Abstract
Pathogenic bacteria sense environmental cues, including the local temperature, to control the production of key virulence factors. Thermal regulation can be achieved at the level of DNA, RNA or protein and although many virulence factors are subject to thermal regulation, the exact mechanisms of control are yet to be elucidated in many instances. Understanding how virulence factors are regulated by temperature presents a significant challenge, as gene expression and protein production are often influenced by complex regulatory networks involving multiple transcription factors in bacteria. Here we highlight some recent insights into thermal regulation of virulence in pathogenic bacteria. We focus on bacteria which cause disease in mammalian hosts, which are at a significantly higher temperature than the outside environment. We outline the mechanisms of thermal regulation and how understanding this fundamental aspect of the biology of bacteria has implications for pathogenesis and human health.
Introduction
Pathogenic bacteria must sense and respond to changes in their local environment in order to successfully colonise and invade their hosts. Microbes achieve this by continually monitoring multiple characteristics of their surroundings, including the ambient temperature, pH, osmotic pressure and nutrient availability. These signals are then integrated to fine-tune gene expression and protein production. Bacteria possess specific virulence factors that enable them to attach to host cell surfaces, evade immune defenses or obtain nutrients which might be otherwise inaccessible. Unregulated expression of these virulence factors can be detrimental for bacteria, through wasteful expenditure of metabolic resources and inappropriate induction of inflammatory and immune responses. Therefore, bacteria possess regulatory mechanisms that restrain production of virulence factors until they detect appropriate environmental cues present in their hosts.Citation1
As the core temperature of mammalian hosts is usually higher than that of their surroundings, an increase in temperature is a change consistently experienced by pathogens when they migrate from the external environment to their host. As a result, many bacterial virulence factors are thermally regulated. The various mechanisms which the bacteria employ to sense host temperatures and control their virulence factors are usually considered distinct from the heat shock response, which is a homeostatic collection of processes shared by both commensal and pathogenic bacteria. However, there are examples of heat shock proteins also playing a role in virulence, which suggests there is a significant degree of overlap between these responses.
Here we review recent advances in understanding the underlying mechanisms of how bacterial pathogens regulate virulence factors in relation to temperature and provide examples of how this contributes to the development of disease (). We focus on examples of how microbes sense the elevated temperatures once they enter a mammalian host, even though pathogens of plants and insects can regulate gene expression in response to lower temperatures.Citation2-4
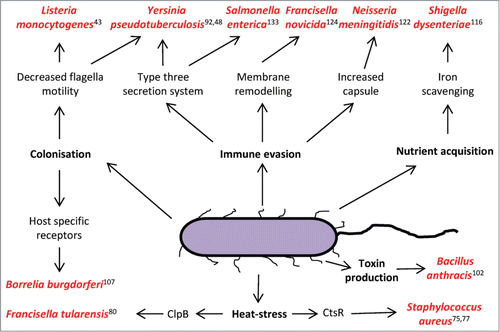
Figure 1. Thermo-regulation of important virulence factors in pathogenic bacteria species. An increase to 37°C (mammalian body temperature) is a universal invasion signal to the bacteria and enables fine tuning of virulence factor expression, promoting survival and proliferation in its host.
Mechanisms of thermo-sensing: exerting multi-level control Regulation at the level of DNA
Changes in the local temperature are detected by bacteria through multiple mechanisms; temperature can influence the topology of DNA, affect RNA structuring and metabolism, and change the activity and processing of proteins (). At the DNA level, temperature influences the extent of DNA supercoiling which can affect the rate of transcription,Citation5 providing a mechanism for thermal regulation of gene expression.Citation6 For example, super-coiling is involved in the expression of genes encoding Type I fimbriae in Escherichia coliCitation7 and a type III secretion system (T3SS) in Salmonella enterica,Citation8 which are both required for virulence. Additionally, alterations in DNA curvature can modulate gene expression in a temperature-dependent manner. This is illustrated by the transcription of virF, which encodes a regulator of genes on the large virulence plasmid in Shigella spp.Citation9 At non-permissive lower temperatures, DNA at the virF promoter assumes a bent configuration, which is facilitated by the histone-like nucleoid structuring protein, H-NS. This impairs binding of RNA polymerase at the promoter, resulting in reduced transcription of genes encoding virulence factors outside the host.
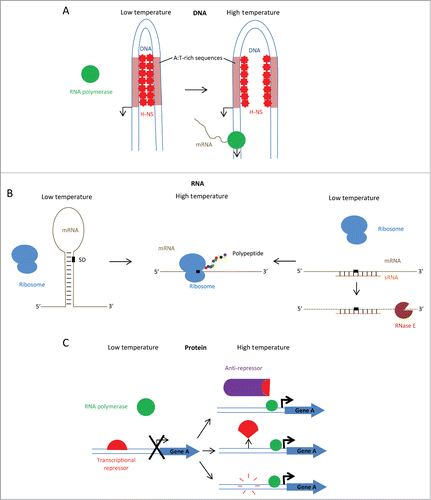
Figure 2. Mechanisms of thermo-regulation (A) At low temperature, the DNA at the promoter region of some virulence genes favors a bent conformation and is stabilised by H-NS, which binds curved DNA at A:T rich sites, blocking access of RNA polymerase to the promoter. At high temperature, homotypic H-NS binding is reduced by relaxation of the DNA bend, enabling access of RNA polymerase to the promoter (e.g. virF in Shigella). (B) At low temperature, mRNA thermometer adopts a stem-loop structure at the 5ʹ position, sequestering the Shine-Dalgarno (SD) sequence from ribosome binding and preventing translation. At high temperature, stem-loop melts to expose the Shine-Dalgarno sequence to ribosome binding and translation occurs. Alternatively, trans-acting small RNAs act by sequestering the SD sequence at low temperature but not at high temperature. Double stranded RNAs are targeted for degradation by RNases. (C) At low temperature, repressor is bound to promoter of Gene A and transcription is prohibited. At high temperature, either: the anti-repressor is stabilised and removes the repressor from the promoter (e.g., GmaR in Listeria monocytogenes), a conformational change causes the repressor to dissociate from the DNA (e.g. RovA in Yersinia spp.) or the repressor is degraded by temperature-dependant proteases which permits the transcription of Gene A (e.g., RovA in Yersinia spp).
H-NS is found in several Gram-negative pathogensCitation10 in which it can repress expression of virulence genes.Citation11-14 H-NS binds AT-rich, curved stretches of DNA,Citation15 and silences genes by oligomerising to form a repression complex that blocks access of RNA polymerase to promoters.Citation16 As formation of the active, tetrameric form of H-NS does not change at temperatures above 28°C,Citation17 the oligomerization state of H-NS cannot fully account for the de-repression of gene expression above 30°C. The importance of temperature-dependent H-NS regulation in bacteria is illustrated by Salmonella enterica in which over 200 genes are upregulated at 37°C in an H-NS dependent fashion; many of these genes are within Salmonella SPI-1 pathogenicity island.Citation18 Furthermore, the binding of H-NS at the hilC promoter decreases between 25°C and 37°C,Citation18 allowing expression of this activator of SPI-1 genes at higher temperatures;Citation19 similar H-NS dependent thermoregulation also occurs at the E. coli hemolysin (hly) locus.Citation20 In both instances, local changes in DNA conformation might contribute to the increased affinity of H-NS for its target sequence at lower temperatures.
Regulation at the level of RNA
mRNA can assume a variety of secondary and tertiary structures which have consequences on protein translation by altering interactions of the mRNA with ribosomes.Citation6,21 For example, RNA thermosensors are typically found in the 5′-untranslated region (5′-UTR) of an mRNA molecule and control translation of that specific message, in which they are said to be cis-acting.Citation22 cis-acting RNA thermosensors were initially discovered in genes encoding heat shock proteins (HSP), and include the hspA ROSE1 element in Bradyrhizobium japonicum,Citation23 the agsA fourU element in Salmonella enterica,Citation24 and the hsp17 bi-partite hairpin found in Synechocystis sppCitation25 In each case, the RNA thermosensor is located in the 5′-UTR of the mRNA with the 3′-end of the stem-loop sequestering the ribosome binding site (Shine-Dalgarno sequence, SD) and preventing translation at lower temperatures. In a stem-loop structure, the length of the hairpin and the presence of non-canonical base pairing define the thermodynamic properties of melting and annealing, and thus determine the relationship between protein translation and temperature.Citation22,26 RNA thermosensors offer a particularly rapid response to a change in temperature, as they do not require a signaling pathway or de novo mRNA transcription. RNA thermosensors can also induce protein expression following cold shock.Citation27 This occurs through the formation of two discrete structures: one which sequesters the SD site from ribosomes, while another structure forms at lower temperature that exposes the SD, enabling translation. Given the complexity of RNA structures, predicting RNA thermosensors using genome-wide bioinformatics has become a significant obstacle and requires further understanding of the dynamics of RNA secondary structures,Citation28 computer modeling,Citation29 and extending databases of known RNA thermometers.
