined as BiTE on Fc(K) and BiTE-KIHr is as BiTE on Fc(H). The pairing Fc(H) or Fc(K) contains the hinge, CH2 and CH3 domains only. Examples are shown for diL2K and 5–10.
Results
KIH construction
Three well-established antibodies, diL2K (αCD3), 5-10 (αEpCAM)Citation56 and 4D5 (αHER2)Citation57 were chosen to construct a total of 8 bispecific antibodies in 4 different KIH scaffolds, namely scFv-KIH (αCD3 on knob), scFv-KIHr (αCD3 on hole), BiTE-KIH (BiTE on knob) and BiTE-KIHr (BiTE on hole) (, ). diL2K engages T-cells via interaction with CD3 signaling complex, a T-cell receptor. 5–10 and 4D5 recognize tumor antigen epithelial cell adhesion molecule (EpCAM) and human epidermal growth factor receptor (HER)-2, respectively. Inspired by catumaxomab, we anchored diL2K to Fc(K) and 5–10 or 4D5 to Fc(H), resulting in an scFv-KIH. A reversed arrangement with diL2K on Fc(H) and 5-10/4D5 on Fc(K) was defined as scFv-KIHr to distinguish the 2 forms, but viewed as an equivalent molecule to scFv-KIH. scFvs were used instead of Fab to circumvent LC mispairing.
Table 1. KIH molecules expressed in Xpress CF
Given that tandem scFv BiTEs are another promising class of bispecific antibodies currently in clinical development,Citation28 we constructed BiTE-KIH in the attempt to extend the half-life of BiTEs. BiTE 5-10xdiL2K (solitomab)Citation56 and BiTE diL2Kx4D5 were constructed and fused with either Fc(K) or Fc(H), resulting in BiTE-KIH and BiTE-KIHr, respectively ().
KIH assembly as function of plasmid ratio between knob and hole
Similar to KIH production in E. coli or mammalian cell expression systems, efficient KIH production in Xpress CF also depends on approximately equal expression of knob and hole polypeptides. While balancing knob and hole polypeptide expression can be challenging in conventional expression systems, this can be quickly addressed in Xpress CF. By simply adjusting the plasmid ratio between knob and hole DNA added into the cell-free reaction, high-quality KIH production can be achieved with maximal assembled KIH and minimal unassembled knob or hole. As shown in , excessive knob or hole is observed at extreme plasmid ratios between 4D5-Fc(K) and diL2K-Fc(H). Shifting the plasmid ratio to 1:1, the majority of knob and hole assemble into KIH with negligible amount of unassembled chains. Such an optimization process is particularly efficient when the expression levels between the 2 participants are different. For example, the expression titer of 5-10xdiL2K-Fc(H) is only about 1/5 of its partner Fc(K) (Fig. S1). The highest titer of this BiTE-KIHr is obtained at knob:hole ratio of 1:5 ().
Figure 2. KIH assembly at various plasmid ratios between knob and hole. Soluble fractions from cell-free reaction were visualized by SDS-PAGE with 14C autoradiography of incorporated leucine at 14 h that measures cell-free produced proteins only. Representative titrations were shown for (A, C) 4D5-Fc(K)+diL2K-Fc(H) (scFv-KIHr) and (B, D) Fc(K)+diL2Kx5-10-Fc(H) (BiTE-KIHr). The plasmid ratios between knob and hole for each lane in (A) and (B) are indicated in (C) and (D), respectively. Optimal plasmid ratio (knob:hole) were identified as 1:1 for scFv-KIHr (C) and 1:5 for BiTE-KIHr (D), giving rise to highest yield and assembly.
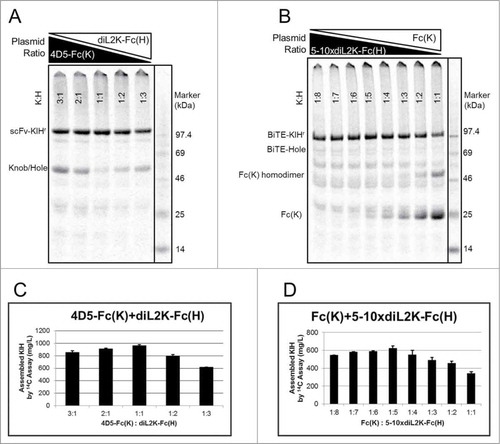
Due to the large difference in size between knob and hole for BiTE-KIH and BiTE-KIHr, SDS-PAGE is a useful tool to evaluate the KIH assembly in these 2 scaffolds. However, it lacks the resolution to distinguish between homodimer and heterodimer for scFv-KIH or scFv-KIHr. As such, LC-MS was applied for assembly assessment. For example, there is minimal knob or hole homodimers observed by LC-MS for 5-10xdiL2K in scFv-KIHr when being co-expressed at optimal K:H plasmid ratio (Fig. S2).
Comparison of KIH scaffolds
BiTE-KIH and BiTE-KIHr give rise to an asymmetric molecule, which combines the structural features and biological functions of the BiTE with the half-life extending properties of the Fc. Despite the small difference, BiTE-KIHr outperforms BiTE-KIH in terms of expression titer (). BiTE-Fc(K) and BiTE-Fc(H) exhibit comparable expression titers on their own, but Fc(K) is greater than Fc(H) with regards to expression titer (Fig. S1). Therefore, it seems that Fc(K) is a better folding partner for BiTE-Fc(H) than Fc(H) is for BiTE-Fc(K).
Figure 3. Expression titer comparison of all 4 KIH scaffolds. Expression titers are reported for the co-expression conditions at optimal plasmid ratio between knob and hole. scFv-KIH subgroup (scFv-KIH and scFv-KIHr) expresses much better than BiTE-KIH subgroup (BiTE-KIH and BiTE-KIHr), except for 5-10xdiL2K-BiTE-KIHr which is comparable to its scFv-KIH or scFv-KIHr counterparts due to its well-behaved BiTE conformation. BiTE-KIHr in general outperformed BiTE-KIH at least 2 folds.
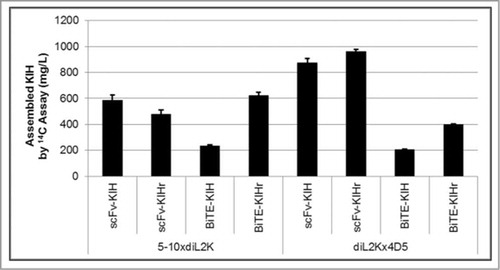
Overall, BiTE-KIH expression is less robust than the scFv-KIH. With only one exception, 5-10xdiL2K-Fc(H)+Fc(K) BiTE-KIHr, the expression titers of scFv-KIHs are at least 2-fold higher than those of the BiTE-KIHs ( and Table S1). The difference may stem from the fact that BiTE molecules are structurally more complex than scFvs and therefore may not fold well when fused to an Fc domain. However, some very well-behaved BiTEs such as 5-10xdiL2K can result in comparable expression titers between BiTE-KIHr and scFv-KIH scaffolds.
