Abstract
In addition to antibodies with the classical composition of heavy and light chains, the adaptive immune repertoire of sharks also includes a heavy-chain only isotype, where antigen binding is mediated exclusively by a small and highly stable domain, referred to as vNAR. In recent years, due to their high affinity and specificity combined with their small size, high physicochemical stability and low-cost of production, vNAR fragments have evolved as promising target-binding scaffolds that can be tailor-made for applications in medicine and biotechnology. This review highlights the structural features of vNAR molecules, addresses aspects of their generation using immunization or in vitro high throughput screening methods and provides examples of therapeutic, diagnostic and other biotechnological applications.
Abbreviations
| CDR | = | complementarity-determining region |
| HV | = | hypervariable region |
| IgNAR | = | immunoglobulin new antigen receptor |
| IgNAR V domain | = | variable domain of IgNAR |
| mAbs | = | monoclonal antibodies |
| scFv | = | single chain variable fragment |
| VL | = | variable domain of the light chain |
| VH | = | variable domain of the heavy chain |
| VHH | = | variable domain of camelid heavy chain antibodies |
| vNAR | = | variable domain of IgNAR |
Introduction
Today, biological entities are one of the main drivers of the pharmaceutical industry as exemplified by their current and predicted market growth rates, which substantially exceed those of the overall sector. Within this group of biologic drugs, monoclonal antibodies (mAbs) are the highest selling class, followed by hormones, growth factors and then fusion proteins.Citation1,Citation2 The high specificity for a cognate antigen combined with Fc-mediated immune effector functions have underpinned the success of antibodies as effective tools for medical applications. With ∼40 antibodies marketed and hundreds of mAbs in clinical development, the therapeutic and economic value of mAbs is evident.Citation1,Citation3-Citation5 Antibodies are structurally complex, large hetero-tetrameric proteins that consist of 2 heavy chains and 2 light chains (). The 2 identical antigen-binding sites, i.e., paratopes, are composed of one variable domain of the light chain and one variable domain of the heavy chain, respectively. However, under certain circumstances the therapeutic and diagnostic efficacy of antibodies might be limited due to inherent attributes, e.g., structural complexity, large size. The paratope of conventional antibodies can be restricted in its ability to access certain epitopes, e.g.,, recessed cryptic epitopes, active sites of enzymes.Citation6-Citation8 Furthermore, the mobility, i.e., tissue penetration, of classical antibody molecules is constrained by their large size.Citation9 For in vivo tumor imaging purposes, the slow blood clearance of conventional antibodies poses a problem due to their extended plasma half-life.Citation10 Slow tumor penetration as well as nonspecific uptake by healthy tissues may represent further drawbacks of conventional antibodies in molecular imaging.Citation11-Citation14 To address these issues and to increase the overall therapeutic efficacy, next-generation-antibodies, antibody fragments and non-immunoglobulin based protein scaffolds have been engineered and developed, as extensively described elsewhere.Citation15-Citation24
Figure 1. Structural features of intact IgG (A), camelid hclgG (B), and shark IgNAR (C) antibody formats shown as surface representation (top) as well as ribbon and schematic representations (bottom). Individual domains are colored as indicated in the schematic representation; hinge regions are colored yellow; glycans not shown. (A) IgG model is based on pdb entry 1IGT.Citation95 (B) Model of hclgG was generated by molecular replacement based on pdb entries 1IGT (for Fc region) and 1IEH (for VHH domain).Citation95,Citation96 1IEH was aligned to the CH1 domain of 1IGT with YASARA structure.Citation97 After deletion of absent domains (VL, CL, VH, and CH1), the VHH section was connected to the Fc region via a short camelid hinge sequence (see Hamers-Casterman et al.).Citation25 Then, a 2-step energy minimization using YASARA2 force field was conducted to yield the depicted structure. (C) Coordinates of intact IgNAR including the hypothetical structure of IgNAR C5 domain were generously provided by Prof. Dr. Michael Sattler and Dr. Janosch Hennig (see Feige et al.).Citation39 Picture rendered with POV-Ray (www.povray.org/).
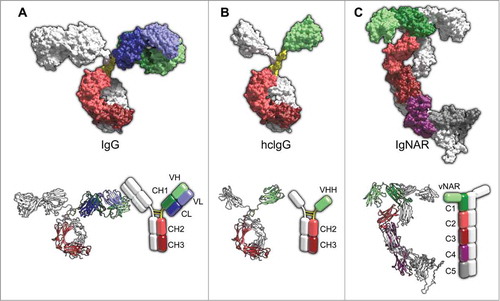
Camelids and the cartilaginous fish possess natural antibodies composed only of heavy chains ().Citation25,Citation26 The antigen binding site is formed by only one single domain, referred to as VHH and vNAR, respectively. Due to an increased frequency for polar and charged amino acids at the solvent-exposed regions corresponding to the hydrophobic VH-VL interface of conventional antibodies, vNAR and VHH domains are highly soluble.Citation27 Antigen‑binding domains of heavy chain only antibodies (HCAbs) combine most of the beneficial features of non-immunoglobulin-based protein scaffolds, e.g., small size, high stability, coupled with the advantageous characteristics of classical antibody molecules, most strikingly the feasibility to generate highly specific and high-affinity binders through immunization.Citation6,Citation28
Even more interestingly, HCAbs naturally complement the conventional repertoire of the aforementioned species. Whereas classical antibodies usually have planar or concave antigen binding sites, vNAR- and VHH-domains possess a wide variety of additional (in the case of vNARs) and different loop structures. This leads to a drastically expanded repertoire of available paratopes capable of accessing and binding to more cryptic epitopes and catalytic clefts of enzymes that are intractable to classical antibodies.Citation29-Citation31 Hence, vNAR and VHH domains could add considerable value to the therapeutic development pipeline by broadening the range of druggable targets.
While camelid VHH domains have proven to be successful in early phase clinical trials,Citation32 the engineering of vNAR domains for biomedical applications is at an earlier stage. However, significant progress demonstrating the therapeutic utility of these domains has been made in the last years. The purpose of this review is to summarize the research conducted to date of IgNARs, placing the emphasis on recent progress of the development of vNAR domains for therapeutic, diagnostic and other biotechnological applications.
