Abstract
Periodontal infection involves a complex interplay between oral biofilms, gingival tissues and cells of the immune system in a dynamic microenvironment. A humanized in vitro model that reduces the need for experimental animal models, while recapitulating key biological events in a periodontal pocket, would constitute a technical advancement in the study of periodontal disease. The aim of this study was to use a dynamic perfusion bioreactor in order to develop a gingival epithelial-fibroblast-monocyte organotypic co-culture on collagen sponges. An 11 species subgingival biofilm was used to challenge the generated tissue in the bioreactor for a period of 24 h. The histological and scanning electron microscopy analysis displayed an epithelial-like layer on the surface of the collagen sponge, supported by the underlying ingrowth of gingival fibroblasts, while monocytic cells were also found within the sponge mass. Bacterial quantification of the biofilm showed that in the presence of the organotypic tissue, the growth of selected biofilm species, especially Campylobacter rectus, Actinomyces oris, Streptococcus anginosus, Veillonella dispar, and Porphyromonas gingivalis, was suppressed, indicating a potential antimicrobial effect by the tissue. Multiplex immunoassay analysis of cytokine secretion showed that interleukin (IL)-1 β, IL-2, IL-4, and tumor necrosis factor (TNF)-α levels in cell culture supernatants were significantly up-regulated in presence of the biofilm, indicating a positive inflammatory response of the organotypic tissue to the biofilm challenge. In conclusion, this novel host-biofilm interaction organotypic model might resemble the periodontal pocket and have an important impact on the study of periodontal infections, by minimizing the need for the use of experimental animal models.
Introduction
The two major forms of periodontal diseases are gingivitis and periodontitis. Approximately 80% of the world population exhibit gingivitis,Citation1 while 15% may exhibit severe destructive periodontitis.Citation2 Periodontitis is the primary cause of adult tooth loss due to its destructive effect on the periodontal connective tissues and underlying alveolar bone.Citation3 Besides, recent evidence shows that the effects of periodontal infections may well expand beyond the oral cavity, to be implicated in systemic diseases, such as diabetes mellitus and cardiovascular diseases.Citation4,5
The causation of these infections involves the complex interaction between biofilm communities forming on the tooth surface, and the neighboring host periodontal tissues.Citation6 Biofilms are polymicrobial communities embedded in a dense extracellular matrix produced by the different constituent species.Citation6 More than 700 speciesCitation7-9 have already identified in the oral cavity that may colonize the biofilms, but the actual number could be even higher, due to the limitations in detection by the contemporary technologies.Citation10 As constituents of the biofilm, bacteria can better withstand environmental stresses and receive metabolic benefits from each other.Citation6 The health-associated biofilms do not trigger a strong inflammatory response. In fact, some studies indicate that these biofilms actually benefit gingival health, by locally boosting the immune systems.Citation9 However, a biofilm could start to trigger inflammation when the composition of bacterial community shifts to a more pathogenic one.Citation11,12
In response to the biofilm accumulation, the host tissue produces an inflammatory response for the recruitment of immune cells (e.g. polymorphonuclear leukocytes, monocytes),Citation3 and secretes a number of small compounds of the immune system (e.g antibodies, cytokines and prostaglandins). If the inflammation tends to become chronic and the biofilm fails to be eliminated, the network of inflammatory molecules that has been established can cause connective tissue damage and bone destruction.Citation3,13 In some severe periodontitis cases, this may eventually lead to tooth loss if the inflammation remains unresolved.Citation14
Several models have been used to understand mechanisms underlying periodontal infections, however, none of them appears to be reliable and reproducible. Experimental animals models of oral infection cannot efficiently represent human oral pathogenic bacteria,Citation15,16 whereas human experimental studies may answer questions regarding the initiation of these diseases,Citation17 but difficult to identify mechanisms that convert protective inflammation to tissue destructive lesion, due to ethical considerations. Most studies using in vitro models have only employed single oral bacterial species to challenge 2-dimensional monolayer cells,Citation18,19 or cells in suspension,Citation20-22 despite that periodontal infections are biofilm-related.Citation23 Moreover, monolayer cell culture systems do not adequately mimic the morphological and functional features of primary gingival tissues.Citation24