“Add-back” approach for KIH expression using prefabricated knob or hole
Previously, Yin et al. have shown that Fab production in Xpress CF can be dramatically improved with production of LC for one hour prior to HC production.Citation46 Thus, we adapted a similar concept for KIH production, suspecting prefabricated knob or hole polypeptide might serve as a chaperon to facilitate the folding of its partner and promote KIH assembly. Using 5-10-Fc(K)+diL2K-Fc(H) (scFv-KIHr) as an example, indeed more 5-10-Fc(K) expressed in the presence of its partner diL2K-Fc(H) protein, and vice versa. As a result, the expression titer of the scFv-KIHr doubled from 500 mg/L in co-expression to 950 mg/L using 1 mg/mL of prefabricated 5-10-Fc(K), and tripled to 1300 mg/L using 1 mg/mL of diL2K-Fc(H) ( and Table S1). Moreover, titrating in diL2K-Fc(H) from 0.75 mg/mL to 1 mg/mL continuously increased the titer of the scFv-KIHr from 800 mg/L to 1300 mg/L (Table S1), suggesting that the expression is probably limited by the amount of diL2K-Fc(H) added to the cell-free reaction, and could potentially be further increased with additional diL2K-Fc(H).
Figure 4. Titration of prefabricated knobs or holes in cell-free KIH expression. In the case of 5-10-Fc(K)CdiL2K-Fc(H) (scFv-KIHr), prefabricated diL2K-Fc(H) and 5-10-Fc(K) at 1 mg/mL approximately increases the expression titer of KIH by 3 folds and 2 folds, respectively, in comparison of co-expression titer (control at optimal plasmid ratio K:H D 1:2) (A, C). For diL2Kx4D5-Fc(K)CFc(H) (BiTE-KIH), 0.25 mg/mL of prefabricated Fc (H) can already increase the BiTE-KIH expression titer by ∼75% compared to co-expression titer (control at optimal plasmid ratio K:H D 2:1). However, no further improvement was observed with increased amount of Fc(H) (B, D). Soluble fractions from cell-free reaction were visualized by SDS-PAGE with 14C autoradiography of incorporated leucine at 14 h that measures cell-free produced proteins only. The concentrations of prefabricated protein added in cell-free reactions in (A) and (B) are indicated in (C) and (D), respectively.
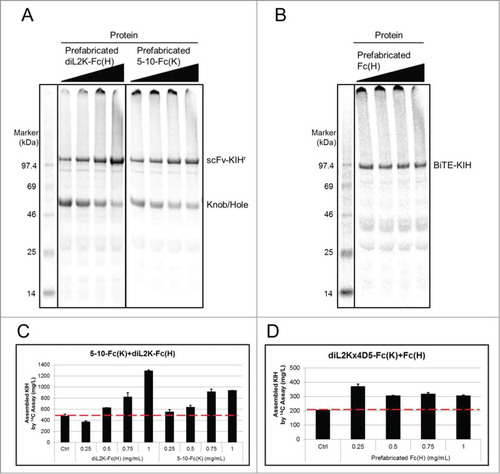
Another case suggested that addition of more prefabricated knob or hole may not always be beneficial. For instance, 0.25 mg/mL of prefabricated Fc(H) increased expression of diL2Kx4D5-Fc(K)+Fc(H) (BiTE-KIH) from 200 mg/L in co-expression to 350 mg/L, but there is no further improvement when more Fc(H) was provided ( and Table S1). Thus, it is most likely that the BiTE-KIH expression plateau is determined by the expression of diL2Kx4D5-Fc(K) itself, which is already at its apex in the presence of 0.25 mg/mL of Fc(H). Taken together, the effect of the “add-back” approach must depend on the expression level of the plasmid in the cell-free expression system, which is governed by the primary sequence.
During the preparation of individual knob or hole for “add-back” expressions, we noticed that concentrated knob tends to precipitation at higher concentrations. The propensity to aggregate is expected for knobs, because the mutant tryptophan (T366W) is more exposed in knob monomers than the mutants (T366S, L368A and Y407V) in hole monomers. These inherent structural features also manifest in more notable homodimerization for holes than for knobs (Fig. S1). However, the homodimerization is likely in equilibrium, enabling dissociation to form monomers upon being added into cell-free reactions.Citation58
Characterization of KIH molecules
The selection of the KIH scaffold depends on not only the expression titer, but also the performance in various biological function assays. To confirm bispecific KIH molecules produced in Xpress CF are capable of dual targeting and cell killing by engaging T cells, we chose the well-established model bispecific antibody, 5-10xdiL2K, as an example, and expressed and purified 5-10xdiL2K KIHs in all 4 scaffolds, which were then subjected to SDS-PAGE for assembly assessment (Fig. S3) and LC-MS for identity confirmation (Table S2). All showed heterodimers as the major product except for BiTE-KIHr due to non-optimal knob:hole ratio, which showed more unassembled Fc(K) as shown in Figure S3.
5-10xdiL2K KIHs were first examined for their dual specificity to CD3 and EpCAM antigens using CD8 T cells and cells that express various levels of EpCAM on the surface, including HCT-116, JIMT-1 and CHO-hEpCAM. All 4 scaffolds possess comparable affinities toward both CD3 and EpCAM as the control, 5-10xdiL2K (BiTE, 2 scFv in tandem) (). Their ability of T cell activation was also evaluated by incubation with T cells in the presence and absence of tumor cells (HCT-116). Only in the presence of tumor cells, T cell activation took place as early as 4 h () and was continuous at 24 h (). Furthermore, cell killing was monitored at various KIH concentrations, and all KIH scaffolds induced robust cell killing by activated T cells (). BiTE-KIH and BiTE-KIHr yield EC50 values that are highly similar to the BiTE control, suggesting that tethering a KIH-Fc to a BiTE does not interfere with its function, and appears as a sound approach for half-life extension of tandem scFvs. It is also interesting to observe that scFv-KIHr, with the highest affinity to CD3, has the lowest EC50. This observation implies that high antigen affinity might not be desirable because release from the target might be difficult, and therefore deteriorate its dual targeting capability. The KIHs (except for scFv-KIHr due to poor cell killing activity) were also investigated for PK properties, and show comparable in vivo stability. All KIHs (100 kDa) have significantly longer half-life than the BiTE control (50 kDa), but are less stable than trastuzumab IgG (150 kDa) and scFv-Fc (100 kDa) as expected due to destabilizing mutations in the CH3 domains ().Citation40
Figure 5. Relative cell binding affinities of αEpCAMxαCD3 bispecific antibodies. 5-10xdiL2K-KIH bispecific antibodies binding to CD3 T cells (A) and EpCAM+ HCT-116 (B), JIMT-1 (C) and CHO-EpCAM cells (D) was determined by FACS. Xpress CF expressed 5-10xdiL2K is a bispecific molecule with 2 tandem scFv, serving as a positive control. Mean values and standard deviation of duplicate values are shown.