The New Antigen Receptor
The cartilaginous fish (sharks, rays, skates and chimaeras) express 3 different isotypes of antibodies, IgM, IgNAR and IgW.Citation33,Citation34 Recent analysis of the genome of African coelacanth, Latimeria chalumnae indicated that IgM was absent in this species, while IgW was present, supporting the notion that IgW is a primordial immunoglobulin class.Citation35,Citation36 IgNAR was first identified in the serum of the nurse shark (Ginglymostoma cirratum) in 1995 by Flajnik and co-workers.Citation26 It is a homodimer of heavy chains devoid of light chains. Each chain of the secretory form consists of one variable domain followed by 5 constant domains, the last 4 being homologous to IgW constant domains.Citation37 Serum IgNAR levels range from ∼0.1 mg/ml to 1 mg/ml.Citation38
Based on atomic resolution structural data as well as small-angle X-ray scattering, Buchner and coworkers were able to develop a structural model of the complete IgNAR molecule ().Citation39 Within the molecule, domains C1 and C3 of each chain cause dimerization of IgNAR. Despite the lack of a canonical hinge region, the variable domains are spaced sufficiently wide for binding multiple epitopes, facilitated by the wide angle of the C1 dimerization interface. A small angle between both C3 domains induces the formation of a narrow stalk for the IgNAR molecule. However, the flexibility of the stalk is induced by a disulfide-bridged linker that connects domains C3 and C4. The heavy chain-only molecule is kinked approximately in the middle of the molecule, at the location of the flexible linker, causing its characteristic shape. Whether any effector functions are mediated by the constant region of IgNAR is currently unresolved.Citation6 Finally, it is important to note that the structure of C5 shown in is completely hypothetical. This is due to a lack of structural data on this domain, which does not exhibit a fold, neither as isolated recombinant protein nor within a C4-C5 construct.Citation39
Absence of a Light Chain Partner
The homodimer IgNAR displays several unique features that are responsible for the inhibition of a potential light chain pairing. At the typical VH-VL interaction site, there is poor conservation of residues that mediate this association in mammals.Citation40 Instead these typically hydrophobic amino acids are frequently replaced by polar or charged residues.Citation27 For classical antibodies, a special mechanism ensures the formation of heavy- and light-chain pairing. In the endoplasmic reticulum, the heavy chain is trapped by an Ig-binding protein (BiP) via interaction with the CH1 domain. For the release, a light chain must displace BiP, and, consequently, only heavy- and light-chain paired antibodies are secreted.Citation27,Citation41 Flajnik and coworkers hypothesized that during evolution, a vNAR-D-J cluster recombined with an IgW cluster in a way that the IgW cluster lost its V-D-J segments and the first C exon.Citation42 Indeed, the C1 domain of IgNAR is somewhat similar to the CH2 domain of IgW and may be derived from this domain.Citation43 BiP- and L-chain-interactions sites are consistently missing in the C1 domain of IgNAR, as elegantly reviewed by Flajnik and colleagues.Citation27
The Variable Domain of IgNAR – Structural Features
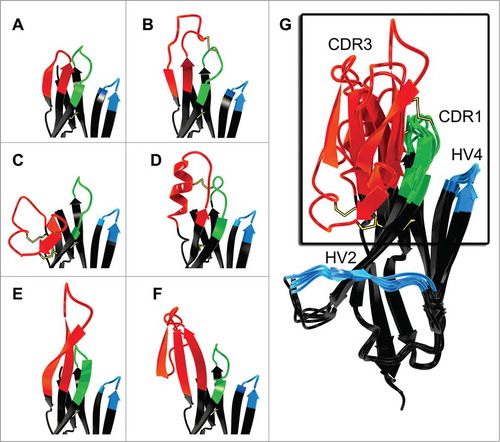
The variable domain of the New Antigen Receptor shows homology to the T-cell receptor (TCR) Vα and also is found as a variable domain in the NAR-TCRδ.Citation44 It also displays sequence homology to immunoglobulin Vκ domains, whereas structurally it is related to Vα, Vλ, and VH domains.Citation30 The evolutionary relationship of IgNAR and TCR and their therapeutic potential was recently reviewed.Citation45 Moreover, since vNAR domains share structural features of cell adhesion molecules, it was suggested that IgNAR evolved from a cell-surface receptor, clearly distinguishing it from VHH, which evidently arose from an IgG lineage.Citation27,Citation46 vNAR belongs to the Ig superfamily, and accordingly it has a β-sandwich fold. However, compared to mammalian V domains, this fold only consists of 8 instead of 10 β-strands due to the deletion in the framework2-CDR2-region ().
Figure 2. Comparison of VH (left; from pdb entry 1IGT) and vNAR (right, from pdb entry 2COQ) binding domains depicted as ribbon representation as well as an overlay of both structures (middle).Citation31,Citation95 CDR1 and CDR3 are shown in gray. Two β strands and CDR2 of the VH domain are highlighted in orange. These structural elements are absent in the vNAR domain which possesses HV2 and HV4 (both highlighted in blue), instead. Disulfide bonds are shown as yellow sticks. Picture rendered with POV-Ray (www.povray.org/).
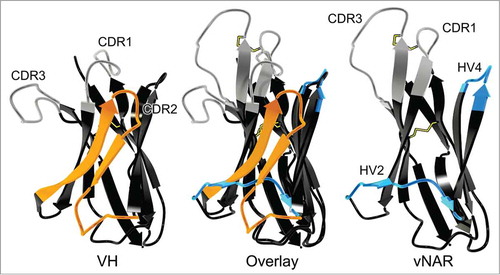
With a molecular mass of ∼12 kDa, the vNAR domain is the smallest antibody-like antigen binding domain in the animal kingdom known to date.Citation6,Citation30 As a consequence, contrary to mammalian variable domains, vNAR domains have only 2 complementarity determining regions CDR1 and CDR3 (). The diversity of the primary vNAR repertoire is predominantly found in CDR3. High rates of somatic mutation after antigen contact are observed in CDR1, at the CDR2 truncation site, where the remaining loop forms a belt-like structure at the bottom of the molecule, and in a loop that corresponds to HV4 in TCRs. Accordingly, these mutation-prone regions have been named HV2 and HV4, respectively ().Citation47 Indeed, it was shown that somatic mutations within HV4 can contribute to antigen binding.Citation48
Despite having a reduced number of possible antigen binding loops (4 across a single chain) compared to conventional antibodies (6 loops across 2 chains), vNAR domains bind antigens with surprisingly high affinities.Citation49,Citation50 Even from primary repertoires, where antigen binding is solely mediated by CDR3, vNAR molecules can be obtained against a given antigen with affinities in the low nanomolar range.Citation48,Citation49 The highest recorded affinities for vNAR domains, however, have been observed after immunization with an anti-albumin binding domain, achieving picomolar levels of affinity.Citation50
Based on the number of non-canonical cysteine residues, which are not found in classical variable domains, vNAR molecules have been categorized into 4 types ().Citation30,Citation31,Citation48,Citation51,Citation52 The classical Ig canonical cysteines, which stabilize the immunoglobulin fold via a disulfide-bond, are common to all types. Type I variable domains carry extra cysteines in framework regions 2 and 4, and, consequently, an even number of partner cysteine residues in CDR3. The determination of the crystal structure of a type I vNAR in complex with lysozyme revealed that both non-canonical framework cysteines each form disulfide-bonds with those of CDR3, causing this loop to be held tightly into the direction of HV2.Citation30 Thus far, type I variable domains of IgNAR have only been identified in the nurse shark, Ginglymostoma cirratum.Citation6
Figure 3. Different types of IgNAR V domains. Variable domains are categorized based on the presence or the absence of non-canonical cysteine residues (black dots). Canonical cysteine residues (white dots) and disulfide bonds (connecting lines), conserved tryptophan (W) as well as complementarity determining regions (CDR) and hypervariable loops (HV) are shown in their relative positions. Ribbon presentations of vNAR domains are depictions of pdb entries 1SQ22 (type I),Citation30 2COQ (type II),Citation31 and 4HGK (type IV)Citation52 as well as a modeled type III structure based on 2COQ. The latter was generated via homology modeling using YASARA structure.Citation97 First, vNAR residues of 2COQ were changed to match a reported type III sequence (AAM76948 from Streltsov et al.).Citation31 Then, side chain geometries were optimized followed by a 2-step energy minimization using the YASARA2 force field.Citation97
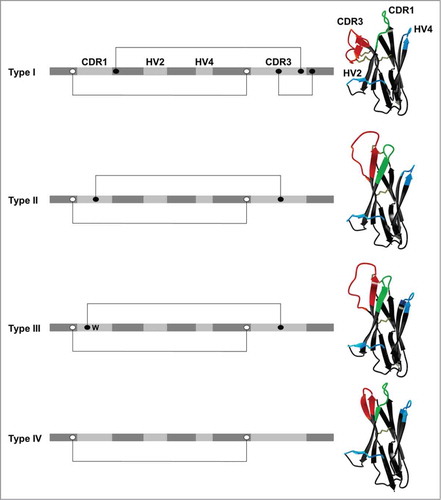
Type II domains differ from type I by means of an additional cysteine in CDR1 and in CDR3, respectively, resulting in an intra-molecular disulfide bond that brings both loops in close vicinity. However, it lacks both cysteine motifs that anchor CDR3 to the framework in type I vNAR. As a consequence, the CDR3 region forms a protrusive ‘finger-like’ structure that is predisposed to binding into pockets or grooves, e.g., the active site of enzymes.Citation31 According to this, it has been shown for both types that active site clefts can be penetrated by the antigen binding loops.Citation30,Citation48
Another type, termed type III, is expressed in neonates at high frequencies.Citation31,Citation51 Akin to type II domains, this isotype is characterized by an additional non-canonical cysteine in CDR1 and CDR3, respectively. However, in contrast to type II, type III domains comprise a restricted CDR3 diversity, highly similar in amino acid composition and length as well as a conserved tryptophan residue in CDR1 positioned adjacent to the disulfide bridge between both loops. Based on the limited CDR3 diversity it is tempting to speculate that type III vNARs evolved as a consequence of exposure to a common pathogen in early development of sharks or that it may play a role in regulatory processes during the development of the shark's immune system.Citation6,Citation31,Citation51
Type IV domains differ from all described vNAR types in that they lack non-canonical disulfide bonds as found in all other vNAR types.Citation50,Citation52 Therefore, the topology of the paratope of type IV variable domains is more flexible and not physically constrained. Type IV domains are also referred to as type IIb, according to Streltsov et al. and Liu et al.Citation46,Citation53 In addition, type IV domains with an invariant tryptophan residue in CDR1, similar to type III, have been identified.Citation54 Besides type III, all types of the vNAR domain give rise to high-affinity binders.Citation30,Citation48,Citation50,Citation55
Diversification of the IgNAR Repertoire
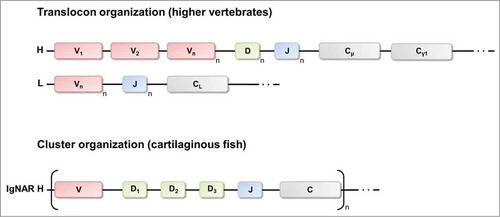
In mammals, antibody diversity is generated by a process referred to as V(D)J-recombination. During B-cell development, one variable (V) segment, one diversity (D) segment and one joining (J) segment are randomly rearranged from a multitude of gene segments of the immunoglobulin heavy chain gene cluster to encode the VH domain, which is fused to a gene segment encoding a constant domain.Citation27 Similarly, for light chain generation, one V segment and one J segment are selected by chance from a pool of gene segments to produce the variable domain of the light chain that is fused to a CL gene. Diversity is further expanded in a process called junctional diversification through imprecise segment joining.Citation56 An additional layer of diversity is introduced by the random arrangement of the heavy chain and light chain to complete the expression of the antibody molecule.