Alternatively, RNAs can regulate other transcripts, when they are trans-acting.Citation30 In this case, a regulatory small RNA (sRNA) is transcribed independently from the target mRNA. Base pairing between the two RNA molecules can result in gene silencing through targeted degradation by ribonucleases (RNases) or sequestration of the ribosome binding site. Alternatively, base pairing can alter the secondary structure of the mRNA to expose the ribosome binding site.Citation31 Another mechanism of thermal regulation at the level of mRNA occurs by changes in the stability of the message. Degradation of specific RNAs is an efficient way of adjusting their abundance and this is brought about by RNases.Citation32,33 For example, RNase E is autoregulated by cleaving a stem-loop structure in its own 5′ UTR,Citation34 and can also be involved in silencing genes by cleaving mRNAs paired with a sRNA.Citation33,35 For example, in pathogenic E. coli, the gene encoding the AfaD adhesin is regulated by the temperature-dependent expression of sRNA, AfaR.Citation36 Approximately 42 bases of AfaR bind to the 5’ UTR of the afaD transcript. Binding is promoted by Hfq, and leads to RNase E degradation of the message. Similarly, in Yersinia pestis and Yersinia pseudotuberculosis, many small RNAs are expressed at 37°C and not at 28°C and have thus been suggested to have roles in regulating virulence.Citation37 The targets of these sRNA have not yet been defined but their function can be modulated by the RNA chaperone Hfq, whose ability to bind RNA is higher at lower temperatures.Citation37,38 Hfq facilitates binding of sRNAs to target mRNAs, leading to their subsequent degradation,Citation39 and has been shown to play an important role in pathogenesis in several organisms.Citation38-41
Regulation at the protein level
At the level of protein activity and processing, thermo-regulation usually results from a conformational change in a thermo-sensitive domain of a protein.Citation6,42 This subsequently modulates the function of the protein or changes its susceptibility to degradation by proteases.Citation43 For example, GmaR, an anti-repressor in Listeria, undergoes a conformational shift at 37°C and is then degraded; loss of GmaR allows MogR to repress genes involved in flagellar based motility within the mammalian host.Citation42 The interaction between the function and stability of several proteins in a network allows fine tuning of virulence factor expression, as well as the integration of inputs from several environmental stimuli,Citation44 permitting a more dynamic response.
Temperature affecting global gene expression through a single transcriptional regulator
Co-ordinating regulation of multiple virulence factors can be efficiently achieved through a single transcription factor that modulates expression of a suite of genes. By placing a single transcription factor under thermal control, a pathogen can adapt to the new environments it encounters when it enters its host. An exemplar is provided by PrfA, a transcriptional regulator in Listeria monocytogenes, that is controlled by an RNA thermosensor.Citation45 At temperatures below 30°C, a stem-loop structure is predicted to form in the 5′-UTR of the prfA mRNA, which occludes the ribosome binding site and prevents protein translation. However, at the higher temperatures found in mammalian hosts, this hairpin melts enabling efficient translation of PrfA, which orchestrates expression of genes involved in intracellular invasion and survival. Analysis of the dynamic response of the PrfA thermosensor demonstrates that it acts as an abrupt, on/off switch; this could be explained by the abundance and clustering of G:C bonds at the base and top of the predicted stem loop.Citation26 Furthermore, a trans-acting riboswitch in the prfA transcript binds S-adenosyl methionine. Binding of this metabolite to the mRNA also prevents translation,Citation30 enabling integration of cues from the local temperature and nutritional state of the bacterium to optimise the expression of virulence factors.
Yersinia spp. provide another example of the diversity of regulatory mechanisms found in bacteria that respond to host temperatures.Citation46,47 LcrF is a global regulator in Y. pseudotuberculosis, the cause of a scarlet fever-like illness in humans. LcrF is regulated by temperature both at the level of transcription and translation.Citation48 This transcriptional activator controls expression of genes encoding a T3SS and secreted proteins, that contribute to pathogenesis.Citation49 The 5′-UTR of the lcrF mRNA harbours a RNA thermosensor similar to prfA. Furthermore, transcription of lcrF is also repressed by the nucleoid-associated protein YmoA, which is selectively degraded at 37°C.Citation48 Yersinia spp also possess RovA which represses the expression of several virulence factors,Citation50-53 and is required for colonisation and adaptation to host-associated stress.Citation54 A shift from external to host physiological temperatures (i.e. to 37°C) results in a conformational change in the DNA-binding domain of RovA, leading to its dissociation from target promoters, and de-repression of gene expression.Citation42,55 Additionally, proteolytic degradation of RovA is enhanced at higher temperatures, further reducing RovA repression regulator within the host.Citation55
In Bordetella spp. many important virulence factors are controlled by the BvgAS two-component system. This system is composed of the histidine kinase BvgS, which is found in the inner membrane of the bacteria, and BvgA, a response regulator. Auto-phosphorylation of the cytoplasmic domain of BvgS in response to specific environmental stimuli leads to phosphorylation and activation of BvgA, which subsequently binds promoter regions and activates transcription.Citation56 Upon shift to 37°C, BvgA also autoregulates its own expression and also controls genes required in the early stage of infection, such as those for cell adhesion.Citation57 Additionally, BvgA activates other genes required later in the infection process, such as ptx encoding pertussis toxin.Citation58 Although the precise mechanism for the temperature dependent activation of BvgAS has yet to be characterized, the linker region between the transmembrane and kinase domains of BvgS is crucial for thermal sensing.Citation59
Feeling the heat in the host: Integration of thermal regulation with stress responses
Pathogens encounter several forms of stress once in a mammalian host including a change in pH, osmotic gradients, exposure to reactive oxygen and nitrogen species, as well as nutrient limitation. As previously discussed, heat shock responses in bacteria are usually considered as distinct from specific, temperature-triggered adaptations for survival in hosts. However overlapping networks of regulation often found in bacteria, and indirect effects of heat shock and other stress responses can influence the production of virulence factors.Citation60,61
In E. coli, the heat shock response is orchestrated by the sigma factor, σCitation32 (RpoH), which controls the expression of a diverse range of heat shock proteins with homeostatic function.Citation62 This transcription factor is subject to several hierarchies of control and functional homologues of RpoH are present in a range of Gram-negative bacteria.Citation63 Translation of rpoH mRNA itself is modulated in E. coli by an RNA thermometerCitation64 which, following an increase in temperature to 42°C, enhances the efficiency of ribosome binding leading to an increase in translation. The activity of RpoH is also regulated post-translationally by the chaperones DnaK and GroEL,Citation65 and its turnover is controlled by specific proteases such as FtsH.Citation66 RpoH indirectly influences the transcription of virulence gene regulators.Citation67,68 In uropathogenic E. coli, RpoH controls expression of leuXCitation68 which encodes tRNA5leu which is important for the synthesis of virulence factors encoded on the pathogenicity island, PAIII356.Citation69
Gram-positive bacteria possess heat shock proteins, but the way in which the heat shock response is co-ordinated is distinct. In Bacillus subtilis, for example, there are three distinct classes of heat-inducible proteins.Citation70 Class I heat shock genes are the most highly induced following heat stress, and include those encoding the major chaperones DnaK and GroEL.Citation71 Class II heat shock genes are subject to regulation by σB which is upregulated in response to several types of stress.Citation71,72 Finally, class III heat shock genes include those encoding ATP-dependent proteases, with expression occurring independent of σB.Citation71 Of note, σB positively regulates expression of sar, which encodes winged helix-turn-helix transcriptional regulator which influences the production of many virulence factors in Staphylococcus aureus.Citation73,74 Additionally, CtsR, which is a transcriptional repressor that is part of the σB-independent heat shock response in several Gram-positive pathogens,Citation75 also controls a wide range of target proteins involved in virulence.Citation76 CtsR exhibits high affinity DNA binding to target promoters at low temperature (30°C) but lower affinities at normal host temperature (37°C) and, in addition, is preferentially degraded at the higher temperature.Citation77
Several ATP-dependent proteases that are integral to heat shock responses also contribute to virulence.Citation61 One example is ClpB, an ATP-dependent chaperone part of the Clp family of proteins.Citation78,79 ClpB is crucial for replication of the human pathogen Francisella tularensisis in host tissuesCitation80 and involved in pathogenesis of L. monocytogenes.Citation81 Finally, there a examples of chaperones contributing to virulence which are induced during heat shock.Citation61 In particular, the major chaperone DnaK has been shown to be important for during phagocytosis of Listeria by macrophages but not for intracellular survival.Citation82
Temperature influencing bacterial motility
Motility is a common feature of many pathogens and permits migration to a specific niche at mucosal surfaces. Thermotaxis, i.e., motility in response to temperature, has been observed in several bacteria including E. coli.Citation83,84 Using flagellar based motility, E. coli swims along temperature gradients to warmer sites at low culture densities.Citation85 This is achieved by varying swimming behavior at different temperatures: at warmer temperatures, the bacteria generally swim undirectionally, but at low temperature the bacteria tumble and change direction.Citation85 Mutants which are incapable of chemotaxis are generally found to be devoid of thermotaxis,Citation86 indicating that these two responses share some common mechanism. Chemotaxis has been well-characterized and requires membrane bound receptors which transduce environmental stimuli into a phosphorylation cascade that modifies flagellar assembly and rotation.Citation87,88 Of note, the direction of migration reverses at higher bacterial densities,Citation83 and this reversal of motility is affected by the nutritional state of the microbe.Citation89 As a consequence, in situations where there is nutrient limitation, E. coli migrates to cooler areas where the metabolic demands on the bacterium may be reduced.Citation90
L. monocytogenes also possesses a switch-like mechanism which ensures that motility is limited to environments which are below physiological temperatures in hosts (i.e. below 37°C).Citation43 In this case, a thermo-sensing anti-repressor protein (GMaR) binds to and inhibits the activity of the flagella motility transcriptional repressor MogR at 30°C, but not at 37°C. Similarly a proteomic analysis of Clostridium difficile demonstrated that flagellin levels are reduced in response to heat shock,Citation91 which could affect motility.