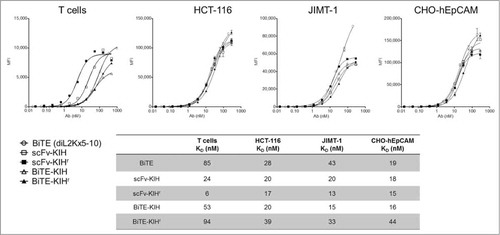
Figure 6. T cell activation induced by αEpCAMxαCD3 bispecific antibodies. CD8 T cells were treated with 5-10xdiL2K-KIH bispecific antibodies (1 μg/mL), either alone or with HCT-116 cells at an E:T ratio of 15:1. Surface expression of immediate/early T cell activation marker CD69 after 4 h treatment (A) or co-expression of CD69 and CD25 after 24 h treatment (B) on CD8 T cells was assessed by FACS. Results were from 2 independent experiments. Xpress CF expressed BiTEs αCD3xαCD19 and 5-10xdiL2K serve as negative and positive controls, respectively.
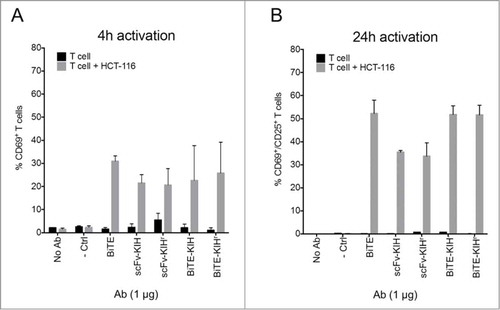
Figure 7. Redirected lysis of EpCAM+ human carcinomas by αEpCAMxαCD3 bispecific antibodies. Redirected lysis of HCT-116 and JIMT-1 cells by 5-10xdiL2K-KIH bispecific antibodies was measured by a FACS-based cytotoxicity assay. CD8 T cell donors, donor 1 and donor 2, were used for both cell line killing assessments at an E:T ratio of 15:1. Representative of 3 independent experiments is shown. Cell-free expressed BiTEs αCD3xαCD19 and 5-10xdiL2K serve as negative and positive controls, respectively.
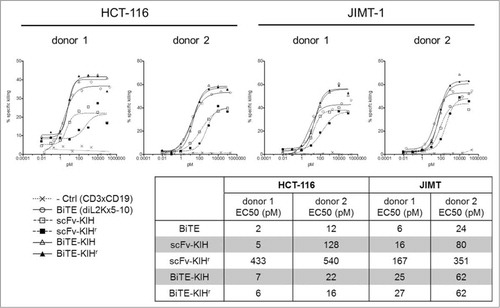
Figure 8. Pharmacokinetics of αEpCAMxαCD3 bispecific antibodies. 5–10xdiL2K-KIH bispecific antibodies were intravenously injected at a dose of 5 mg/kg in mice and plasma samples were collected at different time points. Bispecific antibody plasma levels were determined by EpCAM-binding ELISA for 5-10xdiL2K (control) or hIgG-Fc ELISA for Fc-fusion bispecific antibodies. Trastuzumab-scFv-Fc (red) and trastuzumab-IgG (blue) were made using Xpress CF, and evaluated for their PK using immunodeficient mice in a separate experiment. Mean values and standard deviation of triplicate plasma samples are shown.
Discussion
We used an E. coli based cell-free expression system (Xpress CF) to produce KIH molecules. Taking advantage of its high manipulability, we were able to perform quick optimization processes on various molecules in multiple scaffolds. Furthermore, we developed an “add-back” approach that improves titer in comparison to co-expression.
Consideration of configuration in KIH design
Four KIH scaffolds that are categorized into scFv-KIH, BiTE-KIH and their reverse orientation were evaluated for their expression titers. Without adding another dimension of complexity to the study, we constructed the scFvs and BiTEs to adopt a single configuration. For example, diL2K scFv is constructed as VH-VL and 5-10xdiL2K BiTE as 5-10(VL-VH)xdiL2K(VH-VL). However, other factors should also be taken into consideration during novel KIH design such as 1) the orientation within the scFv (VL-VH vs. VH-VL),Citation59,Citation60 2) the order of 2 scFvs on BiTE construct,Citation61,Citation62 and 3) the length and composition of the linkers (between 2 scFv as well as between VL and VH),Citation63,Citation64 all of which likely contribute to KIH folding and assembly.
Comparison among 4 KIH scaffolds indicates that scFv-KIH and scFv-KIHr appear to be superior scaffolds to BiTE-KIH and BiTE-KIHr to develop a novel KIH styled bispecific antibody, since they offer better expression. However, for the development of an established BiTE with extended half-life in serum, the BiTE-KIHr scaffold would offer a viable approach that is more suitable than the BiTE-KIH scaffold.
It is intriguing to observe the significant difference in expression between BiTE-KIH and BiTE-KIHr, given that they are considered to be conceptually equivalent, indicating that Fc(H) is more tolerant of a hard-to-express sequence, such as a BiTE, than Fc(K). This is in agreement with the observation that holes are generally more stable than knobs. The same strategy can be adopted by scFv-KIH design when the arrangement on the KIH needs to be made for 2 scFv that have significantly different expression levels. Assuming equal biological efficacy, we suggest that it is generally preferable to anchor the hard-to-express sequence to Fc(H) rather than Fc(K) in order to achieve the highest expression titers.
Realistically, subtle differences in KIH scaffolds may manifest unique biological properties. Therefore, it is critical to review both expression titers and bifunctional performance to determine the best KIH scaffold for any given application.
Application of “add-back” approach in high-throughput screening of KIH
For development, the “add-back” strategy may seem labor intensive, with additional expression and purification steps. But it may prove to be efficient and cost effective in high-throughput screening, because there is no need to optimize the plasmid ratio in order to achieve high expression of KIHs. Such an application is especially useful when producing KIHs that share the same knob or hole, for example, BiTE-KIHr production using prefabricated Fc(K). With an expression titer of 1.4 g/L, prefabricated Fc(K) can be easily stocked and used for pairing with different BiTE-Fc(H)s.
Expression of conventional KIH in Xpress CF
Conventional KIH molecules contain 2 distinctive Fab instead of scFv, which adds complexity to KIH production, specifically LC pairing. Practices developed to avoid this problem include the utilization of 1) common LC,Citation32,Citation33 2) Fab with orthogonal LC-HC interface,Citation34 and 3) CrossMab with switched VH-VL or CH1-CL.Citation35 Because of its high manipulability, we are confident that the cell-free expression system is capable of producing conventional KIH. A more sophisticated optimization process is required to determine the optimal plasmid ratio among all participants. If common LC is the approach taken to produce KIH, “add-back” strategy with prefabricated LC might be beneficial.