As IgNARs are devoid of light chains, they lack H-L combinatorial diversification. Correspondingly, one would expect a dramatically restricted primary repertoire. However, this lack of diversification process is at least partially compensated through the diversity achieved within the CDR3 region. Whereas mammalian antibody genes are organized in the translocon-format, shark antibody genes are exclusively arranged in the cluster-organization ().Citation6,Citation30,Citation42,Citation48 Nurse sharks comprise 4 IgNAR gene clusters, though only 2 are expressed in adult life, one encodes type I and one encodes type II IgNARs.Citation30,Citation48 Each IgNAR cluster comprises one V segment, 3 D segments and one J segment and a single set of C segments. Rearrangement occurs solely within this cluster resulting in a VD1D2D3J assembly. Hence, 4 rearrangement processes generate the complete vNAR domain. However, the order of rearrangements remains to be determined.Citation42 The interfaces between the V segment, the 3 D segments and the J segment encode for CDR3. Consequently, diversity in both sequence and length of the primary repertoire is nearly entirely found in CDR3.Citation51,Citation57,Citation58 Extensive junctional diversification through N-region addition, P-nucleotide addition, trimming and D-region rearrangement further expands the heterogeneity of the primary repertoire of IgNAR.Citation6,Citation30,Citation42.Citation59 The type III gene cluster represents an exception within the recombination process. As a result of the fusion of the D1 segment and the D2 segment in the nurse shark, only 3 rearrangement events occur, explaining the restricted diversity of this type. In contrast to this, in the spiny dogfish the type III IgNAR clusters are not partially germline-joined, indicating that germline-joining of Ig clusters might be a species-specific event.Citation60
Figure 4. Translocon arrangements of immunoglobulin genes in higher vertebrates and cluster configuration of IgNAR genes of cartilaginous fish. In the translocon organization there are many variable (V) segments upstream of many diversity (D, only for heavy chains) and joining (J) segments that recombine randomly to encode the variable domain of the heavy chain or the light chain. IgNAR genes (like all Ig genes of the cartilaginous fish) are organized in the cluster configuration. Each cluster contains for the variable domain one V segment, 3 D segments and one J segment. Recombination occurs exclusively within one cluster. H, heavy chain loci; L, light chain loci, C, constant region.
Sharks do not possess conventional germinal centers. Nevertheless, the initial combinatorial diversity, which is mainly restricted to CDR3, is further expanded by extensive somatic hypermutation in an antigen-driven manner, with mutations clustering to the CDRs.Citation57 The mutational pattern and frequencies of this process are similar to that of mammalian immunoglobulins with a bias for transitions over transversions. The mechanism favors the serine codon AGC/T as a hotspot for mutations and most of the changes are base substitutions. Surprisingly, base changes often occur in tandem, particularly in mutational hotspots and palindromic repeats.Citation58 It was first shown by Flajnik and coworkers, that after immunization, somatic mutations promoted an incremental increase in affinity, giving clear evidence for in vivo affinity maturation in sharks.Citation30,Citation38,Citation47-Citation49 Furthermore, they demonstrated that HV4 is prone to somatic mutations and, even more interesting, these mutations can be involved in antigen binding.Citation48
Consistent with the structure of type I and type II IgNARs (), mutations are favored in CDR1 for type II vNARs and in HV2 for type I vNARs.Citation27,Citation30 It is hypothesized that those mutations could either directly contribute to antigen binding or they could indirectly have an effect on the paratope such that they stabilize and influence the conformation of the antigen-binding CDR3.
Selection of Antigen-Specific vNAR Domains From Shark Immune Repertoires
Antigen-specific vNAR domains have been generated from the immune repertoire of a number of different shark species, including the nurse shark,Citation49 the wobbegong shark,Citation55,Citation61 the spiny dogfish,Citation50,Citation53,Citation62 the banded houndsharkCitation63,Citation64 and the bamboo shark.Citation65 Target-specific clones are generally isolated using different display technologies, such as phage displayCitation49,Citation55 or ribosome display.Citation66 We recently showed that antigen-specific vNAR molecules can also be selected using yeast surface display as platform technology.Citation65 A potential advantage of yeast surface display over the above mentioned display technologies is the amenability of single-cell online and real-time analysis, as well as subsequent characterization of individual library candidates in terms of specificity, affinity and stability.Citation67,Citation68
There are several distinct strategies for library establishment. Binders can be selected from immunized sharks,Citation49,Citation50 from the naïve shark repertoire,Citation53 or from a synthetic vNAR library,Citation55,Citation69 where the vNAR molecule serves as a scaffold with randomized loops and from semi-synthetic repertoires. Here, additional diversity is included through the randomization of one or more antigen-binding loops.Citation62,Citation70 For the most part, immunization is the preferred route to obtain high affinity binders. An additional advantage of immunization is that sharks are evolutionary very distant to humans. This greatly reduces the likelihood of immune tolerance that would reduce the induction of target-specific responses when antigens are well conserved across species. Consequently, antigen-specific vNAR molecules have been generated with impressive affinities against a multitude of different targets via immunization.Citation49,Citation50,Citation71,Citation72 However, the process of immunization of sharks is protracted compared to standard mammalian protocols, and not every species tested has proven successful.Citation50,Citation73 For instance Dooley et al. and also our group (unpublished results) were unable to detect an antigen-specific IgNAR response after the immunization of the small spotted catshark, Scyliorhinus canicula.Citation74
Antigen-specific vNAR fragments have also been isolated from non-immunized libraries against a plethora of different targets, including viral targets, cytokines, proteins involved in cancer and arthritis and toxins.Citation53,Citation62,Citation64,Citation65,Citation70,Citation75,Citation76 illustrates vNARs selected against therapeutically relevant targets from immunized and non-immunized origins (). Surprisingly, binders selected from such libraries often show good affinities to their target.Citation61,Citation62,Citation75,Citation76 Nonetheless, when higher affinities are required, vNARs can be optimized using in vitro affinity maturation. In this respect 3 different methodologies have been established. Nuttall and co-workers were able to improve the affinity for an AMA1-specific IgNAR V domain using error-prone PCR resulting in ∼10-fold enhanced affinity.Citation76 In a more recent approach, the same group employed a mutagenesis system dependent on low fidelity RNA polymerase from Qβ bacteriophage to introduce diversity into IgNAR antibody libraries for affinity maturation. With this novel strategy they were able to select mutated vNAR molecules with a more than 20-fold enhanced affinity compared to the wild type clone.Citation66
Table 1. Published vNAR domains against disease related targets. Adapted from ref. Citation54
We recently established a stepwise in vitro affinity maturation methodology for the generation of high affinity binders derived from shark vNAR domains. To this end, binders were selected from a library in which CDR3 was totally randomized using yeast surface display. Affinities of binders were significantly improved by CDR1 diversification and sublibrary screening, resulting in enhanced molecules with affinities in the low nanomolar range.Citation65 Interestingly, this method resembles the natural strategy of the immune system of nurse sharks to generate high affinity antibodies.