Yersinia is non-motile at 37°C,Citation92 and the expression of genes involved in flagellar biogenesis is orchestrated by several different proteins including the sigma factor FliA (σCitation28), its anti-sigma factor FlgM, and the chaperone FliS.Citation93 At 37°C, transcription of both fliA and flgM is arrested, resulting in a flagella-null phenotype.Citation94 The mechanism is unknown, but might be related to changes in DNA supercoiling.Citation95 In Campylobacter coli the flagellum is composed of two structural proteins, FlaA and FlaB.Citation96 The gene encoding FlaB is regulated by a σCitation54-dependent promoter, and its transcription is higher at 42°C than 37°C which correlates with an increase in flagellar motility at the higher temperature.Citation97 In Campylobacter, the interaction of the anti-sigma factor FlgM with the sigma factor FliA (σCitation28) is temperature dependent and prevents excessive elongation of the flagellum, allowing higher motility at 42°C than 37°C.Citation98 Furthermore FlaA biosynthesis is highest at 42°C, which may contribute to increased motility at 42°C,Citation98 the core temperature of the avian host of Campylobacter spp.
Thermoregulation of exotoxin production
The foodborne pathogen enterohemorrhagic E. coli (EHEC) secretes Shiga-like toxins that contribute to the development of hemolytic uraemic syndrome and hemorrhagic colitis in humans.Citation99 Shiga-like toxin 2 (Stx-2), the more potent of the two toxins produced by EHEC, is subject to thermoregulation,Citation14 although the precise mechanism(s) for this are unknown. Thermoregulation of toxin production is also seen in other pathogenic bacteria including B. pertussis (see above) and Y. enterocolitica.Citation100 In Yersina, production of the enterotoxin, Yst, is higher at 28°C (i.e., sub-host temperature).Citation101 The reasons for this thermal regulation in light of this remain unclear. Furthermore in Bacillus anthracis, synthesis of the Anthrax Toxin Activator (AtxA) is temperature-dependent.Citation102 AtxA regulates genes required for synthesis of both the edema and lethal toxins, which are necessary for virulenceCitation103 and have highest expression at 37°C.Citation102 This corresponds to an increase in the transcription of all three toxin structural genes at the permissive temperature.Citation104 To date, the mechanisms by which AtxA regulates the expression of other genes or by which its synthesis is thermally regulated it unknown.Citation105
Temperature as a signal for adaptation between different hosts
Some bacteria colonise both a vertebrate and an invertebrate host, and so are exposed to entirely different sets of conditions including temperature. Borrelia burgdorferi, which causes Lyme disease, must deal with conditions within both its tick vector and its mammalian host. One of the ways this is accomplished is by expressing the outer surface protein OspA or OspC which bind distinct ligands in the invertebrate and vertebrate host, respectively. The alternative sigma factor RpoS (σCitation38), which regulates several virulence factors under heat stress,Citation106 is responsible for the differential expression of OspA/C via a temperature-dependent small RNA, DsrABb.Citation107 DsrABb was predicted to bind to the 5′-UTR of rpoS mRNA through complementary base-paring and was hypothesized to be a trans-acting sRNA that liberates the SD site of rpoS mRNA, permitting translation.Citation107 This finding is supported by translational fusion studies in vivo demonstrating a higher translational efficiency of RpoS mRNA at 37°C compared to the low temperature 26°C,Citation108 comparable to the conditions experienced by the bacteria in its tick vector. However the level of DsrABb transcript was found to be similar at both temperatures, suggesting that DsrABb is not the key temperature-regulatory molecule.Citation107 Recent evidence in E. coli suggests that the RNA chaperone Hfq is required for efficient translation of RpoS in conjunction with DsrABbCitation109 and may confer thermal regulation.Citation37,38 Other virulence factors in Borrelia spp., such as the complement regulator-acquiring surface proteins (CRASPs) are also regulated in response to temperatureCitation110 and the mammal-tick infection cycle,Citation111 indicating that temperature is the primary cue by which this pathogen senses transmission between their hosts.
The haemin uptake system (Hms) in Y. pestis is another example of thermoregulation that occurs during transmission from vector to host.Citation46 Interestingly, the Hms is not essential for pathogenesis in mammalsCitation112 but allows iron acquisition from a blood meal and proliferation of bacteria in fleas that leads to blockage of the proventricular valve,Citation113 regurgitation and the spread of infection when the flea feeds on a human. Hms has been shown to be thermoregulated at the level of protein stability,Citation114 with bacteria containing higher levels of Hms at temperatures found in fleas (i.e. below 26°C).
Thermal control of nutrient acquisition
The availability of nutrients within hosts is dramatically different from the external environment. Several micronutrients, for example iron, are sequestered from invading microbes in mammalian hosts, limiting bacterial proliferation. Many pathogens have developed strategies that allow them to acquire nutrients which would otherwise be inaccessible in their host.Citation115 Enteropathogenic E. coli and Shigella dysenteriae both scavenge iron in the form of heme present in the human gastrointestinal tract using the TonB-dependent receptors, ChuA and ShuA respectively. Two distinct regulatory mechanisms permit iron scavenging within the host where there is both physiological temperature (37°C) and low levels of free iron.Citation116 There is a fourU-type RNA thermosensor located in the 5′-UTR region of the mRNA of chuA and shuA, which forms a hairpin enabling the 4 U nucleotides to pair the RBS, and preventing translation at lower temperatures.Citation117
Similarly, a recent transcriptome analysis of Pseudomonas aeruginosa highlighted that genes encoding enzymes required for the biosynthesis of the siderophores, pyochelin and pyoverdine,Citation118 are differentially regulated at 22°C compared to 37°C.Citation119 In general, genes involved in pyoverdine biosynthesis are upregulated at the low temperature, while those for pyochelin biosynthesis are upregulated at 37°C.Citation119 Expression of ppyR, which encodes an activator of the genes required for pyoverdine synthesis, is also upregulated at 22°CCitation119 by an unknown mechanism. These findings suggest that the bacterium switches strategies for iron utilization from outside to inside its host.
Immune evasion mechanisms
Once inside their mammalian host, bacteria must defend themselves against the immune system as molecules secreted or localized on the surface of microbes can elicit potent inflammatory and immune responses. Therefore bacteria carefully control expression of immune-stimulatory molecules to avoid being detected and eliminated. There are several instances in which temperature plays a key role in regulating bacterial immune evasion.
Neisseria meningitidis is a major cause of meningitis and septicaemia in children and young adults that normally resides as a commensal of the human nasopharynx. It has been found that temperature acts as a danger signal for this important pathogen, as an elevation in temperature leads to an increase in its suite of immune evasion molecules. Production of factor H binding protein, fHbp (which recruits the human complement regulator human factor H at high affinity),Citation120,121 lipopolysaccharide (LPS) sialylation, and the polysaccharide capsule (made by the css operon) protect the pathogen from complement-mediated lysis; these factors appear to be controlled by independent RNA thermosensors.Citation122 This strategy is distinct from those seen in other pathogens, in which multiple effector molecules are controlled by thermo-regulation of a pleiotropic transcription factor. In the meningococcus, these three mechanisms of immune evasion have their own dedicated RNA thermosensor; this may reflect the constant association between the meningococcus and its human host, and its habitat in the thermal gradients of the nasopharynx. The meningococcal capsule thermosensor shows a gradual response to a rise in temperature unlike the Listeria PrfA thermosensor which acts like an on:off switch, and common polymorphisms in the 5′ UTR of the css mRNA do not affect thermoregulation.Citation122 In both the css and fHbp thermosensors, the SD sequence is predicted to be included in a hairpin structure at lower temperatures. Following entry of the bacterium across the epithelial barrier into the bloodstream, increased deployment of immune avoidance mechanisms at the elevated temperature in the body will promote survival in the systemic circulation and thence virulence. However, as virulence is an evolutionary dead end for this microbe, thermosensing of immune defense will be an adaptation for survival in the upper airway. Here, temperature may be acting as a danger signal for the meningococcus, by indicating the inflammatory status of the nasopharyngeal mucosa.