Aglycosylated KIH produced in Xpress CF
As the first approved bispecific antibody, catumaxomab is equipped with 2 critical features to enable efficient cell killing, 1) effective colocalization of tumor cells and activated T-cells, and 2) recruitment of effector cells such as natural killer cells and macrophages.Citation6 The effector functions include antibody-dependent cell-mediated cytotoxicity, antibody-dependent cellular phagocytosis and complement-dependent cytotoxicity, and are only achievable via interactions between glycosylated Fc and Fc receptors on the effector cells.Citation65,Citation66 As an E. coli-based cell-free expression system, the Xpress CF platform produces KIH molecules that are aglycosylated and therefore lack effector function. If effector function is desired, compensatory mutations could be made to the CH2 domain to enable effector function in aglycosylated Fc.Citation67,Citation68 Furthermore, glycosylated proteins could possibly be produced by including appropriate enzymes in cell-free reactions.Citation69 Alternatively, a non-natural amino acid can be incorporated at the glycosylation site N297,Citation49 followed by site-specific conjugation of glycans in a post-translation manner.Citation70-Citation72
Materials and Methods
KIH constructs
All DNA sequences used in this work were codon optimized and inserted in the previously described pYD317 vector between NdeI and SalI cloning sites.Citation46,Citation47 Using site-directed mutagenesis, the Fc region of trastuzumabCitation41,Citation46 was engineered to render the knob (T366W) or the hole (T366S, L368A and Y407V).Citation36 4D5 scFv was generated based on trastuzumab IgG by overlapping PCR in a VL-VH format, and diL2K scFv, 5-10 scFv, 5-10xdiL2K BiTE and diL2Kx4D5 BiTE were synthesized by DNA 2.0 (Menlo Park, CA). Three linkers were used to ensure flexibility to maintain antigen binding capabilities, a 15-25mer between VL and VH, a pentamer between 2 scFv in BiTEs, and a 11mer between scFv or BiTE and Fc(K) or Fc(H).
Cell-free expression
Cell-free reactions were performed as previously described.Citation46,Citation49 A few parameters were fixed to simplify the optimization process of KIH expression conditions, including temperature at 30°C, total plasmid concentration at 10 μg/mL (4 nM), and 30% extract SBDG108. Extract SBDG108 was chosen because it expresses both DsbC and FkpA, 2 chaperons that are essential for IgG folding and assembly.Citation48 Small scale reactions at 100 μL were performed in a 96-well microtiter plate with a breathable cover while shaking at 650 rpm for 14 hours in an Eppendorf Thermomixer R. For protein production purposes, larger scale reactions (20-50 mL) were set up in a thin-film petri dish format without shaking.
Two approaches were employed for KIH expression, 1) co-expression and 2) expression with prefabricated knob or hole. In co-expression reactions, both plasmids encoding either knob or hole were provided at various ratios. Alternatively, either knob or hole proteins were individually expressed and purified in advance, and added into cell-free reactions expressing the complementary partners. The best expression conditions that gave rise to the most assembled KIH and least unassembled single chains were identified.
Protein titer quantification
Protein titers were quantified by monitoring the incorporation of L-[14C(U)]-leucine (NEC279E001MC, PerkinElmer) as previously described.Citation47 Briefly, soluble fractions were isolated and analyzed on SDS-PAGE (Life TechnologiesTM), in terms of fully and partially assembled KIHs, and folded and clipped knobs and holes. The SDS-PAGE gels were visualized by autoradiography using a STORM 840 PhosphoImager, and quantified by ImageQuantTL software (GE Life Sciences, NJ). KIH titers were therefore determined based on the pixel ratio between the fully assembled KIH under non-reducing condition and all soluble protein produced under reducing condition. Titers of individual knob and hole were determined in a similar manner with the exception that both homodimer and monomer under non-reducing condition were taken into account as folded proteins.
Purification of assembled KIH and KIH single chains
Both intact KIH molecules and KIH single chains (individual knobs and holes) were purified using either a ProteinMakerTM system (Emerald) or AKTA FPLC system (GE Healthcare). Briefly, harvested cell-free reaction was diluted with 0.1 M NaH2PO4, pH 7.3 at 1:2 ratio, and centrifuged at 10,000 g for 30 min. The supernatant was filtered through a 0.45 μm filter, and then loaded onto pre-equilibrated MabSelect SuRe column (GE Healthcare). After washing with 10 CV of 0.1 M NaH2PO4, 800 mM arginine, pH 7.0, the desired protein was eluted with 5 CV of 0.1 M citric acid, 300 mM arginine, pH 3.0, and immediately neutralized by ∼17% of 1 M Tris, pH 9. To prevent buffer components from interfering with cell-free reactions, KIH single chains were further dialyzed against 10 mM Tris-acetate, pH 8.2, 60 mM potassium acetate, and concentrated to 5–10 mg/mL with Amicon Ultra-15 Centrifugal Filter Units 10K MWCO (EMD Millipore).
LC-MS
Non-reduced samples were diluted to 10–20 μM in phosphate buffered saline, (PBS; pH 7.4 from 10x stock). Reduced samples were incubated with 4 M guanidine HCL, 20 mM dithiothreitol, and 1x PBS for 45 min at 46°C. After reduction the samples were cooled to room temperature and trifluoroacetic acid added to 0.5% (from 10% stock) and then stored at less than 10°C until analyzed.
All samples were run on an PLRP-S column (5 μm, 1000 Å, 2.1 × 50 mm, Agilent Technologies, Santa Clara, CA) using 0.1% formic acid water (mobile phase A) and 20:80, 2-propanol:acetonitrile with 0.1% formic acid (mobile phase B) on an Agilent 1200 binary SL pump at 0.4 mL/min. Column temperature was 80°C for non-reduced samples and 50°C for reduced samples. The gradients for both non-reduced and reduced samples were the same: washing on column for 5 min at 5% B with diversion to waste post column, followed by a gradient to 90% B over 20 min. To clean the column, mobile phase B was switched between 95% and 10% every 30 sec for a total of 3 times. Lastly, the column was equilibrated for 5 min at 5% B before each sample or blank injection. Sample loadings were approximately 15 pmol of total protein (as determined by A280) per injection.
All samples were run on an Agilent Accurate Mass 6520 QTOF using a dual spray source. The capillary voltage was at 4000 V, source gas temperature at 325°C, skimmer at 65 V, drying gas at 8 L/min, and nebulizer at 30 psig. Spectra were acquired every one second from 500–3200 m/z in extended dynamic range mode. Non-reduced sample spectra were acquired with a Fragmentor voltage of 370 V. Reduced samples were acquired at 200 V and 300 V (using 2 events in the same experiment time segment). The lower Fragmentor voltage was optimized for a protein of around 50 kDa. All runs were internally calibrated using the HP-922 reference compound sprayed in on the reference nebulizer of the dual spray source with averaging every 5 scans.
All data were analyzed using MassHunter Qualitative B.06 from Agilent. All peaks in the total ion chromatogram and UV-Vis chromatograms (214 nm and 280 nm) were interrogated by combining their respective mass spectra. Where an expected species was not observed, extracted ion chromatograms, based on theoretical m/z values, were used to search for ions. Spectra were deconvoluted using the Maximum Entropy algorithm in Bioconfirm (add-on to MassHunter Qualitative). More accurate masses were assigned to wider peaks using the pMod deconvolution algorithm. Theoretical averages masses were calculated with GPMAW (Lighthouse Data, Odense, Denmark).