Therapeutic and Diagnostic Attributes of vNAR Domains
The tremendous diversity found at the sequence-level of the CDR3-loop of IgNAR, as well as the multiplicity of the structural topologies formed by the antigen-binding site of the vNAR domain (), render IgNARs promising alternatives to conventional antibodies.Citation30,Citation31,Citation48 As described above, the different types of vNAR domains form, if any, a very diverse set of disulfide-bridges. Consequently, antigen-specific clones can be selected from a very large, unprecedented repertoire of different loop structures.Citation77 Moreover this unique paratope-architecture of shark domains seems to be predisposed to target clefts of the antigen, whereas recessed epitopes are usually not antigenic for conventional antibodies.Citation27,Citation31,Citation46,Citation78 Indeed, it has been shown that the active site of enzymes and clefts can be targeted by vNAR domains.Citation30,Citation39,Citation48
Figure 5. Examples of CDR3 variability in vNAR domains depicted in ribbon representation. (A) Short loop (type IV, pdb entry 4HGK).Citation52 (B) Large loop with one disulfide constraint (type II, pdb entry 2COQ).Citation31 (C) Highly constrained loop tethered by 2 cystine motifs (type I, pdb entry 1SQ2).Citation30 (D) Extended CDR3 forming an α- helical motif (type II, pdb entry 2I25).Citation48 (E) Extended CDR3 forming a 2-stranded β-sheet (type IV, pdb entry 2Z8V).Citation78 (F) Extended CDR3 incorporating an amyloid-β p3 fragment (type IV, pdb entry 3MOQ).Citation94 (G) Overlay of structures A–F. Disulfide bonds are shown in yellow. Picture rendered with POV-Ray (www.povray.org/).
Above all, vNARs exhibit many additional properties that render them interesting for diagnostic and therapeutic applications. It has been demonstrated that vNARs are extraordinarily stable proteins,Citation49,Citation53,Citation62,Citation69,Citation72 which is probably a consequence of the harsh physiological environment - the blood of sharks contains 350 mM urea - those molecules are exposed to.Citation54 The superior thermal stability and tolerance to irreversible thermal denaturation compared to scFv- and mAb-formats was elegantly demonstrated by Lonsdale and colleagues, as well as by Goldman and coworkers.Citation53,Citation79 Even type IV domains, which lack the non-canonical loop stabilizing disulfide-bond, show superior thermal stability, with Tm values that for the most part exceed 70°C.Citation65 In contrast to this, scFv fragments often show Tm values in the range between 50°C and 65°C.Citation80,Citation81 However, Tm may sometimes be below 40°C, requiring optimization of their thermal stability.Citation82,Citation83 Concordantly, it has been shown that the C2 and C4 domains of IgNAR are very stable. Buchner and coworkers were able to identify structural elements that contribute to their high stability. Compared to mammalian constant domains, C2 and C4 domains of IgNAR contain an additional salt bridge and an extended hydrophobic core. The transfer of these key elements of enhanced stability to a human antibody domain improved its stability significantly.Citation39
The inherent small size of the IgNAR V domain is an additional therapeutic and diagnostic attribute. It can be hypothesized that this property leads to a greater mobility with reference to tissue penetration. Especially for in vivo imaging, where a high contrast to background ratio is crucial, this feature is beneficial, due to an advantageous pharmacokinetic profile, i.e., a much shorter residence time in the blood compared to classical antibodies.Citation10 Furthermore it is assumed that the small molecular weight of the vNAR domain implicates the opportunity to target epitopes otherwise only accessible to small molecules.Citation6
Re-Formatting of vNAR Domains
The simple single chain molecular architecture of vNAR domains affords the benefit of multiple re-formatting opportunities to tailor the final product for purpose. Many formats have been successfully proven, including monomeric, dimeric and trimeric (binding more than one target) in addition to Fc-based formats, with all demonstrating the inherent flexibility of these domains.Citation50,Citation52,Citation54,Citation84
The small size of vNAR domains contributes to rapid renal clearance in vivo and represents a major drawback when non-imaging applications such as tumor-targeting are required. However, reduced size can be advantageous with regard to tumor penetration and ultimately the right format for the individual application needs to be determined. Fast glomerular filtration can be circumvented by multimerization of single vNAR domains, as has been shown by Müller et al.Citation50 Their investigations covered the N- as well as C-terminal fusion of a naїve vNAR domain with an anti-human serum albumin (HSA) vNAR originating from an immunized shark and isolated via phage display.Citation85 The fusion constructs retained high-affinity binding to HSA and exhibited significantly increased in vivo half-lifes compared to their unconjugated parental domains. The size of such dimeric formats is in the range of 25 kDa, and, compared to other antibody fragment formats such as scFvs, achieves double the binding site capacity while attaining increased affinities toward demanding, cryptic epitopes, which is a hallmark of vNAR proteins owing to their unique topology. In addition to dimeric fusions, a trimeric construct comprising the naїve vNAR domain at both termini of the anti-HSA vNAR displayed improved pharmacokinetics in in vivo studies in different species.Citation50 In another study conducted by Nuttall and colleagues, several approaches for the generation of bivalent shark antibodies with enhanced functional affinity were investigated.Citation84 Best affinities were obtained through C-terminal covalent or domain mediated linkages.
As has been shown extensively for rodent mAbs, there is a plethora of rational as well as empirical humanization strategies available to reduce immunogenic responses caused by animal-derived immunoglobulins.Citation86-Citation90 Rational design and the grafting of CDR loops of a xenogenic antibody onto a suitable human scaffold exhibiting a similar sequence has culminated in the development of several blockbuster pharmaceuticals routinely used in the clinic (e.g., trastuzumab, bevacizumab).Citation91
The sequence identity of the IgNAR V domain with mammalian VH regions falls as low as 25%.Citation49 In order to minimize the immunogenic potential of vNAR domains, Kovalenko and coworkers were able to engineer the aforementioned anti-HSA shark vNAR domainCitation50 by converting more than half of the framework amino acids to those of the human germline VK1 sequence DPK9.Citation52 This sequence bears the highest structural resemblance to the corresponding vNAR domain, and concomitantly represents one of the most stable human frameworks for downstream development. Determination of the binding constants of humanized vNAR variants yielded antigen affinities similar to those of the parental construct (14.8 nM vs 13.6 nM for the parental molecule). The opposite approach, i.e., starting with a structurally related human VH domain and converting that to a vNAR-like domain by clipping the CDR2 region, extending CDR3 and introducing stabilizing residues and additional disulfides, may also be a viable alternative as already shown for the camelization of human VH domains.Citation92 To enhance the expression of humanized vNARs in mammalian cells, they were C-terminally fused to human Fc domains.Citation52 This conjugation strategy can also contribute to the formation of dimers due to the interactions of 2 vNAR-Fc conjugates at the respective human constant domains. Fusions to human Fc, besides increasing the overall molecular weight and thus counteracting early renal clearance, can elicit in vivo immune effector functions and ultimately intensify the immune response via antibody-dependent cellular cytotoxicity and complement-dependent cytotoxicity. According to Kovaleva et al., the humanized IgNAR V domain variant showed negligible immunogenicity in dendritic cell assays.Citation54 Notably, also no significant immunogenic effects were observed after subcutaneous application of the parental, non-humanized anti-HSA vNAR in rodents and non-human primates.Citation50,Citation54 However, it remains to be scrutinized more meticulously how these proteins will behave when administered to patients in the scope of clinical trials.
