Abstract
Biofilms are polymicrobial communities that grow on surfaces in nature. Oral bacteria can spontaneously form biofilms on the surface of teeth, which may compromise the health of the teeth, or their surrounding (periodontal) tissues. While the oral bacteria exhibit high tropism for their specialized ecological niche, it is not clear if bacteria that are not part of the normal oral microbiota can efficiently colonize and grow within oral biofilms. By using an in vitro “supragingival” biofilm model of 6 oral species, this study aimed to investigate if 3 individual bacterial species that are not part of the normal oral microbiota (Eschericia coli, Staphylococcus aureus, Enterococcus faecails) and one not previously tested oral species (Aggregatibacter actinomycetemcomitans) can be incorporated into this established supragingival biofilm model. Staphylococcus aureus and A. actinomycetemcomitans were able to grow efficiently in the biofilm, without disrupting the growth of the remaining species. They localized in sparse small aggregates within the biofilm mass. Enterococcus faecalis and E. coli were both able to populate the biofilm at high numbers, and suppressed the growth of A. oris and S. mutants. Enterococcus faecalis was arranged in a chain-like conformation, whereas E. coli was densely and evenly spread throughout the biofilm mass. In conclusion, it is possible for selected species that are not part of the normal oral microbiota to be introduced into an oral biofilm, under the given experimental micro-environmental conditions. Moreover, the equilibrated incorporation of A. actinomycetemcomitans and S. aureus in this oral biofilm model could be a useful tool in the study of aggressive periodontitis and peri-implantitis, in which these organisms are involved, respectively.
Abbreviations
| CFUs | = | colony forming units |
| CLSM | = | confocal laser scanning microscopy |
| FISH | = | fluorescence in situ hybridization |
Introduction
Oral bacteria tend to form complex biofilm communities on the tooth surfaces. An oral biofilm is a polymicrobial consortium of numerous bacterial species, embedded in a polymeric matrix that derives either from their own metabolic products or from components of the host trapped into it, such as salivary glycoproteins.Citation1 Depending on the location of the biofilm in relation to the free gingival margin, a biofilm can be supragingival or subgingival. A supragingival biofilm is one that grows on the surface of the tooth enamel above the gingival margin. A subgingival biofilm grows below the gingival margin and into the periodontal pocket, which is a pathological feature of periodontal disease. The special micro-ecological conditions that prevail in each of these niches will favor the colonization and growth of species most adapted to the established conditions.Citation1,2 For instance, a supragingival biofilm consists of aerobic or facultative anaerobic species, whereas a subgingival biofilm is predominated by anaerobic species, as it grows in an oxygen-restricted environment. Biofilm formation is initiated on a pellicle of salivary proteins that coats the surface of the tooth, which is initially occupied by early colonizing species, such as Streptococcus sp. and Actinomyces spCitation3
The oral microbiome has a huge diversity spanning to more than 700 microbial taxa,Citation4,5 which can account for more than 10,000 phylotypes.Citation6 Despite this enormous diversity, the oral microbiome exhibits a strong tropism. In other terms, the microorganisms that colonize the oral cavity (also characterized as “resident oral microbiota”) have co-evolved with the host, and are highly specialized and adapted to survive in a specific ecological niche.Citation7 Therefore, due to this ecological adaptation, it may be difficult for bacteria that are not typical colonizers of the oral cavity to survive and constitute part of the oral microbiota. However, there is little experimental evidence demonstrating whether species that are not part of the normal oral microbiota can be successfully incorporated into an oral biofilm, under given experimental conditions that favor the formation of oral biofilms.
The hypothesis of this in vitro experimental study is that microorganisms that are not typically found as part of the normal oral microbiota may not be able to colonize and grow within an oral biofilm, during the course of its formation. Therefore, the aim of this study was to investigate if 3 different bacterial species that are not part of the normal oral microbiota (Eschericia coli, Staphylococcus aureus, and Enterococcus faecalis) can be incorporated into an established in vitro 6-species “supragingival” biofilm model,Citation8-12 also known as the “Zürich” biofilm model. Aggregatibacter actinomycetemcomitans, an oral species that has not been tested in this model before has been used as a positive control.