Cell culture and transfection
HCT-116, JIMT-1 and CHO-K cells were obtained from the American Type Culture Collection. HCT-116 and JIMT-1 were cultured in DMEM/F12 media supplemented with 10% heat-inactivated FBS and CHO-K cells were maintained in RPMI media supplemented with 10% heat-inactivated FBS. Stable CHO-K cells expressing human EpCAM were generated by transfecting cells with a human EpCAM-puromycin expression construct and selection with 30 μg/mL puromycin (Clonetech). Stable cell lines with high EpCAM expression were isolated by limiting dilution after 5 d of selection. CD8 T cells were isolated from peripheral blood mononuclear cells (PBMC) of healthy donors (Stanford Blood Center). PBMCs were prepared from enriched lymphocyte preparations (buffy coats) diluted 2-fold with PBS and 30 mL of the diluted buffy coat was layered over 15 mL NycoPrep 1.077 (Cosmo Bio) in a 50 mL Leucosep centrifuge tube (Greiner Bio-One). Lymphocyte layer was collected, washed twice with PBS and red blood cells were depleted with ACK lysing buffer (Lonza). CD8 T cells were isolated by CD8 microbeads (Miltenyi Biotec) using the autoMACS Pro Separator (Miltenyi Biotec). Purified CD8 T cells were cultured in RPMI media containing 10% heat-inactivated FBS, penicillin-streptomycin (Corning) and GlutaMAX (Life Technologies).
Cell binding assay
Bispecific antibody binding affinities to cell-surface expressed CD3 and EpCAM were determined by flow cytometry. Antibodies were serial diluted 3-fold in fluorescence-activated cell sorting (FACS) binding/wash buffer (PBS containing 0.5% BSA and 0.05% sodium azide) and all steps were performed at 4°C. Human CD8 T cells, HCT-116, JIMT-1 or CHO-hEpCAM cells (2e5 cells/96 well) were incubated with diluted antibodies for 1 h and cells were washed twice, followed by antibody detection with 5 μg/mL biotinylated Protein L (Pierce) and 2 μg/mL streptavidin-PE (eBioscience). Cells were fixed with 4% PFA in PBS and analyzed with a BD Bioscience LSR II flow cytometer. Data was analyzed using FlowJo software (Treestar) and Prism (GraphPad) using the one site-specific binding with Hill slope equation.
T cell killing assay
BiTE activity for αEpCAMxαCD3 bispecific antibodies was measured by flow cytometry. EpCAM-expressing HCT-116 or JIMT-1 cells were pre-labeled with 10 μM CellTrace Oregon Green 488 carboxy-DFFDA SE (Life Technologies) for 30 min, washed 3 times and plated at a density of 2e5 cells/96-well in DMEM/F12 media. The following day, cell media was exchanged for RPMI media and 3-fold serial diluted antibodies and CD8 T cells at 15:1 effector-to-target (E:T) cell ratio were added in a total volume of 150 μL. After 24 h co-culture, all cells were harvested with Accutase (BD Biosciences), washed once with FACS wash buffer and resuspended in FACS wash buffer supplemented with 5 μg/mL propidium iodide (PI) (Sigma-Aldrich) to stain dead cells. Cells were analyzed with a BD Bioscience LSR II flow cytometer using the FITC and PE-Cy5 channels to identify labeled target cells and dead cells, respectively. Using FlowJo software (Treestar), the percentage of dead target cells were calculated by the number of PI+/Oregon Green 488+ cells divided by the total number of Oregon Green 488+ cells. Antibody-mediated specific killing (%) was determined by subtracting background killing in the absence of antibody. Dose-response EC50 was calculated using the log (agonist) vs. response - variable slope (4 parameters) equation in Prism (GraphPad). To monitor the extent of T-cell activation, CD8 T cells were treated with 1 μg/mL bispecific antibody in the absence or presence of HCT-116 cells at an E:T ratio of 15:1 for 4 h and 24 h at 37°C. T cells were collected and stained for CD69-APC (FN50, eBioscience) and CD25-PE (2A3, BD Biosciences). CD8 T cells, identified as Oregon Green- cells, were analyzed for CD69 or CD69 and CD25 reactivity. T cell activation after 4 h and 24 h was reported as percent CD69+ and CD69+/CD25+ T cells, respectively.
Pharmacokinetic analysis of bispecific αEpCAMxαCD3 antibodies
Mice were intravenously injected in the tail vein with 100 μg antibody (5 mg/kg dose) and randomized into 4 different sub-groups of 3 mice each. Blood samples were collected at each time point (5 m, 30 m, 1 h, 2 h, 4 h, 6 h and 1 day for 5-10xdiL2K or 5 m, 30 m, 1 h, 2 h, 4 h, 6 h, 1 d, 2 d, 3 d, 5 d and 7 d for KIH antibodies) from alternating sub-groups and antibody plasma concentrations were determined by ELISA. For 5-10xdiL2K, plasma concentrations were determined by EpCAM-binding ELISA. Nunc MaxiSorp plates (Thermo Scientific) were coated with 1 μg/mL recombinant EpCAM-His (ACROBiosystems) diluted in carbonate/bicarbonate buffer pH 9.6 and incubated overnight at 4°C. Following steps were performed at room temperature. Plates were washed twice with PBS + 0.05% Tween, blocked with 0.5% ovalbumin in PBS for 1 h and washed again twice before adding serial diluted plasma samples in diluent buffer (0.5% ovalbumin, 0.05% Tween in PBS) for 2 h. Plates were washed 5 times and HRP-conjugated Protein L (Thermo Scientific) was added for 1 h at a dilution of 1:1,000 in diluent buffer. For KIH scaffolds, plasma concentrations were determined by anti-human Fcγ-specific ELISA. ELISA plates were coated with 0.5 μg/mL goat anti-human IgG, Fcγ-fragment specific (Jackson ImmunoResearch) diluted in carbonate/bicarbonate buffer pH 9.6 and incubated overnight at 4°C. Plates were washed twice, blocked with 0.5% BSA in PBS for 1 h and washed again twice before adding serial diluted plasma samples in diluent buffer (0.5% BSA, 0.05% Tween in PBS) for 2 h. Plates were washed 5 times and HRP-conjugated goat anti-human IgG, Fcγ-fragment specific was added for 1 h at a dilution of 1:10,000 in diluent buffer. For both ELISA formats, secondary-bound plates were washed 5 times and 3,3′,5,5′-tetramethylbenzidine substrate (SureBlue Reserve, KPL) was added for 30 min before quenching the reaction with 1 N phosphoric acid. Absorbance at 450 nm was measured using a SpectraMax M5 microplate reader (Molecular Devices) and data was analyzed with Prism (Graphpad) using log OD versus log Ab concentration linear regression analysis. Plasma samples were diluted within a specified range without plasma interference.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental_Material.zip
Download Zip (274.3 KB)Acknowledgments
We would like to thank Marissa L. Matsumoto for initial work on Fc(K) and Fc(H) construction, Dan Groff and Patrick Rivers for construction and production of extract SBDG108, and Qi Cai and colleagues in Sutro Biopharma, Inc. for scientific discussion and support.
Supplemental Material
Supplemental data for this article can be accessed on thepublisher's website.