Previous studies have shown 3D organotypic culture systems to mimic the in vivo situation better than 2D monolayer cultures.Citation25,26 It is known that epithelial cells differentiate to their nature state in primary tissue with the support of underlying fibroblasts in the connective tissue.Citation25,27 Therefore, 3D models using both epithelial cells and fibroblasts are preferable for the study of gingival tissues.Citation28-32 Besides, during chronic periodontal inflammation, it is possible that part of the epithelial layer of the tissue is degraded and the underlying fibroblast-comprised connective tissue may directly confront the oral biofilm. Indeed, fibroblasts are also actively participating in bacterially-induced inflammation,Citation33,34 and there are studies available that investigate their interaction with biofilms in vitro.Citation35-37
It is technically very challenging to reconstitute multi-tissue structures, such as gingiva, in in vitro cultures. Bioreactors, including perfusion systems, have been developed and used for in vitro modeling of various other infections including the lung,Citation38 the intestine,Citation39,40 and the colon.Citation41,42 Such a system could mimic the natural environment at local sites (e.g., temperature, atmosphere, shear stress), allowing for the study of biofilm-related mucosal disease under more realistic conditions. Yet, such a system has not been so far implemented in the field of periodontal infections. Apart from structural cells (i.e. fibroblasts and epithelial cells), cells of the immune system, such as monocytes can be readily incorporated into a perfusion bioreactor system, for the study of the biofilm challenge in an environment simulating the established periodontal pocket.
The aim of this study was to develop a gingival organotypic tissue consisting of gingival epithelial cells, fibroblasts and monocytic cells, in a dynamic environment, which was achieved with the use of a perfusion bioreactor system. An 11-oral (subgingival) species biofilm grown on hydroxyapatite discs, which was established in our laboratory based on the previous models,Citation36,43-46 provides a model closer to the in vivo situation than single or few bacterial species. This was used to challenge the generated organotypic gingival tissue within the bioreactor, in a low oxygen environment, thus validating the usability of this organotypic tissue. It is anticipated that this novel dynamic tissue-biofilm interaction model may mimic the periodontal pocket environment and thus provide a more accurate in vitro experimental platform for the study of periodontal infections.
Results
Histological analysis of the organotypic tissue on the collagen sponge
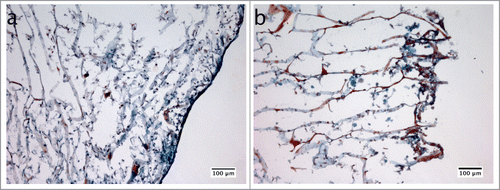
The GFB-16, HGEK-16, and Mono-Mac-6 cells were seeded on the collagen sponges under the U-CUP perfusion bioreactor to construct 3D in vitro gingival tissue as described in materials and methods. The biofilm challenge was later introduced to the system for 24 h, to mimic periodontal infection. The morphology of the 3D model was evaluated by Masson's Trichrome stain (). In the control group whereby the biofilm was absent (), there was a consistent epithelial-like layer of cells formed on the surface of the collagen, while inside the collagen structure, cells with long protrusions spread along the collagen fibers were observed. In the presence of biofilms (), the epithelial-like layer was disrupted, with the appearance of many rounded big cells next to the collagen surface, that were not detectable in the control group.
Scanning electron microscopy (SEM) analysis of the organotypic tissue surface
SEM was used to analyze the structure of the collagen sponge surfaces in contact with the biofilms. As seen from this analysis, the collagen sponge that served as a scaffold displayed a regular mesh of layers (). When seeded onto the collagen sponge, cells filled most gaps between collagen fibers and formed a dense structure (). In the collagen sponge, on which the biofilm was seeded (in the absence of host cells), clumps of predominately coccoid bacteria were observed in conjunction to the collagen fibers, and all over the mass of the sponge (; Fig. S1). Such clumps of bacterial colonies were not found when the biofilms were co-cultured in the presence of the 3D gingival tissue, despite the presence of many large, monocyte-like cells (), rather than the layered cells that were previously observed when the cells were seeded alone ().