Results
The in vitro model used in this study consisted of 6 different oral species characteristic of the supragingival microbiota, co-cultured for 64 h in order to form a biofilm. During the initiation of biofilm formation, 4 individual bacterial species were newly introduced into this model, in order to evaluate if they are able to incorporate into the biofilm mass. When the total bacterial CFUs were counted, it was found that there were no differences between biofilm groups, irrespective of which additional bacterial species was introduced to the standard 6-speices model. The incorporation of each newly introduced species was thereafter evaluated, as well as the quantitative differences in CFU that this might cause to the “standard” 6 microorganisms in this in vitro model.
Starting with A. actinomycetemcomitans, this was found to be incorporated into the biofilm structure after 64 h, and was detected at CFU levels of 5-log. Notably, the quantitative composition of the other 6 species of the biofilm was not affected by the addition of A. actinomycetemcomitans, compared to the control group, with the exception of C. albicans, which was increased. Hence this oral species was successfully incorporated into the supragingival biofilm, without affecting the quantitative bacterial composition of the biofilm.
Enterococcus faecalis was also able to colonize the biofilm, at rather high levels of 7-log to 8-log. This high presence of E. faecalis did not affect the numerical levels of V. dispar, F. nucleatum, S. oralis, but increased that of C. albicans, compared to the 6-species control group. Remarkably however, it caused a reduction of A. oris numbers below the detection levels, and S. mutants close to the lowest detection limits ().
Figure 1. Colony forming units (CFUs) of the 6 species biofilm (control group; green), containing additionally Aggregatibacter actinomycetemcomitans (light blue), or Enterococcus faecalis (orange), or Staphylococcus aureus (blue), or Escherichia coli (red). Data derives from 3 independent experiments, in which every group was represented in triplicate biofilm cultures. Box plots represent the CFUs determined by selective agar plating, while horizontal lines indicate their median values. Undetectable values were ascribed the lowest detection limit value of the assay to allow for log transformation. *Significant difference compared with the control group (P < 0.05). Statistically significant differences compared with the control group are indicated with asterisks (* P < 0.05; ** P < 0 .01; *** P < 0 .001).
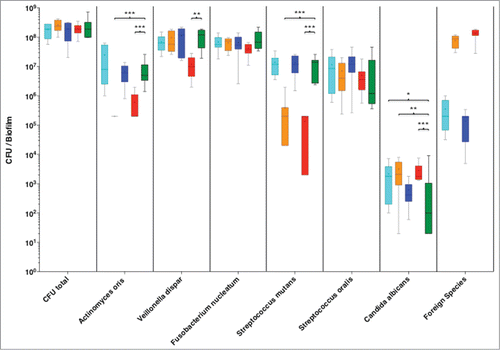
A similar trend to that of E. faecalis was observed by the addition of E. coli into the biofilm. This species was able to establish into the biofilm at 8-log levels and simultaneously caused a significant reduction in the numbers of S. mutans, A. oris and V. dispar, while it increased slightly the numbers of C. albicans.
Finally, S. aureus was also incorporated into the biofilm, with CFU levels of 4-log to 5-log. This was the lowest incorporation level compared to any of the other newly-introduced species. None of the remaining 6 species was quantitatively affected by the presence of S. aureus in the biofilm.
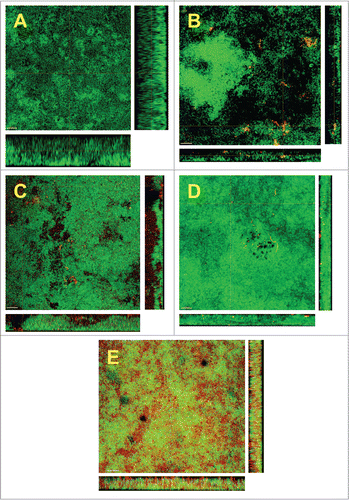
The structural arrangement of the newly introduced species within the biofilm was also considered. For this, CLSM was used in combination with DNA-specific staining (green) and FISH-staining with species-specific 16S rRNA oligonucleotide probes (red), for each individual species tested (). Aggregatibacter actinomycetecmomitans appeared to localize within small secluded cell clusters of its own species, through the biofilm mass (). A comparable pattern of small aggregates within the biofilm was also observable in the case of S. aureus (). In the case of E. faecalis, this species did not form distinctive clusters within the biofilm, but it was arranged in a chain-like pattern of single bacterial cells (). The most striking observation, however, was that after the addition of E. coli. This species was grossly distributed throughout the whole mass of the biofilm and scattered among the other species present ().