LPS is an essential component of the outer membrane of Gram negative bacteria, and required for membrane integrity and protection against exogenous bactericidal agents, such as anti-microbial peptides.Citation123 LPS also acts as a microbe associated molecular pattern molecule (MAMP), with the lipid A portion of LPS being a potent stimulator of inflammatory responses by recognition by Toll-like receptor-4 (TLR-4) on host cells. The human pathogen Francisella novicida can cause septicaemia and remodels its LPS in response to temperature.Citation124 At environmental temperatures (i.e., 18°C), lipid A is acetylated at the 3-OH C16 position by LpxD2. However at host temperatures (37°C), LPS is acetylated at the 3-OH C18 position by LpxD1 which confers increased resistance to anti-microbial peptides, antibodies and in vivo killing. Transcriptional and post-translational regulation of the LpxD enzymes increases their activity at corresponding temperatures but these mechanisms have not been characterized. This demonstrates that subtle changes to components of bacteria, such as modification of acyl chains in LPS in response to temperature, are sufficient to alter innate immune signaling.
Another example of changes in LPS in response to temperature is in Y. enterocolitica; modification with of lipid A with aminoarabinose and palmitate is reduced at 37°C, which contribute to antimicrobial peptide resistance at low temperature (21°C).Citation123 Genes involved in these modifications are transcriptionally regulated the two-component systems, PhoPQ and PmrAB. The expression of both these two component systems is temperature dependent, through relief of H-NS repression.Citation123 Additionally another important component of LPS, the O-antigen, which also plays a role in host-pathogen interactions, is regulated by temperature in Y. enterocolitica.Citation125 The activity of the two promoters, PWb1 and PWb2, which control expression of the genes required for O-antigen synthesis, are repressed at 37°C.Citation125
Polysaccharide capsules offer bacteria protection from harsh external environments and host immunity. However capsule expression can also be detrimental by impairing interactions between microbial outer membrane ligands and their host cell receptors for attachment and invasion.Citation126 Capsule biosynthesis in Streptococcus pyogenes, a cause of tonsillitis in humans, has been shown to be thermoregulated.Citation127 At lower temperatures (i.e. 25°C) capsule expression increases though an undefined, post-transcriptional mechanism; this is thought to confer anti-phagocytic properties to the bacterium. Since the main route of infection is through the skin where the temperature is lower than core body temperature, a thicker capsule may condition the bacterium prior to penetrating the epithelial layer. The membrane-associated endoribonuclease, CvfA (which regulates virulence genes in response to nutrient availability)Citation128 is responsible for the thermo-regulation at a post-transcriptional level although the mechanism is still to be elucidated.
T3SSs are complex multi-protein machineries which allow secretion of proteins directly from the cytoplasm of bacteria into host cells; T3SSs can contribute to intracellular invasion, and thence immune evasion. Temperature can be an important cue to modulate the assembly and activity of T3SSs to precise sites in the host where they are required.Citation129 In Salmonella enterica, there are two T3SSs encoded on two distinct pathogenicity islands, SPI-1 and SPI-2. SPI-1 contains a set of virulence genes specific for invasion into host cells, whereas SPI-2 contains genes required for intracellular replication and survival.Citation130 The expression of genes encoding the SPI-2 T3SS has been shown to be dependent upon local pH,Citation131 concentrations of phosphate and magnesiumCitation132 and, more recently by temperature.Citation133 Activation of SPI-2 T3SS genes requires the SsrAB two-component regulatory system, although the response to temperature is governed by two nucleoid-associated proteins, H-NS and Hha which contribute to the absence of this T3SS on the surface of bacteria located outside the host.Citation133 This highlights the importance of integrating temperature as a regulatory stimulus in virulence factor expression, since other environmental factors (nutrient availability, osmolality and pH) are alone, not host-specific. Similarly, the regulator InvE in Shigella sonnei is required for the transcription of T3SS components.Citation134 invE mRNA has recently been demonstrated to be negatively regulated at the post-transcriptional level by the RNA chaperone Hfq, which, as mentioned previously, is dependent on temperature.Citation37,38 At 37°C there is reduced binding of Hfq to invE mRNA compared to 30°C, leading to increased stability of invE mRNA at the higher temperature and increased activity of the T3SS. Since VirF is the positive regulator of invECitation135 and its synthesis is up-regulated at higher temperatures,Citation9 this is an elegant example in how temperature mediates exquisite control of virulence gene expression through multiple integrated mechanisms.
Leptospira interrogans causes a zoonotic infection, transmitted to humans through broken skin upon contact with infected animal urine, which can cause serious kidney failure and liver damage. The two cell surface lipoproteins LigA and LigB can bind to a multitude of host proteins, contribute to avoidance of the complement systemCitation136 and to colonisation by binding extracellular matrix proteins.Citation137 Recently, mRNA thermosensors have been identified in the 5′-UTR of both ligA and ligB,Citation138 which are predicted to form a double stem loop structure, with the second hairpin occluding the SD sequence at 30°C and melting to enable translation at 37°C.
Conclusions
Understanding how temperature and other environmental cues act on regulatory networks highlights key aspects of the evolution in pathogenic bacteria, and provides insights into the habitats they occupy during steps in the disease process. While cold and heat shock proteins protect bacteria from sudden fluctuations in temperature through a homeostatic mechanism, many virulence factors use the signal provided by temperature, in addition to other cues, to enhance production of virulence factors within their hosts. Besides shedding light on how pathogens cause disease, understanding the molecular mechanisms underlying thermosensing has potential benefits for human health. For example, the production of many molecules that have been exploited as vaccine antigens is under temperature control. Examples include bacterial capsules and fHbp (from the meningococcus), and several toxins; modified Ptx is a component of the licensed acellular pertussis vaccines, while there have been efforts to use detoxified toxin from anthrax as an immunogen. Additionally components of T3SS have been proposed as a vaccine candidate against Shigella spp. Therefore, defining the sequences and motifs that allow thermosensing in microbes and how these signals are linked with other host signals, should allow the rapid identification of other thermally regulated molecules. This could provide the next generation of vaccine candidates and potentially drug targets.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Marteyn B, West NP, Browning DF, Cole JA, Shaw JG, Palm F, Mounier J, Prévost M-C, Sansonetti P, Tang CM. Modulation of Shigella virulence in response to available oxygen in vivo. Nature 2010; 465:355-8; PMID:20436458; http://dx.doi.org/10.1038/nature08970
- Smirnova A V, Wang L, Rohde B, Budde I, Weingart H, Ullrich MS. Control of temperature-responsive synthesis of the phytotoxin coronatine in Pseudomonas syringae by the unconventional two-component system CorRPS. J Mol Microbiol Biotechnol 2002; 4:191-6; PMID:11931546.
- Starke M, Fuchs TM. YmoA negatively controls the expression of insecticidal genes in Yersinia enterocolitica. Mol Microbiol 2014; 92:287-301; PMID:24548183; http://dx.doi.org/10.1111/mmi.12554
- Starke M, Richter M, Fuchs TM. The insecticidal toxin genes of Yersinia enterocolitica are activated by the thermolabile LTTR-like regulator TcaR2 at low temperatures. Mol Microbiol 2013; 89:596-611; PMID:23772992; http://dx.doi.org/10.1111/mmi.12296
- Ye F, Brauer T, Niehus E, Drlica K, Josenhans C, Suerbaum S. Flagellar and global gene regulation in Helicobacter pylori modulated by changes in DNA supercoiling. Int J Med Microbiol 2007; 297:65-81; PMID:17276136; http://dx.doi.org/10.1016/j.ijmm.2006.11.006
- Eriksson S, Hurme R, Rhen M. Low-temperature sensors in bacteria. Philos Trans R Soc Lond B Biol Sci 2002; 357:887-93; PMID:12171652; http://dx.doi.org/10.1098/rstb.2002.1077
- Kelly A, Conway C, O Cróinín T, Smith SGJ, Dorman CJ. DNA supercoiling and the Lrp protein determine the directionality of fim switch DNA inversion in Escherichia coli K-12. J Bacteriol 2006; 188:5356-63; PMID:16855224; http://dx.doi.org/10.1128/JB.00344-06
- O Cróinín T, Carroll RK, Kelly A, Dorman CJ. Roles for DNA supercoiling and the Fis protein in modulating expression of virulence genes during intracellular growth of Salmonella enterica serovar Typhimurium. Mol Microbiol 2006; 62:869-82; http://dx.doi.org/10.1111/j.1365-2958.2006.05416.x
- Prosseda G, Falconi M, Giangrossi M, Gualerzi CO, Micheli G, Colonna B. The virF promoter in Shigella: more than just a curved DNA stretch. Mol Microbiol 2004; 51:523-37; PMID:14756791; http://dx.doi.org/10.1046/j.1365-2958.2003.03848.x
- Tendeng C, Bertin PN. H-NS in Gram-negative bacteria: a family of multifaceted proteins. Trends Microbiol 2003; 11:511-8; PMID:14607068; http://dx.doi.org/10.1016/j.tim.2003.09.005
- Tobe T, Yoshikawa M, Mizuno T, Sasakawa C. Transcriptional control of the invasion regulatory gene virB of Shigella flexneri: activation by virF and repression by H-NS. J Bacteriol 1993; 175:6142-9; PMID:7691791.