Besides diagnostic and therapeutic applications, vNAR domains, due to their small size, high stability and their ability to sustain repeated cycles of unfolding and folding are also promising biomolecules for biotechnological applications to serve for example as high affinity capturing agents for purification of biomolecules or as tools for diagnostic applications. It was recently shown that vNAR fragments can be coupled covalently and site-specifically onto crystalline nanocellulose that serves as a protein-capturing nanoscaffold.Citation93 This evidence, coupled with the demonstrated stability and flexibility of vNAR domains, would predict more biotechnological applications can be expected to show up in the next years. Additionally, the vNAR domain can be utilized to gain information about pathological processes that at present are not completely understood. This was exemplified by Nuttall and colleagues, who grafted parts of the Aβ-peptide involved in Alzheimer's disease into CDR3 of a vNAR domain, and thus solved the crystal structure of the amyloid-β p3 fragment ().Citation94
Conclusion
Antibodies are essential molecules for biomedical and biotechnological applications. As described above, shark IgNARs differ greatly from conventional antibodies in many respects. Because of their unique structural features, these molecules have emerged as promising candidates for therapeutic, diagnostic and biotechnological applications. One striking example is that vNAR domains may complement classical antibodies in terms of druggable antigens. IgNAR V domains possess the potential to access cryptic and recessed epitopes that are usually not antigenic for conventional antibodies, e.g., hydrophobic clefts, active sites. However, this has only been shown for targeting the active site of lysozyme, as well as for targeting the hydrophobic cleft of AMA1, and therefore requires further exploration.Citation30,Citation48,Citation78 Their small size, high stability and the feasibility to re-format those molecules are additional desirable attributes. During the last years, constant progress on shark IgNAR research has been made. Related to this, vNAR domains targeting a plethora of therapeutically relevant antigens were generated and several methodologies for affinity-maturation of target-specific vNAR domains, as well as for re-formatting, e.g., humanization, multimerization, have been established. Moreover, a structural model of the complete IgNAR molecule revealed deeper insights into the extraordinary shape, stability and mode of action of this unique binding domain. Information gained from this might pave the way for the next generation of classical antibody variants showing improved stability. Fundamentally, it can be expected that this exceptional molecule, which evolved several hundred million years ago, might add value to the continuously evolving and exciting field of biologic drug development. Finally, it needs to be emphasized that, in view of future clinical applications, the immunogenic potential of vNAR domains requires further in depth investigation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Prof. Dr. Michael Sattler and Dr. Janosch Hennig for generously providing the structural data of the intact IgNAR molecule.
Funding
This work was supported in part by Federal Ministry of Education and Research (BMBF) in frame of the consortium Nanokat.
References
- Aggarwal RS. What's fueling the biotech engine-2012 to 2013. Nat Biotechnol 2014; 32:32-9; PMID:24406926; http://dx.doi.org/10.1038/nbt.2794
- Aggarwal SR. What's fueling the biotech engine-2011 to 2012. Nat Biotechnol 2012; 30:1191-7; PMID:23222785; http://dx.doi.org/10.1038/nbt.2437
- Buss NA, Henderson SJ, McFarlane M, Shenton JM, de Haan L. Monoclonal antibody therapeutics: history and future. Curr Opin Pharmacol 2012; 12:615-22; PMID:22920732; http://dx.doi.org/10.1016/j.coph.2012.08.001
- Reichert JM. Marketed therapeutic antibodies compendium. MAbs 2012; 4:413-5; PMID:22531442; http://dx.doi.org/10.4161/mabs.19931
- Reichert JM. Which are the antibodies to watch in 2013? MAbs 2013; 5:1-4; PMID:23254906; http://dx.doi.org/10.4161/mabs.22976
- Barelle C, Gill DS, Charlton K. Shark novel antigen receptors–the next generation of biologic therapeutics? Adv Exp Med Biol 2009; 655:49-62; PMID:20047035; http://dx.doi.org/10.1007/978-1-4419-1132-2_6
- Nuttall SD, Walsh RB. Display scaffolds: protein engineering for novel therapeutics. Curr Opin Pharmacol 2008; 8:609-15; PMID:18619558; http://dx.doi.org/10.1016/j.coph.2008.06.007
- Wesolowski J, Alzogaray V, Reyelt J, Unger M, Juarez K, Urrutia M, Cauerhff A, Danquah W, Rissiek B, Scheuplein F, et al. Single domain antibodies: promising experimental and therapeutic tools in infection and immunity. Med Microbiol Immunol 2009; 198:157-74; PMID:19529959; http://dx.doi.org/10.1007/s00430-009-0116-7
- Mordenti J, Cuthbertson RA, Ferrara N, Thomsen K, Berleau L, Licko V, Allen PC, Valverde CR, Meng YG, Fei DT, et al. Comparisons of the intraocular tissue distribution, pharmacokinetics, and safety of 125I-labeled full-length and Fab antibodies in rhesus monkeys following intravitreal administration. Toxicol Pathol 1999; 27:536-44; PMID:10528633; http://dx.doi.org/10.1177/019262339902700507
- Vaneycken I, D'Huyvetter M, Hernot S, De Vos J, Xavier C, Devoogdt N, Caveliers V, Lahoutte T. Immuno-imaging using nanobodies. Curr Opin Biotechnol 2011; 22:877-81; PMID:21726996; http://dx.doi.org/10.1016/j.copbio.2011.06.009
- Banta S, Dooley K, Shur O. Replacing antibodies: engineering new binding proteins. Annu Rev Biomed Eng 2013; 15:93-113; PMID:23642248; http://dx.doi.org/10.1146/annurev-bioeng-071812-152412
- Huang L, Gainkam LO, Caveliers V, Vanhove C, Keyaerts M, De Baetselier P, Bossuyt A, Revets H, Lahoutte T. SPECT imaging with 99mTc-labeled EGFR-specific nanobody for in vivo monitoring of EGFR expression. Mol Imaging Biol 2008; 10:167-75; PMID:18297364; http://dx.doi.org/10.1007/s11307-008-0133-8
- Kenanova V, Wu AM. Tailoring antibodies for radionuclide delivery. Expert Opin Drug Deliv 2006; 3:53-70; PMID:16370940; http://dx.doi.org/10.1517/17425247.3.1.53
- Van de Wiele C, Revets H, Mertens N. Radioimmunoimaging. Advances and prospects. Q J Nucl Med Mol Imaging 2004; 48:317-25; PMID:15640795
- Elia G, Fugmann T, Neri D. From target discovery to clinical trials with armed antibody products. J Proteomics 2014; 107c:50-5; PMID:24631826; http://dx.doi.org/10.1016/j.jprot.2014.02.034
- Enever C, Batuwangala T, Plummer C, Sepp A. Next generation immunotherapeutics–honing the magic bullet. Curr Opin Biotechnol 2009; 20:405-11; PMID:19709876; http://dx.doi.org/10.1016/j.copbio.2009.07.002
- Gebauer M, Skerra A. Engineered protein scaffolds as next-generation antibody therapeutics. Curr Opin Chem Biol 2009; 13:245-55; PMID:19501012; http://dx.doi.org/10.1016/j.cbpa.2009.04.627