Figure 2. Confocal laser scanning microscopy (CLSM) images of the 6 species biofilm (control group; A), containing additionally Aggregatibacter actinomycetemcomitans (B), or Staphylococcus aureus (C), or Enterococcus faecalis (D), or Escherichia coli (E). Due to FISH staining of biofilms in B-F using Cy3-labeled probes (see), the newly added bacteria appear red. Non-hybridized bacteria appear green due to DNA staining (YoPro 1 + Sytox). The biofilm base in the cross sections is directed toward the top view. Scales = 10 μm.
Discussion
In the present study, 4 different bacterial species were individually introduced into a well-established 6-species supragingival biofilm. One of them, namely A. actinomycetecomitans, comprises part of the normal oral microbiota. After 64 h of biofilm growth, this species was detectable at levels comparable to those of the other oral species, and did not affect their quantitative composition. As a facultative anaerobe, A. actinomycetemcomitans can be part of supragingival biofims in clinical samples.Citation13 In a recent study A. actinomycetemcomitans has been successfully grown as part of a 4 species biofilm, including also Streptococcus gordonii, F. nucleatum and Porphyromonas gingivalis,Citation14 or a 6 species biofilm, including also S. oralis, A. naeslundii, V. parvula, F. nucleatum, and P. gingivalis.Citation15 Therefore, the successful and homogenous incorporation of A. actinomycetemcomitans in the present in vitro biofilm model could be expected. This revised variant of the supragingival biofilm that includes A. actinomycetemcomitans could be further implemented, for instance in research questions pertinent to localized aggressive periodontitis, an entity of periodontal disease in which this species is highly prevalent.Citation16 It should also be noted that the oral buccal epithelium may serve as a reservoir for A. actinomycetemcomitans, which can be translocated from the epithelium to hard surfaces, but not the other way around.Citation17
The other 3 species tested in this model are not major constituents of the oral microbiota. Therefore, it was of interest to evaluate whether they can be incorporated into an oral biofilm during a standard period of growth. Enterococcus faecalis is not readily detected as part of the oral microbiota, and its prevalence in all intraoral microenvironments is reported to range from 3.5% to 13.5%.Citation18 However, in light of studies using metagenomic approaches, there is moderate evidence to support E. faecalis as a candidate periodontal pathogen, with a potential geographic specificity.Citation19 Moreover, it is frequently isolated from the root canal system of teeth with failed endodontic treatments.Citation20,21 In the present experimental model, E. faecalis was successfully incorporated into the biofilm and, interestingly, caused a significant reduction in the numbers of A. oris and S. mutans. This indicates a spatial or nutritional competition between these 2 early colonizers and E. faecalis. This may be in line with recent studies demonstrating that E. faecalis dominates numerically over S. mutans in dual-species biofilms.Citation22,23 Another study also demonstrated that Enterococcus faecium was able to inhibit biofilm formation by oral streptococci, including S. mutans.Citation24 It is of interest that E. faecalis was distributed in a chain-like, rather than cluster-like pattern, within the supragingival biofilm. This chain-like pattern resembles that acquired by Prevotella intermedia in a subgingival biofilm model, once streptococci and A. oris are excluded from its composition.Citation25 As oral streptococci tend to form long filamentous structures, which are important for multi-cellularity,Citation26 their reduction in a biofilm may necessitate that other species compensate for their structural conformation. The survival of E. faecalis into this oral biofilm model shows that it is possible to co-exist among oral species, and justifies its occasional presence in the root-canal system of endodontically-involved teeth. It is also noteworthy that E. faecalis is detected with high prevalence in biofilms of HIV-infected patients with necrotizing or chronic periodontal diseases,Citation27,28 denoting the opportunistic nature of this pathogen under immunocompromised conditions.