- Müller CM, Dobrindt U, Nagy G, Emödy L, Uhlin BE, Hacker J. Role of histone-like proteins H-NS and StpA in expression of virulence determinants of uropathogenic Escherichia coli. J Bacteriol 2006; 188:5428-38; PMID:16855232; http://dx.doi.org/10.1128/JB.01956-05
- Nye MB, Pfau JD, Skorupski K, Taylor RK. Vibrio cholerae H-NS silences virulence gene expression at multiple steps in the ToxR regulatory cascade. J Bacteriol 2000; 182:4295-303; PMID:10894740; http://dx.doi.org/10.1128/JB.182.15.4295-4303.2000
- Mühldorfer I, Hacker J, Keusch GT, Acheson DW, Tschäpe H, Kane A V, Ritter A, Olschläger T, Donohue-Rolfe A. Regulation of the Shiga-like toxin II operon in Escherichia coli. Infect Immun 1996; 64:495-502; PMID:8550198.
- Dame RT, Wyman C, Goosen N. Structural basis for preferential binding of H-NS to curved DNA. Biochimie 2001; 83:231-4; PMID:11278073; http://dx.doi.org/10.1016/S0300-9084(00)01213-X
- Dame RT, Wyman C, Wurm R, Wagner R, Goosen N. Structural basis for H-NS-mediated trapping of RNA polymerase in the open initiation complex at the rrnB P1. J Biol Chem 2002; 277:2146-50; PMID:11714691; http://dx.doi.org/10.1074/jbc.C100603200
- Stella S, Falconi M, Lammi M, Gualerzi CO, Pon CL. Environmental control of the in vivo oligomerization of nucleoid protein H-NS. J Mol Biol 2006; 355:169-74; PMID:16303134; http://dx.doi.org/10.1016/j.jmb.2005.10.034
- Ono S, Goldberg MD, Olsson T, Esposito D, Hinton JCD, Ladbury JE. H-NS is a part of a thermally controlled mechanism for bacterial gene regulation. Biochem J 2005; 391:203-13.; .
- Akbar S, Schechter LM, Lostroh CP, Lee CA. AraC/XylS family members, HilD and HilC, directly activate virulence gene expression independently of HilA in Salmonella typhimurium. Mol Microbiol 2003; 47:715-28; PMID:12535071; http://dx.doi.org/10.1046/j.1365-2958.2003.03322.x
- Madrid C, Nieto JM, Paytubi S, Falconi M, Gualerzi CO, Juárez A. Temperature- and H-NS-dependent regulation of a plasmid-encoded virulence operon expressing Escherichia coli hemolysin. J Bacteriol 2002; 184:5058-66; PMID:12193622; http://dx.doi.org/10.1128/JB.184.18.5058-5066.2002
- Johansson J. RNA thermosensors in bacterial pathogens. Contrib Microbiol 2009; 16:150-60; PMID:19494584; http://dx.doi.org/10.1159/000219378
- Kortmann J, Narberhaus F. Bacterial RNA thermometers: molecular zippers and switches. Nat Rev Microbiol 2012; 10:255-65; PMID:22421878; http://dx.doi.org/10.1038/nrmicro2730
- Chowdhury S, Maris C, Allain FH-T, Narberhaus F. Molecular basis for temperature sensing by an RNA thermometer. EMBO J 2006; 25:2487-97; PMID:16710302; http://dx.doi.org/10.1038/sj.emboj.7601128
- Waldminghaus T, Heidrich N, Brantl S, Narberhaus F. FourU: a novel type of RNA thermometer in Salmonella. Mol Microbiol 2007; 65:413-24; PMID:17630972; http://dx.doi.org/10.1111/j.1365-2958.2007.05794.x
- Kortmann J, Sczodrok S, Rinnenthal J, Schwalbe H, Narberhaus F. Translation on demand by a simple RNA-based thermosensor. Nucleic Acids Res 2011; 39:2855-68; PMID:21131278; http://dx.doi.org/10.1093/nar/gkq1252
- Loh E, Memarpour F, Vaitkevicius K, Kallipolitis BH, Johansson J, Sondén B. An unstructured 5’-coding region of the prfA mRNA is required for efficient translation. Nucleic Acids Res 2012; 40:1818-27; PMID:22053088; http://dx.doi.org/10.1093/nar/gkr850
- Giuliodori AM, Di Pietro F, Marzi S, Masquida B, Wagner R, Romby P, Gualerzi CO, Pon CL. The cspA mRNA is a thermosensor that modulates translation of the cold-shock protein CspA. Mol Cell 2010; 37:21-33; PMID:20129052; http://dx.doi.org/10.1016/j.molcel.2009.11.033
- Mao Y, Najafabadi HS, Salavati R. Genome-wide computational identification of functional RNA elements in Trypanosoma brucei. BMC Genomics 2009; 10:355; PMID:19653906; http://dx.doi.org/10.1186/1471-2164-10-355
- Rabani M, Kertesz M, Segal E. Computational prediction of RNA structural motifs involved in posttranscriptional regulatory processes. Proc Natl Acad Sci U S A 2008; 105:14885-90; PMID:18815376; http://dx.doi.org/10.1073/pnas.0803169105
- Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, Repoila F, Buchrieser C, Cossart P, Johansson J. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell 2009; 139:770-9; PMID:19914169; http://dx.doi.org/10.1016/j.cell.2009.08.046
- Waters LSS. Regulatory RNAs in Bacteria. Cell 2009; 136:615-28; PMID:19239884; http://dx.doi.org/10.1016/j.cell.2009.01.043
- Laalami S, Zig L, Putzer H. Initiation of mRNA decay in bacteria. Cell Mol Life Sci 2014; 71:1799-828; PMID:24064983; http://dx.doi.org/10.1007/s00018-013-1472-4
- Gripenland J, Netterling S, Loh E, Tiensuu T, Toledo-Arana A, Johansson J. RNAs: regulators of bacterial virulence. Nat Rev Microbiol 2010; 8:857-66; PMID:21079634; http://dx.doi.org/10.1038/nrmicro2457
- Schuck A, Diwa A, Belasco JG. RNase E autoregulates its synthesis in Escherichia coli by binding directly to a stem-loop in the rne 5’ untranslated region. Mol Microbiol 2009; 72:470-8; PMID:19320830; http://dx.doi.org/10.1111/j.1365-2958.2009.06662.x
- Caron M-P, Lafontaine DA, Massé E. Small RNA-mediated regulation at the level of transcript stability. RNA Biol 7:140-4; PMID:20220305; http://dx.doi.org/10.4161/rna.7.2.11056
- Pichon C, du Merle L, Lequeutre I, Le Bouguénec C. The AfaR small RNA controls expression of the AfaD-VIII invasin in pathogenic Escherichia coli strains. Nucleic Acids Res 2013; 41:5469-82; PMID:23563153; http://dx.doi.org/10.1093/nar/gkt208
- Beauregard A, Smith EA, Petrone BL, Singh N, Karch C, McDonough KA, Wade JT. Identification and characterization of small RNAs in Yersinia pestis. RNA Biol 2013; 10:397-405; PMID:23324607; http://dx.doi.org/10.4161/rna.23590
- Mitobe J, Morita-Ishihara T, Ishihama A, Watanabe H. Involvement of RNA-binding protein Hfq in the post-transcriptional regulation of invE gene expression in Shigella sonnei. J Biol Chem 2008; 283:5738-47; PMID:18156173; http://dx.doi.org/10.1074/jbc.M710108200
- Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol 2011; 9:578-89; PMID:21760622; http://dx.doi.org/10.1038/nrmicro2615
- Chao Y, Vogel J. The role of Hfq in bacterial pathogens. Curr Opin Microbiol 2010; 13:24-33; PMID:20080057; http://dx.doi.org/10.1016/j.mib.2010.01.001
- Fantappiè L, Metruccio MME, Seib KL, Oriente F, Cartocci E, Ferlicca F, Giuliani MM, Scarlato V, Delany I. The RNA chaperone Hfq is involved in stress response and virulence in Neisseria meningitidis and is a pleiotropic regulator of protein expression. Infect Immun 2009; 77:1842-53; http://dx.doi.org/10.1128/IAI.01216-08
- Quade N, Mendonca C, Herbst K, Heroven AK, Ritter C, Heinz DW, Dersch P. Structural basis for intrinsic thermosensing by the master virulence regulator RovA of Yersinia. J Biol Chem 2012; 287:35796-803; PMID:22936808; http://dx.doi.org/10.1074/jbc.M112.379156
- Kamp HD, Higgins DE. A protein thermometer controls temperature-dependent transcription of flagellar motility genes in Listeria monocytogenes. PLoS Pathog 2011; 7:e1002153; PMID:21829361; http://dx.doi.org/10.1371/journal.ppat.1002153
- Rothenbacher FP, Zhu J. Efficient responses to host and bacterial signals during Vibrio cholerae colonization. Gut Microbes 5:120-8; PMID:24256715; http://dx.doi.org/10.4161/gmic.26944
- Johansson J, Mandin P, Renzoni A. An RNA Thermosensor Controls Expression of Virulence Genes in Listeria monocytogenes. Cell 2002; 110:551-61; PMID:12230973; http://dx.doi.org/10.1016/S0092-8674(02)00905-4
- Marceau M. Transcriptional regulation in Yersinia: an update. Curr Issues Mol Biol 2005; 7:151-77; PMID:16053248.