Quantification of biofilms
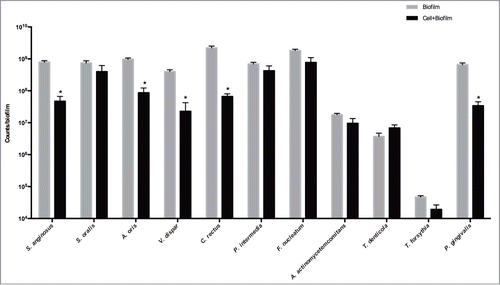
After 24 h in co-culture with the organotypic gingival tissue, the bacterial composition of the biofilm was evaluated and compared to that of the biofilm in culture with the collagen sponge alone (). Significantly different numbers between the two groups were detected for C. rectus, A. oris, S. anginosus, V. dispar, and P. gingivalis (33.28-fold, P = 0.0002; 11.09-fold, P = 0.0067; 16.65-fold, P = 0.0036; 17.46-fold, P = 0.0266; and 19.28- fold, P = 0.0014; respectively). Other differences did not prove to be statistically significant, hence the numbers of the remaining species were not affected.
Figure 3. SEM images of original collagen sponge structures (a), collagen sponges on which the 3D in vitro gingival tissue had been reconstructed (b), collagen sponges in the presence of biofilms only (c), or collagen sponges on which the 3D in vitro gingival tissue had been reconstructed and challenged with the biofilms for 24 h (d).
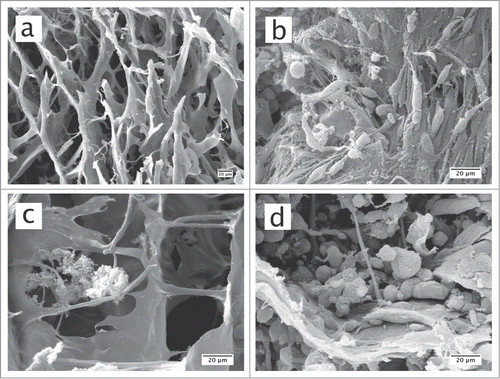
Figure 4. Quantification of bacterial numbers in biofilms. The biofilms were either cultured on blank collagen sponges (biofilm), or co-cultured with the 3D in vitro gingival tissue for 24 h (biofilm + cell). These values represent mean values of triplicate experiments ± standard deviation (SD). The results are presented on a logarithmic scale. Asterisk (*) represents the significance of differences (P ≤ 0.05) between the 2 groups.
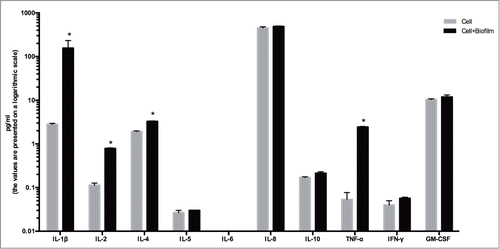
Quantification of cytokines secretion
Finally, the level of cytokines in the culture medium was investigated by the use of a 10-plex immunoassay platform (). After 24 h of challenge, the biofilm caused a significant increase in the production of IL-1β, IL-2, IL-4, and TFN-α by the organotypic gingival tissue, compared to the unchallenged control tissue alone (56.01-fold, P = 0.0011; 6.94-fold, P < 0.0001; 1.70-fold, P < 0.0001; and 45.63- fold, P = 0.0006; respectively). This finding confirms the inflammatory-inducing capacity of the biofilm on the organotypic gingival tissue.
Figure 5. Quantification of cytokine secretion. The 3D in vitro gingival tissue were cultured either in the presence of only pellicle coated hydroxyapatite discs (cell) or co-cultured with biofilm-grown hydroxyapatite discs (biofilm + cell) for 24 h. These values represent mean values of triplicate experiments ± standard deviation (SD). The results are presented on a logarithmic scale. Asterisk (*) represents the significance of differences (P ≤ 0.05) between the 2 groups.