A successful colonization of the biofilm was also evident in the case of S. aureus. Its incorporation was at relatively low numbers, did not affect the quantitative composition of the other species, and was regularly distributed in small clusters of its own species within the biofilm. Although the oral cavity is not its typical habitat, S. aureus has occasionally been isolated from dental plaque,Citation29-31 particularly of patients with respiratory infection.Citation32 The present findings indicate that, given the appropriate micro-environmental conditions, S. aureus can indeed constitute part of a supragingival biofilm microbiota. This is particularly important, as there is growing evidence of an association between S. aureus and peri-implantitis, an emerging oral infection.Citation33
The capacity of E. coli to colonize and grow in this supragingival biofilm model was also evaluated. Under the present experimental conditions, E. coli growth was exacerbated, and was detectable in a perfuse pattern throughout the biofilm mass. These in vitro results were striking, as there has only been circumstantial clinical evidence documenting the presence of E. coli in dental plaque. Nevertheless, a very recent systematic meta-analysis of studies using metagenomic approaches indicates that E. coli can be detected in subgingival plaque of periodontitis patients.Citation19 The dominance of E. coli in the present model shows that, given the appropriate nutritional and environmental conditions, it has the capacity to survive and even dominate among oral species, in a polymicrobial biofilm. In the in vivo situation, the host immune defenses may control and prevent its routine colonization of oral sites. This is further supported by its increased prevalence in supragingival dental plaque of elderly institutionalized patients.Citation34
In conclusion, E. faecalis and E. coli may successfully colonize and grow in a biofilm consisting of supragingival species. They also exert antagonistic interactions upon S. mutans or A. oris. While the study proves that it is possible for bacterial species that are not part of the normal oral microbiota to be incorporated into an oral biofilm model, additional (potentially host-related) factors may account for their absence, or infrequent presence, in dental plaque in vivo. The study also documents that A. actinomycetemcomitans and S. aureus can be part of a supragingival biofilm, without affecting the composition of the remaining bacterial species. Hence, they can be adjunctively used in this model, for the study of questions related to the etiology of aggressive periodontitis and peri-implantitis, respectively. Finally, one should consider that only single strains of each bacterial species were used in this experimental system. This may pose a limitation in the interpretation of the results since there can be great genetic variation at the strain level, even for the same species.
Materials and Methods
Formation of “supragingival” biofilm in vitro
The in vitro biofilm model used in this study consisted of 6 microorganisms that can be typically found as part of the supragingival microbiota, and has been described earlier.Citation35 Briefly, the standard supragingival in vitro biofilm contained Actinomyces oris (formerly Actinomyces naeslundii) OMZ 745, Veillonella dispar OMZ 493 (ATCC 17748T), Fusobacterium nucleatum OMZ 598 (KP-F2), Streptococcus mutans OMZ 918 (UA159), Streptococcus oralis OMZ 607 (SK 248) and Candida albicans OMZ 110. This standard biofilm was supplemented with either Aggregatibacter actinomycetemcomitans (OMZ 295 (JP2)), Enterococcus faecalis (OMZ 422 (ATCC 29212)), Staphylococcus aureus (OMZ 1122 (ATCC 25923)), or Escherichia coli (OMZ 1123 (Nissle 1917)). Biofilms were grown in 24-well polystyrene cell culture plates on hydroxyapatite discs (Ø 9 mm; Clarkson Chromatography Products, DCHAP-.38”) that had been preconditioned (pellicle-coated) in 1 ml of pasteurized whole un-stimulated saliva, pooled from individual donors, and incubated for 4 h at room temperature. The same saliva batch was used in all experimentations. To initiate biofilm formation, the discs were covered with 1 ml of growth medium containing saliva and modified fluid universal medium (mFUM), and 200 μl of a microbial suspension prepared from equal volumes and densities of each strain, corresponding to OD550 = 1.0. mFUM is a well-established tryptone-yeast-based broth medium designated as FUMCitation36 and modified by supplementing 67 mM Sørensen's buffer (final pH 7.2). The carbohydrate concentration in mFUM was 0.3% (w/v), and consisted of glucose for the first 16 h and from then on of a 1:1 (w/w) mixture of glucose and sucrose. Biofilms were incubated anaerobically at 37 °C for 64 h. After inoculation, the discs remained for 45 min in the feeding solution containing 0.3% glucose, and were subjected to 3 consecutive 1 min dip-washes in 2 ml 0.9% NaCl to remove growth medium and free floating cells. The biofilms were then further incubated in new wells containing 1 ml of saliva only. After 16 h, 20 h, 24 h, 40 h, 44 h and 48 h biofilms were pulse-fed by transferring the discs for 45 min into medium containing 30% saliva and 70% mFUM with 0.15% glucose and 0.15% sucrose. They were washed again as described above and re-incubated in saliva. Fresh saliva was provided after 16 h and 40 h. After 64 h the biofilms were dip-washed again prior to harvesting for culture analyses or processing for fluorescence in situ hybridization (FISH) staining and confocal laser scanning microscopy (CLSM) analyses, as described below.