- Han Y, Zhou D, Pang X, Song Y, Zhang L, Bao J, Tong Z, Wang J, Guo Z, Zhai J, et al. Microarray analysis of temperature-induced transcriptome of Yersinia pestis. Microbiol Immunol 2004; 48:791-805; PMID:15557737; http://dx.doi.org/10.1111/j.1348-0421.2004.tb03605.x
- Böhme K, Steinmann R, Kortmann J, Seekircher S, Heroven AK, Berger E, Pisano F, Thiermann T, Wolf-Watz H, Narberhaus F, et al. Concerted Actions of a Thermo-labile Regulator and a Unique Intergenic RNA Thermosensor Control Yersinia Virulence. PLoS Pathog 2012; 8:e1002518; PMID:22359501; http://dx.doi.org/10.1371/journal.ppat.1002518
- Grosdent N, Maridonneau-Parini I, Sory M-P, Cornelis GR. Role of Yops and adhesins in resistance of Yersinia enterocolitica to phagocytosis. Infect Immun 2002; 70:4165-76; PMID:12117925; http://dx.doi.org/10.1128/IAI.70.8.4165-4176.2002
- Cathelyn JS, Ellison DW, Hinchliffe SJ, Wren BW, Miller VL. The RovA regulons of Yersinia enterocolitica and Yersinia pestis are distinct: evidence that many RovA-regulated genes were acquired more recently than the core genome. Mol Microbiol 2007; 66:189-205; PMID:17784909; http://dx.doi.org/10.1111/j.1365-2958.2007.05907.x
- Wagner NJ, Lin CP, Borst LB, Miller VL. YaxAB, a Yersinia enterocolitica pore-forming toxin regulated by RovA. Infect Immun 2013; 81:4208-19; PMID:24002058; http://dx.doi.org/10.1128/IAI.00781-13
- Nagel G, Lahrz A, Dersch P. Environmental control of invasin expression in Yersinia pseudotuberculosis is mediated by regulation of RovA, a transcriptional activator of the SlyA/Hor family. Mol Microbiol 2001; 41:1249-69; PMID:11580832; http://dx.doi.org/10.1046/j.1365-2958.2001.02522.x
- Cathelyn JS, Crosby SD, Lathem WW, Goldman WE, Miller VL. RovA, a global regulator of Yersinia pestis, specifically required for bubonic plague. Proc Natl Acad Sci U S A 2006; 103:13514-9; PMID:16938880; http://dx.doi.org/10.1073/pnas.0603456103
- Heroven AK, Böhme K, Tran-Winkler H, Dersch P. Regulatory elements implicated in the environmental control of invasin expression in enteropathogenic Yersinia. Adv Exp Med Biol 2007; 603:156-66; PMID:17966412; http://dx.doi.org/10.1007/978-0-387-72124-8_13
- Herbst K, Bujara M, Heroven AK, Opitz W, Weichert M, Zimmermann A, Dersch P. Intrinsic thermal sensing controls proteolysis of Yersinia virulence regulator RovA. PLoS Pathog 2009; 5:e1000435; PMID:19468295; http://dx.doi.org/10.1371/journal.ppat.1000435
- Capra EJ, Laub MT. Evolution of two-component signal transduction systems. Annu Rev Microbiol 2012; 66:325-47; PMID:22746333; http://dx.doi.org/10.1146/annurev-micro-092611-150039
- Scarlato V, Aricò B, Prugnola A, Rappuoli R. Sequential activation and environmental regulation of virulence genes in Bordetella pertussis. EMBO J 1991; 10:3971-5; PMID:1718746.
- Mishra M, Parise G, Jackson KD, Wozniak DJ, Deora R. The BvgAS signal transduction system regulates biofilm development in Bordetella. J Bacteriol 2005; 187:1474-84; PMID:15687212; http://dx.doi.org/10.1128/JB.187.4.1474-1484.2005
- Manetti R, Aricò B, Rappuoli R, Scarlato V. Mutations in the linker region of BvgS abolish response to environmental signals for the regulation of the virulence factors in Bordetella pertussis. Gene 1994; 150:123-7; PMID:7959037; http://dx.doi.org/10.1016/0378-1119(94)90870-2
- Ingmer H, Brøndsted L. Proteases in bacterial pathogenesis. Res Microbiol 2009; 160:704-10; PMID:19778606; http://dx.doi.org/10.1016/j.resmic.2009.08.017
- Gophna U, Ron EZ. Virulence and the heat shock response. Int J Med Microbiol 2003; 292:453-61; PMID:12635928; http://dx.doi.org/10.1078/1438-4221-00230
- Guisbert E, Yura T, Rhodius V a, Gross C a. Convergence of molecular, modeling, and systems approaches for an understanding of the Escherichia coli heat shock response. Microbiol Mol Biol Rev 2008; 72:545-54; PMID:18772288; http://dx.doi.org/10.1128/MMBR.00007-08
- Nakahigashi K, Yanagi H, Yura T. Isolation and sequence analysis of rpoH genes encoding sigma 32 homologs from gram negative bacteria: conserved mRNA and protein segments for heat shock regulation. Nucleic Acids Res 1995; 23:4383-90; PMID:7501460.
- Morita MT, Tanaka Y, Kodama TS, Kyogoku Y, Yanagi H, Yura T. Translational induction of heat shock transcription factor sigma32: evidence for a built-in RNA thermosensor. Genes Dev 1999; 13:655-65; PMID:10090722; http://dx.doi.org/10.1101/gad.13.6.655
- Guisbert E, Herman C, Lu CZ, Gross C a. A chaperone network controls the heat shock response in E. coli. Genes Dev 2004; 18:2812-21; PMID:15545634; http://dx.doi.org/10.1101/gad.1219204
- Herman C, Thévenet D, D’Ari R, Bouloc P. Degradation of sigma 32, the heat shock regulator in Escherichia coli, is governed by HflB. Proc Natl Acad Sci U S A 1995; 92:3516-20; PMID:7724592; http://dx.doi.org/10.1073/pnas.92.8.3516
- Parsot C, Mekalanos JJ. Expression of ToxR, the transcriptional activator of the virulence factors in Vibrio cholerae, is modulated by the heat shock response. Proc Natl Acad Sci U S A 1990; 87:9898-902; PMID:2124707; http://dx.doi.org/10.1073/pnas.87.24.9898
- Dobrindt U, Hacker J. Regulation of tRNA5Leu-encoding gene leuX that is associated with a pathogenicity island in the uropathogenic Escherichia coli strain 536. Mol Genet Genomics 2001; 265:895-904; PMID:11523807; http://dx.doi.org/10.1007/s004380100486
- Dobrindt U, Janke B, Piechaczek K, Nagy G, Ziebuhr W, Fischer G, Schierhorn A, Hecker M, Blum-Oehler G, Hacker J. Toxin genes on pathogenicity islands: impact for microbial evolution. Int J Med Microbiol 2000; 290:307-11; PMID:11111903; http://dx.doi.org/10.1016/S1438-4221(00)80028-4
- Hecker M, Schumann W, Völker U. Heat-shock and general stress response in Bacillus subtilis. Mol Microbiol 1996; 19:417-28; PMID:8830234; http://dx.doi.org/10.1046/j.1365-2958.1996.396932.x
- Völker U, Engelmann S, Maul B, Riethdorf S, Völker A, Schmid R, Mach H, Hecker M. Analysis of the induction of general stress proteins of Bacillus subtilis. Microbiology 1994; 140:741-52; PMID:8012595; http://dx.doi.org/10.1099/00221287-140-4-741
- Maul B, Völker U, Riethdorf S, Engelmann S, Hecker M. sigma B-dependent regulation of gsiB in response to multiple stimuli in Bacillus subtilis. Mol Gen Genet 1995; 248:114-20; PMID:7651322; http://dx.doi.org/10.1007/BF02456620
- Bischoff M, Entenza JM, Giachino P. Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus. J Bacteriol 2001; 183:5171-9; PMID:11489871; http://dx.doi.org/10.1128/JB.183.17.5171-5179.2001
- Chan PF, Foster SJ. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J Bacteriol 1998; 180:6232-41; PMID:9829932.