References
- Byrne H, Conroy PJ, Whisstock JC, O’Kennedy RJ. A tale of two specificities: bispecific antibodies for therapeutic and diagnostic applications. Trends Biotechnol 2013; 31:621-32; PMID:24094861; http://dx.doi.org/10.1016/j.tibtech.2013.08.007
- May C, Sapra P, Gerber H-P. Advances in bispecific biotherapeutics for the treatment of cancer. Biochem Pharmacol 2012; 84:1105-12; PMID:22858161; http://dx.doi.org/10.1016/j.bcp.2012.07.011
- Klein C, Sustmann C, Thomas M, Stubenrauch K, Croasdale R, Schanzer J, Brinkmann U, Kettenberger H, Regula JT, Schaefer W. Progress in overcoming the chain association issue in bispecific heterodimeric IgG antibodies. MAbs 2012; 4:653-63; PMID:22925968; http://dx.doi.org/10.4161/mabs.21379
- Kontermann RE. Dual targeting strategies with bispecific antibodies. MAbs 2012; 4:182-97; PMID:22453100; http://dx.doi.org/10.4161/mabs.4.2.19000
- Zeidler R, Reisbach G, Wollenberg B, Lang S, Chaubal S, Schmitt B, Lindhofer H. Simultaneous activation of T cells and accessory cells by a new class of intact bispecific antibody results in efficient tumor cell killing. J Immunol 1999; 163:1246-52; PMID:10415020
- Maher J, Adami AA. Antitumor immunity: easy as 1, 2, 3 with monoclonal bispecific trifunctional antibodies? Cancer Res 2013; 73:5613-7; PMID:24014596; http://dx.doi.org/10.1158/0008-5472.CAN-13-1852
- Grziano R, Guptill P. Chemical producition of bispecific antibodies. Methods Mol Biol 2004; 283:71-85; PMID:15197303
- Doppalapudi VR, Huang J, Liu D, Jin P, Liu B, Li L, Desharnais J, Hagen C, Levin NJ, Shields MJ, et al. Chemical generation of bispecific antibodies. Proc Natl Acad Sci USA 2010; 107:22611-6; PMID:21149738; 10.1073/pnas.1016478108
- De Meyer T, Muyldermans S, Depicker A. Nanobody-based products as research and diagnostic tools. Trends Biotechnol 2014; 32:263-70; PMID:24698358; http://dx.doi.org/10.1016/j.tibtech.2014.03.001
- Chang C-H, Rossi EA, Goldenberg DM. The dock and lock method: a novel platform technology for building multivalent, multifunctional structures of defined composition with retained bioactivity. Clin Cancer Res 2007; 13:5586s-5591s; PMID:17875793; http://dx.doi.org/10.1158/1078-0432.CCR-07-1217
- Coloma MJ, Morrison SL. Design and production of novel tetravalent bispecific antibodies. Nat Biotechnol 1997; 15:159-63; PMID:9035142; http://dx.doi.org/10.1038/nbt0297-159
- Orcutt KD, Ackerman ME, Cieslewicz M, Quiroz E, Slusarczyk AL, Frangioni J V, Wittrup KD. A modular IgG-scFv bispecific antibody topology. Protein Eng Des Sel 2010; 23:221-8; PMID:20019028; http://dx.doi.org/10.1093/protein/gzp077
- Dong J, Sereno A, Snyder WB, Miller BR, Tamraz S, Doern A, Favis M, Wu X, Tran H, Langley E, et al. Stable IgG-like bispecific antibodies directed toward the type I insulin-like growth factor receptor demonstrate enhanced ligand blockade and anti-tumor activity. J Biol Chem 2011; 286:4703-17; PMID:21123183; http://dx.doi.org/10.1074/jbc.M110.184317
- Schoonjans R, Willems A, Schoonooghe S, Leoen J, Grooten J, Mertens N. A new model for intermediate molecular weight recombinant bispecific and trispecific antibodies by efficient heterodimerization of single chain variable domains through fusion to a Fab-chain. Biomol Eng 2001; 17:193-202; PMID:11337278; http://dx.doi.org/10.1016/S1389-0344(01)00066-1
- Ahmad ZA, Yeap SK, Ali AM, Ho WY, Alitheen NBM, Hamid M. scFv antibody: principles and clinical application. Clin Dev Immunol 2012; 2012:980250; PMID:22474489; http://dx.doi.org/10.1155/2012/980250
- Röthlisberger D, Honegger A, Plückthun A. Domain interactions in the Fab fragment: a comparative evaluation of the single-chain Fv and Fab format engineered with variable domains of different stability. J Mol Biol 2005; 347:773-89; PMID:15769469; http://dx.doi.org/10.1016/j.jmb.2005.01.053
- Digiammarino E, Ghayur T, Liu J. Therapeutic proteins. Methods Mol Biol 2012; 899:145-56; PMID:22735951; http://dx.doi.org/10.1007/978-1-61779-921-1_9
- Miller K, Meng G, Liu J, Hurst A, Hsei V, Wong W-L, Ekert R, Lawrence D, Sherwood S, DeForge L, et al. Design, construction, and in vitro analyses of multivalent antibodies. J Immunol 2003; 170:4854-61; PMID:12728922; http://dx.doi.org/10.4049/jimmunol.170.9.4854
- Goldenberg DM, Rossi EA, Sharkey RM, McBride WJ, Chang C-H. Multifunctional antibodies by the Dock-and-Lock method for improved cancer imaging and therapy by pretargeting. J Nucl Med 2008; 49:158-63; PMID:18077530; http://dx.doi.org/10.2967/jnumed.107.046185
- Christensen EH, Eaton DL, Vendel AC, Wranik B. Coiled coil and/or tether containing protein complexes and uses thereof. WO2011034605. 2011
- Schaefer G, Haber L, Crocker LM, Shia S, Shao L, Dowbenko D, Totpal K, Wong A, Lee CV, Stawicki S, et al. A two-in-one antibody against HER3 and EGFR has superior inhibitory activity compared with monospecific antibodies. Cancer Cell 2011; 20:472-86; PMID:22014573; http://dx.doi.org/10.1016/j.ccr.2011.09.003
- Wozniak-Knopp G, Bartl S, Bauer A, Mostageer M, Woisetschläger M, Antes B, Ettl K, Kainer M, Weberhofer G, Wiederkum S, et al. Introducing antigen-binding sites in structural loops of immunoglobulin constant domains: Fc fragments with engineered HER2/neu-binding sites and antibody properties. Protein Eng Des Sel 2010; 23:289-97; PMID:20150180; http://dx.doi.org/10.1093/protein/gzq005
- Holliger P, Prospero T, Winter G. “Diabodies”: small bivalent and bispecific antibody fragments. Proc Natl Acad Sci USA 1993; 90:6444-8; PMID:8341653
- FitzGerald K, Holliger P, Winter G. Improved tumour targeting by disulphide stabilized diabodies expressed in Pichia pastoris. Protein Eng 1997; 10:1221-5; PMID:9488147; http://dx.doi.org/10.1093/protein/10.10.1221
- Johnson S, Burke S, Huang L, Gorlatov S, Li H, Wang W, Zhang W, Tuaillon N, Rainey J, Barat B, et al. Effector cell recruitment with novel Fv-based dual-affinity re-targeting protein leads to potent tumor cytolysis and in vivo B-cell depletion. J Mol Biol 2010; 399:436-49; PMID:20382161; http://dx.doi.org/10.1016/j.jmb.2010.04.001
- Sabine Brüsselbach TK. Enzyme recruitment and tumor cell killing in vitro by a secreted bispecific single-chain diabody. Tumor Target 1999; 115-23.