Quantitative determination of the biofilm species
After 64 h of biofilm growth, the hydroxyapatite discs were vortexed vigorously for 1 minute in 1 ml of 0.9% NaCl and then sonicated at 25W in a Sonifier B-12 (Branson Sonic Power Company) for 5 sec, to harvest the adherent biofilms. The resulting bacterial suspensions were serially diluted in 0.9% NaCl. Of each serial dilution, 50 μl aliquots were plated on agar plates supplemented with 5% whole human blood to estimate total colony-forming units (CFUs). To determine the species-specific bacterial numbers, 6 different selective agars were used to determine the CFUs for the 6 standard species of the biofilms and the 4 newly introduced species (). Agar plates were incubated at 37°C for 72 h. Species identification was achieved by observation of colony morphology.
Table 1. Selective agars used for quantitative analyses of biofilm organisms
Staining of biofilms
Biofilms were stained by FISH using species-specific Cy3-labeled probes following the protocols described before.Citation37,38 Pre-hybridization (15 min, 46°C) was performed in 500 μl hybridization buffer in the absence of any oligonucleotide probes. Thereafter, 500 μl of hybridization buffer was used for each biofilm, supplemented with probes at a concentration of 10 ng/μl. The incubation time for the hybridization was at least 3 h at 46°C in the dark. After the incubation, biofilms were transferred into washing buffer pre-heated to 48°C and incubated for 20 min at this temperature. Probe sequences and formamide concentrations used for the hybridizations, as well as the NaCl concentrations of the washing buffers are given in . For counterstaining, biofilms were stained using a mixture of 3 μM YoPro 1 iodide (Invitrogen, Y3603) and 15 μM Sytox green (Invitrogen, S7020) (20 min, room temperature, in the dark), following the FISH procedure. After staining, the samples were embedded upside-down on chamber slides in 100 μl of Mowiol 4–88 (Calbiochem-Novabiochem; 475904).Citation39
Table 2. Sequence and formamide concentrations for FISH Probes
Confocal laser scanning microscopy (CLSM)
Stained biofilms were examined by CLSM at randomly selected positions using a Leica TCS SP5 microscope (Leica Microsystems) with a x63/1.4 NA oil immersion objective lens, in conjunction with 488-nm laser excitation and 530-nm emission filters for YoPro 1/Sytox, and 561-nm laser excitation and 640-nm emission filters for Cy3. Image acquisition was performed in x8 line average mode. Scans were recombined and processed using Imaris 7.6.5 software (Bitplane), without any qualitative changes to the raw images.
Statistical analyses
Three independent experiments were performed, and within each experiment every group was represented in triplicate biofilm cultures. The statistical significance of the differences in microbial numbers between the control group (standard 6 species biofilm) and test groups (biofilm with newly introduced species) was evaluated by 2-way analysis of variance (ANOVA), corrected by Tukey's multiple comparisons test (significance level P < 0.05). Undetectable values were ascribed the lowest detection limit value of the assay to allow for log transformation. The data were analyzed using the Prism version 6, statistical analysis software (GraphPad).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Mrs. Ruth Graf and Mr. Andre Meier for their excellent technical assistance with the experimentations. We thank the Center of Microscopy and Image Analysis (ZMB) of the University of Zürich for their support with confocal microscopy.