- Chastanet A, Fert J, Msadek T. Comparative genomics reveal novel heat shock regulatory mechanisms in Staphylococcus aureus and other Gram-positive bacteria. Mol Microbiol 2003; 47:1061-73; PMID:12581359; http://dx.doi.org/10.1046/j.1365-2958.2003.03355.x
- De Oliveira NEM, Abranches J, Gaca AO, Laport MS, Damaso CR, Bastos M do C, de F, Lemos JA, Giambiagi-deMarval M. clpB, a class III heat-shock gene regulated by CtsR, is involved in thermotolerance and virulence of Enterococcus faecalis. Microbiology 2011; 157:656-65; PMID:21148206; http://dx.doi.org/10.1099/mic.0.041897-0
- Elsholz AKW, Michalik S, Zühlke D, Hecker M, Gerth U. CtsR, the Gram-positive master regulator of protein quality control, feels the heat. EMBO J 2010; 29:3621-9; PMID:20852588; http://dx.doi.org/10.1038/emboj.2010.228
- Farrand AJ, Reniere ML, Ingmer H, Frees D, Skaar EP. Regulation of host hemoglobin binding by the Staphylococcus aureus Clp proteolytic system. J Bacteriol 2013; 195:5041-50; PMID:23995637; http://dx.doi.org/10.1128/JB.00505-13
- Knudsen GM, Olsen JE, Aabo S, Barrow P, Rychlik I, Thomsen LE. ClpP deletion causes attenuation of Salmonella Typhimurium virulence through mis-regulation of RpoS and indirect control of CsrA and the SPI genes. Microbiology 2013; 159:1497-509; PMID:23676436; http://dx.doi.org/10.1099/mic.0.065797-0
- Meibom KL, Dubail I, Dupuis M, Barel M, Lenco J, Stulik J, Golovliov I, Sjöstedt A, Charbit A. The heat-shock protein ClpB of Francisella tularensis is involved in stress tolerance and is required for multiplication in target organs of infected mice. Mol Microbiol 2008; 67:1384-401; PMID:18284578; http://dx.doi.org/10.1111/j.1365-2958.2008.06139.x
- Chastanet A, Derre I, Nair S, Msadek T. clpB, a novel member of the Listeria monocytogenes CtsR regulon, is involved in virulence but not in general stress tolerance. J Bacteriol 2004; 186:1165-74; PMID:14762012; http://dx.doi.org/10.1128/JB.186.4.1165-1174.2004
- Hanawa T, Fukuda M, Kawakami H, Hirano H, Kamiya S, Yamamoto T. The Listeria monocytogenes DnaK chaperone is required for stress tolerance and efficient phagocytosis with macrophages. Cell Stress Chaperones 1999; 4:118-28; PMID:10547061.
- Salman H, Libchaber A. A concentration-dependent switch in the bacterial response to temperature. Nat Cell Biol 2007; 9:1098-100; PMID:17694049; http://dx.doi.org/10.1038/ncb1632
- Paster E, Ryu WS. The thermal impulse response of Escherichia coli. Proc Natl Acad Sci U S A 2008; 105:5373-7; PMID:18385380; http://dx.doi.org/10.1073/pnas.0709903105
- Maeda K, Imae Y, Shioi JI, Oosawa F. Effect of temperature on motility and chemotaxis of Escherichia coli. J Bacteriol 1976; 127:1039-46; PMID:783127
- Maeda K, Imae Y. Thermosensory transduction in Escherichia coli: inhibition of the thermoresponse by L-serine. Proc Natl Acad Sci U S A 1979; 76:91-5; PMID:370831; http://dx.doi.org/10.1073/pnas.76.1.91
- Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol 2004; 5:1024-37; PMID:15573139; http://dx.doi.org/10.1038/nrm1524
- Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci 2008; 33:9-19; PMID:18165013; http://dx.doi.org/10.1016/j.tibs.2007.09.014
- Nara T, Kawagishi I, Nishiyama S, Homma M, Imae Y. Modulation of the thermosensing profile of the Escherichia coli aspartate receptor tar by covalent modification of its methyl-accepting sites. J Biol Chem 1996; 271:17932-6; PMID:8663384; http://dx.doi.org/10.1074/jbc.271.30.17932
- Tagkopoulos I, Liu Y-C, Tavazoie S. Predictive behavior within microbial genetic networks. Science 2008; 320:1313-7; PMID:18467556; http://dx.doi.org/10.1126/science.1154456
- Ternan NG, Jain S, Srivastava M, McMullan G. Comparative transcriptional analysis of clinically relevant heat stress response in Clostridium difficile strain 630. PLoS One 2012; 7:e42410; PMID:22860125; http://dx.doi.org/10.1371/journal.pone.0042410
- Xu S, Peng Z, Cui B, Wang T, Song Y, Zhang L, Wei G, Wang Y, Shen X. FliS modulates FlgM activity by acting as a non-canonical chaperone to control late flagellar gene expression, motility and biofilm formation in Yersinia pseudotuberculosis. Environ Microbiol 2014; 16:1090-104; PMID:23957589; http://dx.doi.org/10.1111/1462-2920.12222
- Ding L, Wang Y, Hu Y, Atkinson S, Williams P, Chen S. Functional characterization of FlgM in the regulation of flagellar synthesis and motility in Yersinia pseudotuberculosis. Microbiology 2009; 155:1890-900; PMID:19383707; http://dx.doi.org/10.1099/mic.0.026294-0
- Kapatral V, Olson JW, Pepe JC, Miller VL, Minnich SA. Temperature-dependent regulation of Yersinia enterocolitica Class III flagellar genes. Mol Microbiol 1996; 19:1061-71; PMID:8830263; http://dx.doi.org/10.1046/j.1365-2958.1996.452978.x
- Rohde JR, Fox JM, Minnich SA. Thermoregulation in Yersinia enterocolitica is coincident with changes in DNA supercoiling. Mol Microbiol 1994; 12:187-99; PMID:8057844; http://dx.doi.org/10.1111/j.1365-2958.1994.tb01008.x
- Guerry P, Alm RA, Power ME, Logan SM, Trust TJ. Role of two flagellin genes in Campylobacter motility. J Bacteriol 1991; 173:4757-64; PMID:1856171.
- Alm RA, Guerry P, Trust TJ. The Campylobacter sigma 54 flaB flagellin promoter is subject to environmental regulation. J Bacteriol 1993; 175:4448-55; PMID:8331072.
- Wösten MMSM, van Dijk L, Veenendaal AKJ, de Zoete MR, Bleumink-Pluijm NMC, van Putten JPM. Temperature-dependent FlgM/FliA complex formation regulates Campylobacter jejuni flagella length. Mol Microbiol 2010; 75:1577-91; PMID:20199595; http://dx.doi.org/10.1111/j.1365-2958.2010.07079.x
- Johannes L, Römer W. Shiga toxins–from cell biology to biomedical applications. Nat Rev Microbiol 2010; 8:105-16; PMID:20023663.
- Mikulskis A V, Delor I, Thi VH, Cornelis GR. Regulation of the Yersinia enterocolitica enterotoxin Yst gene. Influence of growth phase, temperature, osmolarity, pH and bacterial host factors. Mol Microbiol 1994; 14:905-15; PMID:7715452; http://dx.doi.org/10.1111/j.1365-2958.1994.tb01326.x
- Delor I, Cornelis GR. Role of Yersinia enterocolitica Yst toxin in experimental infection of young rabbits. Infect Immun 1992; 60:4269-77; PMID:1398938.
- Dai Z, Koehler TM. Regulation of anthrax toxin activator gene (atxA) expression in Bacillus anthracis: temperature, not CO2/bicarbonate, affects AtxA synthesis. Infect Immun 1997; 65:2576-82; PMID:9199422.
- Dai Z, Sirard JC, Mock M, Koehler TM. The atxA gene product activates transcription of the anthrax toxin genes and is essential for virulence. Mol Microbiol 1995; 16:1171-81; PMID:8577251; http://dx.doi.org/10.1111/j.1365-2958.1995.tb02340.x
- Sirard JC, Mock M, Fouet A. The three Bacillus anthracis toxin genes are coordinately regulated by bicarbonate and temperature. J Bacteriol 1994; 176:5188-92; PMID:8051039.
- Fouet A. AtxA, a Bacillus anthracis global virulence regulator. Res Microbiol 2010; 161:735-42; PMID:20863885; http://dx.doi.org/10.1016/j.resmic.2010.09.006
- Kazmierczak MJ, Wiedmann M, Boor KJ. Alternative Sigma Factors and Their Roles in Bacterial Virulence. Microbiol Mol Biol Rev 2005; 69:527-43; PMID:16339734; http://dx.doi.org/10.1128/MMBR.69.4.527-543.2005
- Lybecker MC, Samuels DS. Temperature-induced regulation of RpoS by a small RNA in Borrelia burgdorferi. Mol Microbiol 2007; 64:1075-89; PMID:17501929; http://dx.doi.org/10.1111/j.1365-2958.2007.05716.x
- Archambault L, Linscott J, Swerdlow N, Boyland K, Riley E, Schlax P. Translational efficiency of rpoS mRNA from Borrelia burgdorferi: effects of the length and sequence of the mRNA leader region. Biochem Biophys Res Commun 2013; 433:73-8; PMID:23454119; http://dx.doi.org/10.1016/j.bbrc.2013.02.063
- Hämmerle H, Večerek B, Resch A, Bläsi U. Duplex formation between the sRNA DsrA and rpoS mRNA is not sufficient for efficient RpoS synthesis at low temperature. RNA Biol 2013; 10:1834-41.