- Holt LJ, Herring C, Jespers LS, Woolven BP, Tomlinson IM. Domain antibodies: proteins for therapy. Trends Biotechnol 2003; 21:484-90; PMID:14573361; http://dx.doi.org/10.1016/j.tibtech.2003.08.007
- Lambert JM. Drug-conjugated antibodies for the treatment of cancer. Br J Clin Pharmacol 2013; 76:248-62; PMID:23173552; http://dx.doi.org/10.1111/bcp.12044
- Lofblom J, Frejd FY, Stahl S. Non-immunoglobulin based protein scaffolds. Curr Opin Biotechnol 2011; 22:843-8; PMID:21726995; http://dx.doi.org/10.1016/j.copbio.2011.06.002
- Müller D, Kontermann RE. Bispecific antibodies for cancer immunotherapy: current perspectives. BioDrugs 2010; 24:89-98; PMID:20199124; http://dx.doi.org/10.2165/11530960-000000000-00000
- Nelson AL. Antibody fragments: hope and hype. MAbs 2010; 2:77-83; PMID:20093855; http://dx.doi.org/10.4161/mabs.2.1.10786
- Wozniak-Knopp G, Bartl S, Bauer A, Mostageer M, Woisetschlager M, Antes B, Ettl K, Kainer M, Weberhofer G, Wiederkum S, et al. Introducing antigen-binding sites in structural loops of immunoglobulin constant domains: Fc fragments with engineered HER2/neu-binding sites and antibody properties. Protein Eng Des Sel 2010; 23:289-97; PMID:20150180; http://dx.doi.org/10.1093/protein/gzq005
- Kolmar H. Natural and engineered cystine knot miniproteins for diagnostic and therapeutic applications. Curr Pharm Des 2011; 17:4329-36; PMID:22204431; http://dx.doi.org/10.2174/138161211798999465
- Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, Bendahman N, Hamers R. Naturally occurring antibodies devoid of light chains. Nature 1993; 363:446-8; PMID:8502296; http://dx.doi.org/10.1038/363446a0
- Greenberg AS, Avila D, Hughes M, Hughes A, McKinney EC, Flajnik MF. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature 1995; 374:168-73; PMID:7877689; http://dx.doi.org/10.1038/374168a0
- Flajnik MF, Deschacht N, Muyldermans S. A case of convergence: why did a simple alternative to canonical antibodies arise in sharks and camels? PLoS Biol 2011; 9:e1001120; PMID:21829328; http://dx.doi.org/10.1371/journal.pbio.1001120
- Muyldermans S. Nanobodies: natural single-domain antibodies. Annu Rev Biochem 2013; 82:775-97; PMID:23495938; http://dx.doi.org/10.1146/annurev-biochem-063011-092449
- Desmyter A, Transue TR, Ghahroudi MA, Thi MH, Poortmans F, Hamers R, Muyldermans S, Wyns L. Crystal structure of a camel single-domain VH antibody fragment in complex with lysozyme. Nat Struct Biol 1996; 3:803-11; PMID:8784355; http://dx.doi.org/10.1038/nsb0996-803
- Stanfield RL, Dooley H, Flajnik MF, Wilson IA. Crystal structure of a shark single-domain antibody V region in complex with lysozyme. Science 2004; 305:1770-3; PMID:15319492; http://dx.doi.org/10.1126/science.1101148
- Streltsov VA, Carmichael JA, Nuttall SD. Structure of a shark IgNAR antibody variable domain and modeling of an early-developmental isotype. Protein Sci 2005; 14:2901-9; PMID:16199666; http://dx.doi.org/10.1110/ps.051709505
- Holz JB. The TITAN trial–assessing the efficacy and safety of an anti-von Willebrand factor Nanobody in patients with acquired thrombotic thrombocytopenic purpura. Transfus Apher Sci 2012; 46:343-6; PMID:22475545; http://dx.doi.org/10.1016/j.transci.2012.03.027
- Rumfelt LL, Lohr RL, Dooley H, Flajnik MF. Diversity and repertoire of IgW and IgM VH families in the newborn nurse shark. BMC Immunol 2004; 5:8; PMID:15132758; http://dx.doi.org/10.1186/1471-2172-5-8
- Rumfelt LL, Diaz M, Lohr RL, Mochon E, Flajnik MF. Unprecedented multiplicity of Ig transmembrane and secretory mRNA forms in the cartilaginous fish. J Immunol 2004; 173:1129-39; PMID:15240702; http://dx.doi.org/10.4049/jimmunol.173.2.1129
- Ohta Y, Flajnik M. IgD, like IgM, is a primordial immunoglobulin class perpetuated in most jawed vertebrates. Proc Natl Acad Sci U S A 2006; 103:10723-8; PMID:16818885; http://dx.doi.org/10.1073/pnas.0601407103
- Amemiya CT, Alfoldi J, Lee AP, Fan S, Philippe H, MacCallum I, Braasch I, Manousaki T, Schneider I, Rohner N, et al. The African coelacanth genome provides insights into tetrapod evolution. Nature 2013; 496:311-6; PMID:23598338; http://dx.doi.org/10.1038/nature12027
- Greenberg AS, Hughes AL, Guo J, Avila D, McKinney EC, Flajnik MF. A novel "chimeric" antibody class in cartilaginous fish: IgM may not be the primordial immunoglobulin. Eur J Immunol 1996; 26:1123-9; PMID:8647177; http://dx.doi.org/10.1002/eji.1830260525
- Dooley H, Flajnik MF. Antibody repertoire development in cartilaginous fish. Dev Comp Immunol 2006; 30:43-56; PMID:16146649; http://dx.doi.org/10.1016/j.dci.2005.06.022
- Feige MJ, Grawert MA, Marcinowski M, Hennig J, Behnke J, Ausländer D, Herold EM, Peschek J, Castro CD, Flajnik M, et al. The structural analysis of shark IgNAR antibodies reveals evolutionary principles of immunoglobulins. Proc Natl Acad Sci U S A 2014; 111:8155-60; PMID:24830426; http://dx.doi.org/10.1073/pnas.1321502111
- Roux KH, Greenberg AS, Greene L, Strelets L, Avila D, McKinney EC, Flajnik MF. Structural analysis of the nurse shark (new) antigen receptor (NAR): molecular convergence of NAR and unusual mammalian immunoglobulins. Proc Natl Acad Sci U S A 1998; 95:11804-9; PMID:9751746; http://dx.doi.org/10.1073/pnas.95.20.11804
- Feige MJ, Groscurth S, Marcinowski M, Shimizu Y, Kessler H, Hendershot LM, Buchner J. An unfolded CH1 domain controls the assembly and secretion of IgG antibodies. Mol Cell 2009; 34:569-79; PMID:19524537; http://dx.doi.org/10.1016/j.molcel.2009.04.028
- Hsu E, Pulham N, Rumfelt LL, Flajnik MF. The plasticity of immunoglobulin gene systems in evolution. Immunol Rev 2006; 210:8-26; PMID:16623761
- Berstein RM, Schluter SF, Shen S, Marchalonis JJ. A new high molecular weight immunoglobulin class from the carcharhine shark: implications for the properties of the primordial immunoglobulin. Proc Natl Acad Sci U S A 1996; 93:3289-93; PMID:8622930; http://dx.doi.org/10.1073/pnas.93.8.3289
- Criscitiello MF, Saltis M, Flajnik MF. An evolutionarily mobile antigen receptor variable region gene: doubly rearranging NAR-TcR genes in sharks. Proc Natl Acad Sci U S A 2006; 103:5036-41; PMID:16549799; http://dx.doi.org/10.1073/pnas.0507074103
- Criscitiello MF. What the shark immune system can and cannot provide for the expanding design landscape of immunotherapy. Expert Opin Drug Discov 2014; 9:725-39; PMID:24836096; http://dx.doi.org/10.1517/17460441.2014.920818
- Streltsov VA, Varghese JN, Carmichael JA, Irving RA, Hudson PJ, Nuttall SD. Structural evidence for evolution of shark Ig new antigen receptor variable domain antibodies from a cell-surface receptor. Proc Natl Acad Sci U S A 2004; 101:12444-9; PMID:15304650; http://dx.doi.org/10.1073/pnas.0403509101
- Dooley H, Stanfield RL, Brady RA, Flajnik MF. First molecular and biochemical analysis of in vivo affinity maturation in an ectothermic vertebrate. Proc Natl Acad Sci U S A 2006; 103:1846-51; PMID:16446445; http://dx.doi.org/10.1073/pnas.0508341103