Funding
This study was supported by the Institute of Oral Biology, University of Zurich.
References
- Filoche S, Wong L, Sissons CH. Oral biofilms: emerging concepts in microbial ecology. J Dent Res 2010; 89:8-18; PMID:19918089; http://dx.doi.org/10.1177/0022034509351812
- Marsh PD. Are dental diseases examples of ecological catastrophes? Microbiology 2003; 149:279-94; PMID:12624191; http://dx.doi.org/10.1099/mic.0.26082-0
- Kolenbrander PE, Palmer RJ, Rickard AH, Jakubovics NS, Chalmers NI, Diaz PI. Bacterial interactions and successions during plaque development. Periodontology 2000 2006; 42:47-79; PMID:16930306; http://dx.doi.org/10.1111/j.1600-0757.2006.00187.x
- Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol 2005; 43:5721-32; PMID:16272510; http://dx.doi.org/10.1128/JCM.43.11.5721-5732.2005
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. The human oral microbiome. J Bacteriol 2010; 192:5002-17; PMID:20656903; http://dx.doi.org/10.1128/JB.00542-10
- Zaura E, Keijser BJ, Huse SM, Crielaard W. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol 2009; 9:259; PMID:20003481; http://dx.doi.org/10.1186/1471-2180-9-259
- Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontol 2000 2005; 38:135-87; PMID:15853940; http://dx.doi.org/10.1111/j.1600-0757.2005.00107.x
- Thurnheer, T, Gmür, R, Shapiro, S, Guggenheim, B. Mass transport of macromolecules within an in vitro model of supragingival plaque. Appl Environ Microbiol 2003; 69:1702-9; PMID:12620862; http://dx.doi.org/10.1128/AEM.69.3.1702-1709.2003
- Thurnheer T, Giertsen E, Gmür R, Guggenheim B. Cariogenicity of soluble starch in oral in vitro biofilm and experimental rat caries studies: a comparison. J Appl Microbiol 2008; 105:829-36; PMID:18452534; http://dx.doi.org/10.1111/j.1365-2672.2008.03810.x
- Thurnheer T, Rohrer E, Belibasakis GN, Attin T, Schmidlin PR. Static biofilm removal around ultrasonic tips in vitro. Clin Oral Investig 2013; PMID:24317957
- Guggenheim B, Guggenheim M, Gmür R, Giertsen E, Thurnheer T. Application of the Zürich biofilm model to problems of cariology. Caries Res 2004; 38:212-22; PMID:15153691; http://dx.doi.org/10.1159/000077757
- Sahrmann P, Zehnder M, Mohn D, Meier A, Imfeld T, Thurnheer T. Effect of low direct current on anaerobic multispecies biofilm adhering to a titanium implant surface. Clin Implant Dent Relat Res 2014; 16:552–56.