- Kipriyanov SM, Moldenhauer G, Schuhmacher J, Cochlovius B, Von der Lieth CW, Matys ER, Little M. Bispecific tandem diabody for tumor therapy with improved antigen binding and pharmacokinetics. J Mol Biol 1999; 293:41-56; PMID:10512714; http://dx.doi.org/10.1006/jmbi.1999.3156
- Frankel SR, Baeuerle P A. Targeting T cells to tumor cells using bispecific antibodies. Curr Opin Chem Biol 2013; 17:385-92; PMID:23623807; http://dx.doi.org/10.1016/j.cbpa.2013.03.029
- Olafsen T, Elgqvist J, Wu AM. Protein targeting constructs in alpha therapy. Curr Radiopharm 2011; 4:197-213; PMID:22201709; http://dx.doi.org/10.2174/1874471011104030197
- Hu S, Shively L, Raubitschek A, Shively JE, Wu AM, Sherman M, Williams LE, Wong JYC. Minibody : a novel engineered anti-carcinoembryonic antigen antibody fragment (Single-Chain Fv-CH3) which exhibits rapid , high-level targeting of xenografts minibody : a novel engineered anti-carcinoembryonic high-level targeting of xenografts1. 1996; 56:3055-61; PMID:8674062
- Kontermann RE. Strategies to extend plasma half-lives of recombinant antibodies. BioDrugs 2009; 23:93-109; PMID:19489651; http://dx.doi.org/10.2165/00063030-200923020-00003
- Merchant AM, Zhu Z, Yuan JQ, Goddard A, Adams CW, Presta LG, Carter P. An efficient route to human bispecific IgG. Nat Biotechnol 1998; 16:677-81; PMID:9661204
- Carter P. Bispecific human IgG by design. J Immunol Methods 2001; 248:7-15; PMID:11223065
- Lewis SM, Wu X, Pustilnik A, Sereno A, Huang F, Rick HL, Guntas G, Leaver-Fay A, Smith EM, Ho C, et al. Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface. Nat Biotechnol 2014; 32:191-8; PMID:24463572; http://dx.doi.org/10.1038/nbt.2797
- Schaefer W, Regula JT, Bähner M, Schanzer J, Croasdale R, Dürr H, Gassner C, Georges G, Kettenberger H, Imhof-Jung S, et al. Immunoglobulin domain crossover as a generic approach for the production of bispecific IgG antibodies. Proc Natl Acad Sci U S A 2011; 108:11187-92; PMID:21690412; http://dx.doi.org/10.1073/pnas.1019002108
- Atwell S, Ridgway JB, Wells JA, Carter P. Stable heterodimers from remodeling the domain interface of a homodimer using a phage display library. J Mol Biol 1997; 270:26-35; PMID:9231898; http://dx.doi.org/10.1006/jmbi.1997.1116
- Labrijn AF, Meesters JI, de Goeij BECG, van den Bremer ETJ, Neijssen J, van Kampen MD, Strumane K, Verploegen S, Kundu A, Gramer MJ, et al. Efficient generation of stable bispecific IgG1 by controlled Fab-arm exchange. Proc Natl Acad Sci U S A 2013; 110:5145-50; PMID:23479652; http://dx.doi.org/10.1073/pnas.1220145110
- Moore GL, Bautista C, Pong E, Nguyen D-HT, Jacinto J, Eivazi A, Muchhal US, Karki S, Chu SY, Lazar GA. A novel bispecific antibody format enables simultaneous bivalent and monovalent co-engagement of distinct target antigens. MAbs 2011; 3:546-57; PMID:22123055; http://dx.doi.org/10.4161/mabs.3.6.18123
- Strop P, Ho W-H, Boustany LM, Abdiche YN, Lindquist KC, Farias SE, Rickert M, Appah CT, Pascua E, Radcliffe T, et al. Generating bispecific human IgG1 and IgG2 antibodies from any antibody pair. J Mol Biol 2012; 420:204-19; PMID:22543237; http://dx.doi.org/10.1016/j.jmb.2012.04.020
- Gunasekaran K, Pentony M, Shen M, Garrett L, Forte C, Woodward A, Ng SB, Born T, Retter M, Manchulenko K, et al. Enhancing antibody Fc heterodimer formation through electrostatic steering effects: applications to bispecific molecules and monovalent IgG. J Biol Chem 2010; 285:19637-46; PMID:20400508; http://dx.doi.org/10.1074/jbc.M110.117382
- Ridgway JB, Presta LG, Carter P. “Knobs-into-holes” engineering of antibody CH3 domains for heavy chain heterodimerization. Protein Eng 1996; 9:617-21; PMID:8844834
- Davis JH, Aperlo C, Li Y, Kurosawa E, Lan Y, Lo K-M, Huston JS. SEEDbodies: fusion proteins based on strand-exchange engineered domain (SEED) CH3 heterodimers in an Fc analogue platform for asymmetric binders or immunofusions and bispecific antibodies. Protein Eng Des Sel 2010; 23:195-202; PMID:20299542; http://dx.doi.org/10.1093/protein/gzp094
- Davis S, Smith E, MacDonald D, Olson KL. Readily isolated bispecific antibodies with native immunoglobulin format. WO2010151792. 2010
- Junttila TT, Li J, Johnston J, Hristopoulos M, Clark R, Ellerman D, Wang B-E, Li Y, Mathieu M, Li G, et al. Antitumor Efficacy of a Bispecific Antibody That Targets HER2 and Activates T Cells. Cancer Res 2014; 74(19): 5561-71; PMID:25228655; http://dx.doi.org/10.1158/0008-5472.CAN-13-3622-T
- Jackman J, Chen Y, Huang A, Moffat B, Scheer JM, Leong SR, Lee WP, Zhang J, Sharma N, Lu Y, et al. Development of a two-part strategy to identify a therapeutic human bispecific antibody that inhibits IgE receptor signaling. J Biol Chem 2010; 285:20850-9; PMID:20444694; http://dx.doi.org/10.1074/jbc.M110.113910
- Yin G, Garces ED, Yang J, Zhang J, Tran C, Steiner AR, Roos C, Bajad S, Hudak S, Penta K, et al. Aglycosylated antibodies and antibody fragments produced in a scalable in vitro transcription-translation system. MAbs 2012; 4(2):217-25; PMID:22377750; http://dx.doi.org/10.4161/mabs.4.2.19202
- Zawada JF, Yin G, Steiner AR, Yang J, Naresh A, Roy SM, Gold DS, Heinsohn HG, Murray CJ. Microscale to manufacturing scale-up of cell-free cytokine production–a new approach for shortening protein production development timelines. Biotechnol Bioeng 2011; 108:1570-8; PMID:21337337; http://dx.doi.org/10.1002/bit.23103
- Groff D, Armstrong S, Rivers PJ, Zhang J, Yang J, Green E, Rozzelle J, Liang S, Kittle JD, Steiner AR, et al. Engineering toward a bacterial “endoplasmic reticulum” for the rapid expression of immunoglobulin proteins. MAbs 2014; 6:671-8; PMID:24517929