- Kraiczy P, Skerka C, Brade V, Zipfel PF. Further characterization of complement regulator-acquiring surface proteins of Borrelia burgdorferi. Infect Immun 2001; 69:7800-9; PMID:11705962; http://dx.doi.org/10.1128/IAI.69.12.7800-7809.2001
- Von Lackum K, Miller JC, Bykowski T, Riley SP, Woodman ME, Brade V, Kraiczy P, Stevenson B, Wallich R. Borrelia burgdorferi regulates expression of complement regulator-acquiring surface protein 1 during the mammal-tick infection cycle. Infect Immun 2005; 73:7398-405; PMID:16239539; http://dx.doi.org/10.1128/IAI.73.11.7398-7405.2005
- Lillard JW, Bearden SW, Fetherston JD, Perry RD. The haemin storage (Hms+) phenotype of Yersinia pestis is not essential for the pathogenesis of bubonic plague in mammals. Microbiology 1999; 145(Pt 1):197-209; PMID:10206699; http://dx.doi.org/10.1099/13500872-145-1-197
- Hinnebusch BJ, Perry RD, Schwan TG. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science 1996; 273:367-70; PMID:8662526; http://dx.doi.org/10.1126/science.273.5273.367
- Perry RD, Bobrov AG, Kirillina O, Jones HA, Pedersen L, Abney J, Fetherston JD. Temperature regulation of the hemin storage (Hms+) phenotype of Yersinia pestis is posttranscriptional. J Bacteriol 2004; 186:1638-47; PMID:14996794; http://dx.doi.org/10.1128/JB.186.6.1638-1647.2004
- Porcheron G, Garénaux A, Proulx J, Sabri M, Dozois CM. Iron, copper, zinc, and manganese transport and regulation in pathogenic Enterobacteria: correlations between strains, site of infection and the relative importance of the different metal transport systems for virulence. Front Cell Infect Microbiol 2013; 3:90; PMID:24367764; http://dx.doi.org/10.3389/fcimb.2013.00090
- Kouse AB, Righetti F, Kortmann J, Narberhaus F, Murphy ER. RNA-mediated thermoregulation of iron-acquisition genes in Shigella dysenteriae and pathogenic Escherichia coli. PLoS One 2013; 8:e63781; PMID:23704938; http://dx.doi.org/10.1371/journal.pone.0063781
- Viswanathan VK. Shigella takes the temperature. Gut Microbes 4:267-8; PMID:23851363; http://dx.doi.org/10.4161/gmic.25726
- Xiao R, Kisaalita WS. Iron acquisition from transferrin and lactoferrin by Pseudomonas aeruginosa pyoverdin. Microbiology 1997; 143(Pt 7):2509-15; PMID:9245831; http://dx.doi.org/10.1099/00221287-143-7-2509
- Barbier M, Damron FH, Bielecki P, Suárez-Diez M, Puchałka J, Albertí S, Dos Santos VM, Goldberg JB. From the environment to the host: re-wiring of the transcriptome of Pseudomonas aeruginosa from 22°C to 37°C. PLoS One 2014; 9:e89941; PMID:24587139; http://dx.doi.org/10.1371/journal.pone.0089941
- Schneider M, Exley R. Functional significance of factor H binding to Neisseria meningitidis. J Immunol 2006; 176:7566-75; PMID:16751403; http://dx.doi.org/10.4049/jimmunol.176.12.7566
- Schneider MC, Prosser BE, Caesar JJE, Kugelberg E, Li S, Zhang Q, Quoraishi S, Lovett JE, Deane JE, Sim RB, et al. Neisseria meningitidis recruits factor H using protein mimicry of host carbohydrates. Nature 2009; 458:890-3; PMID:19225461; http://dx.doi.org/10.1038/nature07769
- Loh E, Kugelberg E, Tracy A, Zhang Q, Gollan B, Ewles H, Chalmers R, Pelicic V, Tang CM. Temperature triggers immune evasion by Neisseria meningitidis. Nature 2013; 502:237-40; PMID:24067614; http://dx.doi.org/10.1038/nature12616
- Reinés M, Llobet E, Llompart CM, Moranta D, Pérez-Gutiérrez C, Bengoechea J a. Molecular basis of Yersinia enterocolitica temperature-dependent resistance to antimicrobial peptides. J Bacteriol 2012; 194:3173-88; http://dx.doi.org/10.1128/JB.00308-12
- Li Y, Powell D. LPS remodeling is an evolved survival strategy for bacteria. PNAS 2012; 109:8716-21; PMID:22586119; http://dx.doi.org/10.1073/pnas.1202908109
- Bengoechea JA, Zhang L, Toivanen P, Skurnik M. Regulatory network of lipopolysaccharide O-antigen biosynthesis in Yersinia enterocolitica includes cell envelope-dependent signals. Mol Microbiol 2002; 44:1045-62; PMID:12010497; http://dx.doi.org/10.1046/j.1365-2958.2002.02940.x
- Bartley S, Tzeng Y, Heel K. Attachment and Invasion of Neisseria meningitidis to Host Cells Is Related to Surface Hydrophobicity, Bacterial Cell Size and Capsule. PLoS One 2013; 8:e55798; PMID:23405216; http://dx.doi.org/10.1371/journal.pone.0055798
- Kang SO, Wright JO, Tesorero R a, Lee H, Beall B, Cho KH. Thermoregulation of capsule production by Streptococcus pyogenes. PLoS One 2012; 7:e37367; PMID:22615992; http://dx.doi.org/10.1371/journal.pone.0037367
- Kang SO, Caparon MG, Cho KH. Virulence gene regulation by CvfA, a putative RNase: the CvfA-enolase complex in Streptococcus pyogenes links nutritional stress, growth-phase control, and virulence gene expression. Infect Immun 2010; 78:2754-67; PMID:20385762; http://dx.doi.org/10.1128/IAI.01370-09
- Sturm A, Heinemann M, Arnoldini M. The cost of virulence: retarded growth of Salmonella typhimurium cells expressing type III secretion system 1. PLoS Pathog 2011; 7:1-10; http://dx.doi.org/10.1371/journal.ppat.1002143
- Rhen M, Dorman CJ. Hierarchical gene regulators adapt Salmonella enterica to its host milieus. Int J Med Microbiol 2005; 294:487-502; PMID:15790293; http://dx.doi.org/10.1016/j.ijmm.2004.11.004
- Coombes BK, Brown NF, Valdez Y, Brumell JH, Finlay BB. Expression and secretion of Salmonella pathogenicity island-2 virulence genes in response to acidification exhibit differential requirements of a functional type III secretion apparatus and SsaL. J Biol Chem 2004; 279:49804-15; PMID:15383528; http://dx.doi.org/10.1074/jbc.M404299200
- Deiwick J, Nikolaus T, Erdogan S, Hensel M. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol Microbiol 1999; 31:1759-73; PMID:10209748; http://dx.doi.org/10.1046/j.1365-2958.1999.01312.x
- Duong N, Osborne S, Bustamante VH, Tomljenovic AM, Puente JL, Coombes BK. Thermosensing coordinates a cis-regulatory module for transcriptional activation of the intracellular virulence system in Salmonella enterica serovar Typhimurium. J Biol Chem 2007; 282:34077-84; PMID:17895240; http://dx.doi.org/10.1074/jbc.M707352200
- Beloin C, Dorman CJ. An extended role for the nucleoid structuring protein H-NS in the virulence gene regulatory cascade of Shigella flexneri. Mol Microbiol 2003; 47:825-38; PMID:12535079; http://dx.doi.org/10.1046/j.1365-2958.2003.03347.x
- Watanabe H, Arakawa E, Ito K, Kato J, Nakamura A. Genetic analysis of an invasion region by use of a Tn3-lac transposon and identification of a second positive regulator gene, invE, for cell invasion of Shigella sonnei: significant homology of invE with ParB of plasmid P1. J Bacteriol 1990; 172:619-29; PMID:1688841
- Choy HA. Multiple activities of LigB potentiate virulence of Leptospira interrogans: inhibition of alternative and classical pathways of complement. PLoS One 2012; 7:e41566; PMID:22911815; http://dx.doi.org/10.1371/journal.pone.0041566
- Choy HA, Kelley MM, Croda J, Matsunaga J, Babbitt JT, Ko AI, Picardeau M, Haake DA. The multifunctional LigB adhesin binds homeostatic proteins with potential roles in cutaneous infection by pathogenic Leptospira interrogans. PLoS One 2011; 6:e16879; PMID:21347378; http://dx.doi.org/10.1371/journal.pone.0016879
- Matsunaga J, Schlax PJ, Haake DA. Role for cis-acting RNA sequences in the temperature-dependent expression of the multiadhesive lig proteins in Leptospira interrogans. J Bacteriol 2013; 195:5092-101; PMID:24013626; http://dx.doi.org/10.1128/JB.00663-13