- Moore WEC, Holdeman LV, Smibert RM, Good IJ, Burmeister JA, Palcanis KG, Ranney RR. Bacteriology of experimental gingivitis in young adult humans. Infect Immun 1982; 38:651-67; PMID:7141708
- Park JH, Lee JK, Um HS, Chang BS, Lee SY. A periodontitis-associated multispecies model of an oral biofilm. J Periodontal Implant Sci 2014; 44:79-84; PMID:24778902; http://dx.doi.org/10.5051/jpis.2014.44.2.79
- Blanc V, Isabal S, Sanchez MC, Llama-Palacios A, Herrera D, Sanz M, Leon R. Characterization and application of a flow system for in vitro multispecies oral biofilm formation. J Periodontal Res 2014; 49:323-32; PMID:23815431; http://dx.doi.org/10.1111/jre.12110
- Aberg CH, Sjodin B, Lakio L, Pussinen PJ, Johansson A, Claesson R. Presence of Aggregatibacter actinomycetemcomitans in young individuals: a 16-year clinical and microbiological follow-up study. J Clin Periodontol 2009; 36:815-22; PMID:19678862; http://dx.doi.org/10.1111/j.1600-051X.2009.01457.x
- Fine DH, Markowitz K, Furgang D, Velliyagounder K. Aggregatibacter actinomycetemcomitans as an early colonizer of oral tissues: epithelium as a reservoir? J Clin Microbiol 2010; 48:4464-73; PMID:20881174; http://dx.doi.org/10.1128/JCM.00964-10
- Estrela CR, Pimenta FC, Alencar AH, Ruiz LF, Estrela C. Detection of selected bacterial species in intraoral sites of patients with chronic periodontitis using multiplex polymerase chain reaction. J Appl Oral Sci 2010; 18:426-31; PMID:20835581; http://dx.doi.org/10.1590/S1678-77572010000400018
- Perez-Chaparro PJ, Goncalves C, Figueiredo LC, Faveri M, Lobao E, Tamashiro N, Duarte P, Feres M. Newly identified pathogens associated with periodontitis: a systematic review. J Dent Res 2014; 93:846-58; PMID:25074492; http://dx.doi.org/10.1177/0022034514542468
- Zehnder M, Guggenheim B. The mysterious appearance of enterococci in filled root canals. Int Endod J 2009; 42:277-87; PMID:19220511; http://dx.doi.org/10.1111/j.1365-2591.2008.01537.x
- Rechenberg DK, De-Deus G, Zehnder M. Potential systematic error in laboratory experiments on microbial leakage through filled root canals: review of published articles. Int Endod J 2011; 44:183-94; PMID:21219357; http://dx.doi.org/10.1111/j.1365-2591.2010.01821.x
- Deng DM, Hoogenkamp MA, Exterkate RA, Jiang LM, van der Sluis LW, Ten Cate JM, Crielaard W. Influence of Streptococcus mutans on Enterococcus faecalis biofilm formation. J Endod 2009; 35:1249-52; PMID:19720224; http://dx.doi.org/10.1016/j.joen.2009.05.038
- Li X, Hoogenkamp MA, Ling J, Crielaard W, Deng DM. Diversity of Streptococcus mutans strains in bacterial interspecies interactions. J Basic Microbiol 2014; 54:97-103; PMID:23456658; http://dx.doi.org/10.1002/jobm.201200457
- Suzuki N, Yoneda M, Hatano Y, Iwamoto T, Masuo Y, Hirofuji T. Enterococcus faecium WB2000 Inhibits biofilm formation by oral cariogenic streptococci. Int J Dent 2011; 2011:834151; PMID:22114599; http://dx.doi.org/10.1155/2011/834151
- Ammann TW, Belibasakis GN, Thurnheer T. Impact of Early Colonizers on In Vitro Subgingival Biofilm Formation. PLoS ONE 2013; 8 12:e83090; PMID:24340084; http://dx.doi.org/10.1371/journal.pone.0083090
- Rossetti V, Ammann TW, Thurnheer T, Bagheri HC, Belibasakis GN. Phenotypic diversity of multicellular filamentation in oral streptococci. PLoS ONE 2013; 8:e76221; PMID:24086713; http://dx.doi.org/10.1371/journal.pone.0076221
- Ramos MP, Ferreira SM, Silva-Boghossian CM, Souto R, Colombo AP, Noce CW, Goncalves LS. Necrotizing periodontal diseases in HIV-infected Brazilian patients: a clinical and microbiologic descriptive study. Quintessence Int 2012; 43:71-82; PMID:22259811