- Zimmerman ES, Heibeck TH, Gill A, Li X, Murray CJ, Madlansacay MR, Tran C, Uter NT, Yin G, Rivers PJ, et al. Production of site-specific antibody-drug conjugates using optimized non-natural amino acids in a cell-free expression system. Bioconjug Chem 2014; 25:351-61; PMID:24437342; http://dx.doi.org/10.1021/bc400490z
- Murray CJ, Baliga R. Cell-free translation of peptides and proteins: from high throughput screening to clinical production. Curr Opin Chem Biol 2013; 17:420-6; PMID:23499386; http://dx.doi.org/10.1016/j.cbpa.2013.02.014
- Lu Y, Welsh JP, Swartz JR. Production and stabilization of the trimeric influenza hemagglutinin stem domain for potentially broadly protective influenza vaccines. Proc Natl Acad Sci USA 2014; 111:125-30; PMID:24344259; http://dx.doi.org/10.1073/pnas.1308701110
- Wuu JJ, Swartz JR. High yield cell-free production of integral membrane proteins without refolding or detergents. Biochim Biophys Acta 2008; 1778:1237-50; PMID:18295592; http://dx.doi.org/10.1016/j.bbamem.2008.01.023
- Kuchenreuther JM, Shiigi SA, Swartz JR. Cell-free synthesis of the H-cluster: a model for the in vitro assembly of metalloprotein metal centers. Methods Mol Biol 2014; 1122:49-72; PMID:24639253; http://dx.doi.org/10.1007/978-1-62703-794-5_5
- Bundy BC, Swartz JR. Efficient disulfide bond formation in virus-like particles. J Biotechnol 2011; 154:230-9; PMID:21536082; http://dx.doi.org/10.1016/j.jbiotec.2011.04.011
- Yin G, Swartz JR. Enhancing multiple disulfide bonded protein folding in a cell-free system. Biotechnol Bioeng 2004; 86:188-95; PMID:15052638; http://dx.doi.org/10.1002/bit.10827
- Cioffi M, Dorado J, Baeuerle PA, Heeschen C. EpCAM/CD3-Bispecific T-cell engaging antibody MT110 eliminates primary human pancreatic cancer stem cells. Clin Cancer Res 2012; 18:465-74; PMID:22096026; http://dx.doi.org/10.1158/1078-0432.CCR-11-1270
- Lowman HB, Gerstner RB, Carter PJ. Method of treating breast cancer using anti-HER2 antibody variants. US7850966. 2010
- Elliott JM, Ultsch M, Lee J, Tong R, Takeda K, Spiess C, Eigenbrot C, Scheer JM. Antiparallel Conformation of Knob and Hole Aglycosylated Half-Antibody Homodimers Is Mediated by a CH2-CH3 Hydrophobic Interaction. J Mol Biol 2014; 426(9): 1947-57; PMID:24576605; http://dx.doi.org/10.1016/j.jmb.2014.02.015
- Cheng M, Ahmed M, Xu H, Cheung N-KV. Structural design of disialoganglioside GD2 and CD3-bispecific antibodies to redirect T cells for tumor therapy. Int J Cancer 2015; 136(2): 476-86; PMID:24895182
- Kim Y-J, Neelamegam R, Heo M-A, Edwardraja S, Paik H-J, Lee S-G. Improving the productivity of single-chain Fv antibody against c-Met by rearranging the order of its variable domains. J Microbiol Biotechnol 2008; 18:1186-90; PMID:18600066
- Asano R, Kumagai T, Nagai K, Taki S, Shimomura I, Arai K, Ogata H, Okada M, Hayasaka F, Sanada H, et al. Domain order of a bispecific diabody dramatically enhances its antitumor activity beyond structural format conversion: the case of the hEx3 diabody. Protein Eng Des Sel 2013; 26:359-67; PMID:23468569; http://dx.doi.org/10.1093/protein/gzt009
- Kipriyanov SM, Moldenhauer G, Braunagel M, Reusch U, Cochlovius B, Le Gall F, Kouprianova OA, Von der Lieth CW, Little M. Effect of domain order on the activity of bacterially produced bispecific single-chain Fv antibodies. J Mol Biol 2003; 330:99-111; PMID:12818205; http://dx.doi.org/10.1016/S0022-2836(03)00526-6
- Le Gall F, Reusch U, Little M, Kipriyanov SM. Effect of linker sequences between the antibody variable domains on the formation, stability and biological activity of a bispecific tandem diabody. Protein Eng Des Sel 2004; 17:357-66; PMID:15126676; http://dx.doi.org/10.1093/protein/gzh039
- Whitlow M, Bell BA, Feng SL, Filpula D, Hardman KD, Hubert SL, Rollence ML, Wood JF, Schott ME, Milenic DE. An improved linker for single-chain Fv with reduced aggregation and enhanced proteolytic stability. Protein Eng 1993; 6:989-95; PMID:8309948; http://dx.doi.org/10.1093/protein/6.8.989
- Jefferis R. Glycosylation as a strategy to improve antibody-based therapeutics. Nat Rev Drug Discov 2009; 8:226-34; PMID:19247305; http://dx.doi.org/10.1038/nrd2804
- Ha S, Ou Y, Vlasak J, Li Y, Wang S, Vo K, Du Y, Mach A, Fang Y, Zhang N. Isolation and characterization of IgG1 with asymmetrical Fc glycosylation. Glycobiology 2011; 21:1087-96; PMID:21470983; http://dx.doi.org/10.1093/glycob/cwr047
- Sazinsky SL, Ott RG, Silver NW, Tidor B, Ravetch JV, Wittrup KD. Aglycosylated immunoglobulin G1 variants productively engage activating Fc receptors. Proc Natl Acad Sci USA 2008; 105:20167-72
- Jung ST, Reddy ST, Kang TH, Borrok MJ, Sandlie I, Tucker PW, Georgiou G. Aglycosylated IgG variants expressed in bacteria that selectively bind FcgammaRI potentiate tumor cell killing by monocyte-dendritic cells. Proc Natl Acad Sci USA 2010; 107:604-9
- Guarino C, DeLisa MP. A prokaryote-based cell-free translation system that efficiently synthesizes glycoproteins. Glycobiology 2012; 22:596-601; PMID:22068020; http://dx.doi.org/10.1093/glycob/cwr151
- Gamblin DP, Scanlan EM, Davis BG. Glycoprotein synthesis: an update. Chem Rev 2009; 109:131-63; PMID:19093879; http://dx.doi.org/10.1021/cr078291i
- Wang L-X, Lomino JV. Emerging technologies for making glycan-defined glycoproteins. ACS Chem Biol 2012; 7:110-22; PMID:22141574; http://dx.doi.org/10.1021/cb200429n
- Chalker JM, Bernardes ALOJL, Davis BG. A “tag-and-modify” approach to site-selective protein modification. Acc Chem Res 2011; 44:730-41; PMID:21563755