- Stanfield RL, Dooley H, Verdino P, Flajnik MF, Wilson IA. Maturation of shark single-domain (IgNAR) antibodies: evidence for induced-fit binding. J Mol Biol 2007; 367:358-72; PMID:17258766; http://dx.doi.org/10.1016/j.jmb.2006.12.045
- Dooley H, Flajnik MF, Porter AJ. Selection and characterization of naturally occurring single-domain (IgNAR) antibody fragments from immunized sharks by phage display. Mol Immunol 2003; 40:25-33; PMID:12909128; http://dx.doi.org/10.1016/S0161-5890(03)00084-1
- Müller MR, Saunders K, Grace C, Jin M, Piche-Nicholas N, Steven J, O'Dwyer R, Wu L, Khetemenee L, Vugmeyster Y, et al. Improving the pharmacokinetic properties of biologics by fusion to an anti-HSA shark VNAR domain. MAbs 2012; 4:673-85; PMID:23676205; http://dx.doi.org/10.4161/mabs.22242
- Diaz M, Stanfield RL, Greenberg AS, Flajnik MF. Structural analysis, selection, and ontogeny of the shark new antigen receptor (IgNAR): identification of a new locus preferentially expressed in early development. Immunogenetics 2002; 54:501-12; PMID:12389098; http://dx.doi.org/10.1007/s00251-002-0479-z
- Kovalenko OV, Olland A, Piche-Nicholas N, Godbole A, King D, Svenson K, Calabro V, Müller MR, Barelle CJ, Somers W, et al. Atypical antigen recognition mode of a shark immunoglobulin new antigen receptor (IgNAR) variable domain characterized by humanization and structural analysis. J Biol Chem 2013; 288:17408-19; PMID:23632026; http://dx.doi.org/10.1074/jbc.M112.435289
- Liu JL, Anderson GP, Delehanty JB, Baumann R, Hayhurst A, Goldman ER. Selection of cholera toxin specific IgNAR single-domain antibodies from a naive shark library. Mol Immunol 2007; 44:1775-83; PMID:17007931; http://dx.doi.org/10.1016/j.molimm.2006.07.299
- Kovaleva M, Ferguson L, Steven J, Porter A, Barelle C. Shark variable new antigen receptor biologics - a novel technology platform for therapeutic drug development. Expert Opin Biol Ther 2014; 14(10):1527-1539; PMID:25090369; http://dx.doi.org/10.1517/14712598.2014.937701
- Nuttall SD, Krishnan UV, Hattarki M, De Gori R, Irving RA, Hudson PJ. Isolation of the new antigen receptor from wobbegong sharks, and use as a scaffold for the display of protein loop libraries. Mol Immunol 2001; 38:313-26; PMID:11566324; http://dx.doi.org/10.1016/S0161-5890(01)00057-8
- Jackson KJ, Kidd MJ, Wang Y, Collins AM. The shape of the lymphocyte receptor repertoire: lessons from the B cell receptor. Front Immunol 2013; 4:263; PMID:24032032; http://dx.doi.org/10.3389/fimmu.2013.00263
- Diaz M, Greenberg AS, Flajnik MF. Somatic hypermutation of the new antigen receptor gene (NAR) in the nurse shark does not generate the repertoire: possible role in antigen-driven reactions in the absence of germinal centers. Proc Natl Acad Sci U S A 1998; 95:14343-8; PMID:9826702; http://dx.doi.org/10.1073/pnas.95.24.14343
- Diaz M, Velez J, Singh M, Cerny J, Flajnik MF. Mutational pattern of the nurse shark antigen receptor gene (NAR) is similar to that of mammalian Ig genes and to spontaneous mutations in evolution: the translesion synthesis model of somatic hypermutation. Int Immunol 1999; 11:825-33; PMID:10330287; http://dx.doi.org/10.1093/intimm/11.5.825
- Rumfelt LL, Avila D, Diaz M, Bartl S, McKinney EC, Flajnik MF. A shark antibody heavy chain encoded by a nonsomatically rearranged VDJ is preferentially expressed in early development and is convergent with mammalian IgG. Proc Natl Acad Sci U S A 2001; 98:1775-80; PMID:11172027; http://dx.doi.org/10.1073/pnas.98.4.1775
- Smith LE, Crouch K, Cao W, Muller MR, Wu L, Steven J, Lee M, Liang M, Flajnik MF, Shih HH, et al. Characterization of the immunoglobulin repertoire of the spiny dogfish (Squalus acanthias). Dev Comp Immunol 2012; 36:665-79; PMID:22040740; http://dx.doi.org/10.1016/j.dci.2011.10.007
- Nuttall SD, Krishnan UV, Doughty L, Nathanielsz A, Ally N, Pike RN, Hudson PJ, Kortt AA, Irving RA. A naturally occurring NAR variable domain binds the Kgp protease from Porphyromonas gingivalis. FEBS Lett 2002; 516:80-6; PMID:11959108; http://dx.doi.org/10.1016/S0014-5793(02)02506-1
- Liu JL, Anderson GP, Goldman ER. Isolation of anti-toxin single domain antibodies from a semi-synthetic spiny dogfish shark display library. BMC Biotechnol 2007; 7:78; PMID:18021450; http://dx.doi.org/10.1186/1472-6750-7-78
- Ohtani M, Hikima J, Jung TS, Kondo H, Hirono I, Aoki T. Construction of an artificially randomized IgNAR phage display library: screening of variable regions that bind to hen egg white lysozyme. Mar Biotechnol (NY) 2013; 15:56-62; PMID:22552958; http://dx.doi.org/10.1007/s10126-012-9456-1
- Ohtani M, Hikima J, Jung TS, Kondo H, Hirono I, Takeyama H, Aoki T. Variable domain antibodies specific for viral hemorrhagic septicemia virus (VHSV) selected from a randomized IgNAR phage display library. Fish Shellfish Immunol 2013; 34:724-8; PMID:23257206; http://dx.doi.org/10.1016/j.fsi.2012.11.041
- Zielonka S, Weber N, Becker S, Doerner A, Christmann A, Christmann C, Uth C, Fritz J, Schäfer E, Steinmann B, et al. Shark Attack: High affinity binding proteins derived from shark vNAR domains by stepwise in vitro affinity maturation. J Biotechnol 2014; 10.1016/j.jbiotec.2014.04.023; PMID:24862193
- Kopsidas G, Roberts AS, Coia G, Streltsov VA, Nuttall SD. In vitro improvement of a shark IgNAR antibody by Qβ replicase mutation and ribosome display mimics in vivo affinity maturation. Immunol Lett 2006; 107:163-8; PMID:17069896; http://dx.doi.org/10.1016/j.imlet.2006.09.004
- Doerner A, Rhiel L, Zielonka S, Kolmar H. Therapeutic antibody engineering by high efficiency cell screening. FEBS Lett 2014; 588:278-87; PMID:24291259; http://dx.doi.org/10.1016/j.febslet.2013.11.025
- Gai SA, Wittrup KD. Yeast surface display for protein engineering and characterization. Curr Opin Struct Biol 2007; 17:467-73; PMID:17870469; http://dx.doi.org/10.1016/j.sbi.2007.08.012
- Shao CY, Secombes CJ, Porter AJ. Rapid isolation of IgNAR variable single-domain antibody fragments from a shark synthetic library. Mol Immunol 2007; 44:656-65; PMID:16500706; http://dx.doi.org/10.1016/j.molimm.2006.01.010
- Nuttall SD, Krishnan UV, Doughty L, Pearson K, Ryan MT, Hoogenraad NJ, Hattarki M, Carmichael JA, Irving RA, Hudson PJ. Isolation and characterization of an IgNAR variable domain specific for the human mitochondrial translocase receptor Tom70. Eur J Biochem 2003; 270:3543-54; PMID:12919318; http://dx.doi.org/10.1046/j.1432-1033.2003.03737.x
- Camacho-Villegas T, Mata-Gonzalez T, Paniagua-Solis J, Sanchez E, Licea A. Human TNF cytokine neutralization with a vNAR from Heterodontus francisci shark: a potential therapeutic use. mAbs 2013; 5:80-5; PMID:23221782; http://dx.doi.org/10.4161/mabs.22593
- Goodchild SA, Dooley H, Schoepp RJ, Flajnik M, Lonsdale SG. Isolation and characterisation of Ebolavirus-specific recombinant antibody fragments from murine and shark immune libraries. Mol Immunol 2011; 48:2027-37; PMID:21752470; http://dx.doi.org/10.1016/j.molimm.2011.06.437