- Goncalves LS, Souto R, Colombo AP. Detection of helicobacter pylori, Enterococcus faecalis, and pseudomonas aeruginosa in the subgingival biofilm of HIV-infected subjects undergoing HAART with chronic periodontitis. Eur J Clin Microbiol Infect Dis 2009; 28:1335-42; PMID:19639349; http://dx.doi.org/10.1007/s10096-009-0786-5
- Scannapieco FA, Stewart EM, Mylotte JM. Colonization of dental plaque by respiratory pathogens in medical intensive care patients. Crit Care Med 1992; 20:740-5; PMID:1597025; http://dx.doi.org/10.1097/00003246-199206000-00007
- Scannapieco FA, Mylotte JM. Relationships between periodontal disease and bacterial pneumonia. J Periodontol 1996; 67:1114-22; PMID:8910830; http://dx.doi.org/10.1902/jop.1996.67.10s.1114
- Cuesta AI, Jewtuchowicz V, Brusca MI, Nastri ML, Rosa AC. Prevalence of Staphylococcus spp and Candida spp in the oral cavity and periodontal pockets of periodontal disease patients. Acta Odontol Latinoam 2010; 23:20-6; PMID:20645638
- Didilescu AC, Skaug N, Marica C, Didilescu C. Respiratory pathogens in dental plaque of hospitalized patients with chronic lung diseases. Clin Oral Investig 2005; 9:141-7; PMID:15909174; http://dx.doi.org/10.1007/s00784-005-0315-6
- Belibasakis GN. Microbiological and immuno-pathological aspects of peri-implant diseases. Arch Oral Biol 2014; 59:66-72; PMID:24209597; http://dx.doi.org/10.1016/j.archoralbio.2013.09.013
- Russell SL, Boylan RJ, Kaslick RS, Scannapieco FA, Katz RV. Respiratory pathogen colonization of the dental plaque of institutionalized elders. Spec Care Dentist 1999; 19:128-34; PMID:10860077; http://dx.doi.org/10.1111/j.1754-4505.1999.tb01413.x
- Thurnheer T, van der Ploeg JR, Giertsen E, Guggenheim B. Effects of Streptococcus mutans gtfC deficiency on mixed oral biofilms in vitro. Caries Res 2006; 40:163-71; PMID:16508276; http://dx.doi.org/10.1159/000091065
- Gmür R, Guggenheim B. Antigenic heterogeneity of Bacteroides intermedius as recognized by monoclonal antibodies. Infect Immun 1983; 42:459-70; PMID:6196291
- Thurnheer T, Gmür R, Guggenheim B. Multiplex FISH analysis of a six-species bacterial biofilm. J Microbiol Meth 2004; 56:37-47; http://dx.doi.org/10.1016/j.mimet.2003.09.003
- Ammann TW, Gmur R, Thurnheer T. Advancement of the 10-species subgingival Zurich Biofilm model by examining different nutritional conditions and defining the structure of the in vitro biofilms. BMC Microbiol 2012; 12:227; PMID:23040057; http://dx.doi.org/10.1186/1471-2180-12-227
- Guggenheim M, Shapiro S, Gmür R, Guggenheim B. Spatial arrangements and associative behavior of species in an in vitro oral biofilm model. Appl Environ Microbiol 2001; 67:1343-50; PMID:11229930; http://dx.doi.org/10.1128/AEM.67.3.1343-1350.2001
- Guggenheim B, Giertsen E, Schüpbach P, Shapiro S. Validation of an in vitro biofilm model of supragingival plaque. J Dent Res 2001; 80:363-70; PMID:11269730; http://dx.doi.org/10.1177/00220345010800011201
- Klinke T, Guggenheim B, Klimm W, Thurnheer T. Dental caries in rats associated with candida albicans. Caries Res 2011; 45:100-6; PMID:21412001; http://dx.doi.org/10.1159/000324809
- Baird RM, Lee WH. Media used in the detection and enumeration of Staphylococcus aureus. Int J Food Microbiol 1995; 26:15-24; PMID:7662517; http://dx.doi.org/10.1016/0168-1605(93)E0028-P
- Rechenberg DK, Thurnheer T, Zehnder M. Potential systematic error in laboratory experiments on microbial leakage through filled root canals: an experimental study. Int Endod J 2011; 44:827-35; PMID:21535022; http://dx.doi.org/10.1111/j.1365-2591.2011.01888.x
- Bohnert J, Hübner B, Botzenhart K. Rapid identification of Enterobacteriaceae using a novel 23S rRNA-targeted oligonucleotide probe. Int J Hyg Environ Health 2000; 203:77-82; PMID:10956593; http://dx.doi.org/10.1078/S1438-4639(04)70011-5