- Dooley H, Flajnik MF. Shark immunity bites back: affinity maturation and memory response in the nurse shark, Ginglymostoma cirratum. Eur J Immunol 2005; 35:936-45; PMID:15688348; http://dx.doi.org/10.1002/eji.200425760
- Crouch K, Smith LE, Williams R, Cao W, Lee M, Jensen A, Dooley H. Humoral immune response of the small-spotted catshark, Scyliorhinus canicula. Fish Shellfish Immunol 2013; 34:1158-69; PMID:23439398; http://dx.doi.org/10.1016/j.fsi.2013.01.025
- Walsh R, Nuttall S, Revill P, Colledge D, Cabuang L, Soppe S, Dolezal O, Griffiths K, Bartholomeusz A, Locarnini S. Targeting the hepatitis B virus precore antigen with a novel IgNAR single variable domain intrabody. Virology 2011; 411:132-41; PMID:21239030; http://dx.doi.org/10.1016/j.virol.2010.12.034
- Nuttall SD, Humberstone KS, Krishnan UV, Carmichael JA, Doughty L, Hattarki M, Coley AM, Casey JL, Anders RF, Foley M, et al. Selection and affinity maturation of IgNAR variable domains targeting Plasmodium falciparum AMA1. Proteins 2004; 55:187-97; PMID:14997552; http://dx.doi.org/10.1002/prot.20005
- Simmons DP, Streltsov VA, Dolezal O, Hudson PJ, Coley AM, Foley M, Proll DF, Nuttall SD. Shark IgNAR antibody mimotopes target a murine immunoglobulin through extended CDR3 loop structures. Proteins 2008; 71:119-30; PMID:17932913; http://dx.doi.org/10.1002/prot.21663
- Henderson KA, Streltsov VA, Coley AM, Dolezal O, Hudson PJ, Batchelor AH, Gupta A, Bai T, Murphy VJ, Anders RF, et al. Structure of an IgNAR-AMA1 complex: targeting a conserved hydrophobic cleft broadens malarial strain recognition. Structure 2007; 15:1452-66; PMID:17997971; http://dx.doi.org/10.1016/j.str.2007.09.011
- Goodchild SA, Dooley H, Schoepp RJ, Flajnik M, Lonsdale SG. Isolation and characterisation of Ebolavirus-specific recombinant antibody fragments from murine and shark immune libraries. Mol Immunol 2011; 48:2027-37; PMID:21752470; http://dx.doi.org/10.1016/j.molimm.2011.06.437
- Young NM, MacKenzie CR, Narang SA, Oomen RP, Baenziger JE. Thermal stabilization of a single-chain Fv antibody fragment by introduction of a disulphide bond. FEBS Lett 1995; 377:135-9; PMID:8543036; http://dx.doi.org/10.1016/0014-5793(95)01325-3
- Yasui H, Ito W, Kurosawa Y. Effects of substitutions of amino acids on the thermal stability of the Fv fragments of antibodies. FEBS Lett 1994; 353:143-6; PMID:7926039; http://dx.doi.org/10.1016/0014-5793(94)01027-7
- Miller BR, Demarest SJ, Lugovskoy A, Huang F, Wu X, Snyder WB, Croner LJ, Wang N, Amatucci A, Michaelson JS, et al. Stability engineering of scFvs for the development of bispecific and multivalent antibodies. Protein Eng Des Sel 2010; 23:549-57; PMID:20457695; http://dx.doi.org/10.1093/protein/gzq028
- Worn A, Plückthun A. Stability engineering of antibody single-chain Fv fragments. J Mol Biol 2001; 305:989-1010; PMID:11162109; http://dx.doi.org/10.1006/jmbi.2000.4265
- Simmons DP, Abregu FA, Krishnan UV, Proll DF, Streltsov VA, Doughty L, Hattarki MK, Nuttall SD. Dimerisation strategies for shark IgNAR single domain antibody fragments. J Immunol Methods 2006; 315:171-84; PMID:16962608; http://dx.doi.org/10.1016/j.jim.2006.07.019
- Müller MR, O'Dwyer R, Kovaleva M, Rudkin F, Dooley H, Barelle CJ. Generation and isolation of target-specific single-domain antibodies from shark immune repertoires. Methods Mol Biol 2012; 907:177-94; PMID:22907351; http://dx.doi.org/10.1007/978-1-61779-974-7_9
- Lu ZJ, Deng SJ, Huang DG, He Y, Lei M, Zhou L, Jin P. Frontier of therapeutic antibody discovery: The challenges and how to face them. World J Biol Chem 2012; 3:187-96; PMID:23275803; http://dx.doi.org/10.4331/wjbc.v3.i12.187
- Osbourn J, Groves M, Vaughan T. From rodent reagents to human therapeutics using antibody guided selection. Methods 2005; 36:61-8; PMID:15848075; http://dx.doi.org/10.1016/j.ymeth.2005.01.006
- De Pascalis R, Iwahashi M, Tamura M, Padlan EA, Gonzales NR, Santos AD, Giuliano M, Schuck P, Schlom J, Kashmiri SV. Grafting of "abbreviated" complementarity-determining regions containing specificity-determining residues essential for ligand contact to engineer a less immunogenic humanized monoclonal antibody. J Immunol 2002; 169:3076-84; PMID:12218124; http://dx.doi.org/10.4049/jimmunol.169.6.3076
- Ewert S, Honegger A, Plückthun A. Stability improvement of antibodies for extracellular and intracellular applications: CDR grafting to stable frameworks and structure-based framework engineering. Methods 2004; 34:184-99; PMID:15312672; http://dx.doi.org/10.1016/j.ymeth.2004.04.007
- Roguska MA, Pedersen JT, Keddy CA, Henry AH, Searle SJ, Lambert JM, Goldmacher VS, Blattler WA, Rees AR, Guild BC. Humanization of murine monoclonal antibodies through variable domain resurfacing. Proc Natl Acad Sci U S A 1994; 91:969-73; PMID:8302875; http://dx.doi.org/10.1073/pnas.91.3.969
- Riechmann L, Clark M, Waldmann H, Winter G. Reshaping human antibodies for therapy. Nature 1988; 332:323-7; PMID:3127726; http://dx.doi.org/10.1038/332323a0
- Riechmann L, Muyldermans S. Single domain antibodies: comparison of camel VH and camelised human VH domains. J Immunol Methods 1999; 231:25-38; PMID:10648925; http://dx.doi.org/10.1016/S0022-1759(99)00138-6
- Uth C, Zielonka S, Horner S, Rasche N, Plog A, Orelma H, Avrutina O, Zhang K, Kolmar H. A chemoenzymatic approach to protein immobilization onto crystalline cellulose nanoscaffolds. Angew Chem Int Ed Engl 2014; 53:12618-23; PMID:25070515; http://dx.doi.org/10.1002/anie.201404616
- Streltsov VA, Varghese JN, Masters CL, Nuttall SD. Crystal structure of the amyloid-β p3 fragment provides a model for oligomer formation in Alzheimer disease. J Neurosci 2011; 31:1419-26; PMID:21273426; http://dx.doi.org/10.1523/JNEUROSCI.4259-10.2011
- Harris LJ, Larson SB, Hasel KW, McPherson A. Refined structure of an intact IgG2a monoclonal antibody. Biochemistry 1997; 36:1581-97; PMID:9048542; http://dx.doi.org/10.1021/bi962514+
- Vranken W, Tolkatchev D, Xu P, Tanha J, Chen Z, Narang S, Ni F. Solution structure of a llama single-domain antibody with hydrophobic residues typical of the VH/VL interface. Biochemistry 2002; 41:8570-9; PMID:12093273; http://dx.doi.org/10.1021/bi012169a
- Krieger E, Joo K, Lee J, Lee J, Raman S, Thompson J, Tyka M, Baker D, Karplus K. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins 2009; 77 Suppl 9:114-22; PMID:19768677; http://dx.doi.org/10.1002/prot.22570
- Bojalil R, Mata-Gonzalez MT, Sanchez-Munoz F, Yee Y, Argueta I, Bolanos L, Amezcua-Guerra LM, Camacho-Villegas TA, Sanchez-Castrejon E, Garcia-Ubbelohde WJ, et al. Anti-tumor necrosis factor VNAR single domains reduce lethality and regulate underlying inflammatory response in a murine model of endotoxic shock. BMC Immunol 2013; 14:17; PMID:23548047; http://dx.doi.org/10.1186/1471-2172-14-17