Discussion
In this study, we developed an organotypic co-culture system for the study of interactions between gingival tissues and subgingival biofilms in a U-CUP perfusion bioreactor device. The model shows compatible histological features to a periodontal pocket in vivo and utilizes a biofilm in order to challenge a multi-layered epithelial-fibroblast tissue, constructed using previously established immortalized human gingival cells,Citation32 and enriched further with monocytic cells.
As first line of the host defense, gingival tissue provides an epithelial layer firmly aligned onto the tooth surface.Citation47 Many approaches have been applied to create such an epithelial structure in vitro, for the study of the mechanisms of periodontal diseases. Moreover, the gingival epithelium is supported by the gingival connective tissue,Citation47 which consist of fibroblasts that produce a firm collagen network and produce important growth factors for epithelial survival and growth.Citation33,48 Since Tomakidi et alCitation49 developed an oral gingival model constituted epithelial cell with underlying fibroblasts in the collagen gel, many similar models were established for the study of gingival and oral mucosa.Citation28-30,50,51 However, one of the main challenges to apply such model in the periodontal infection is the incorporation of immune cells in a dynamic environment of continuous flow. It should be noted that at least 2–5% of the total surface of the gingiva that attach to the tooth (i.e., junctional epithelium) is occupied by intercellular spaces.Citation52 This looser structure allows gingival crevicular fluid (GCF) to pass through the periodontal pocket. This carries primarily transmigrating neutrophilic granulocytes and infiltrating mononuclear cells, like macrophages and lymphocytesCitation53,54 to the tissue against which the biofilm is established over time.
In our model, collagen sponges were used to create such a space while allowing gingival fibroblast and epithelial cells to attach onto them. Thus, a 3-phase seeding protocol was developed, ensuring that an epithelial layering and underlying connective tissue is formed, while maintaining constant flow of culture medium and cells of the immune cell. This system was microbially-challenged with an in vitro generated subgingival biofilm, grown onto hydroxyapatite surface (analogous to the tooth surface). Previous experimental models have already used single bacteria species, including A. actinomycetemcomitans, F. nucleatum, to challenge an epithelial-fibroblasts co-cultured model.Citation55-57 Earlier works in our group have used this subgingival biofilm model in static co-culture either with gingival fibroblastsCitation36 or with multilayer gingival organotypic epithelium, in order to study various aspects of host tissue-oral biofilm interaction.Citation58,59
As most subgingival species are strict anaerobes,Citation6 they were grown separately under anaerobic conditions, prior to being introduced into the bioreactor environment along with the organotypic tissue. It should be noted that, in vivo, diseased human gingival tissues present in 2 % oxygen and are exposed to shear forces by saliva and gingival exudate at flow rate of 0.4–2.0 ml/min.Citation60 These microenvironment conditions were also mimicked within the present bioreactor model, demonstrating that both the organotypic tissue and the bacterial biofilm are able to survive, co-exist and interact during the given experimental period of 24 h. The numbers of some bacterial species (namely S. anginosus, A. oris, V. dispar, C. rectus and P. gingivalis) were suppressed in the presence of the organotypic tissue, compared to the collagen sponge alone. Accordingly, bacterial aggregates were sparsely detectable on cells-seeded collagen surfaces. This potentially indicates the antimicrobial capacity of the tissue cells and/or the involvement of the immune cells in controlling bacterial colonization in the present experimental model.
To further evaluate the in vivo relevance of this model, selected secreted cytokines were measured by multiplex immunoassay technology. We detected the up-regulation of the pro-inflammatory cytokines IL-1β, IL-2, IL-6, and TNF-α on the secreted bioreactor medium (analogous to GCF), after 24 h of challenge of the tissue with the biofilm. Indeed, several studies have demonstrated that patients with periodontitis also display higher concentrations of IL-1β,Citation61,62 IL-2,Citation61 IL-6,Citation63,64 and TNF-α62 in GCF, compared to healthy gingiva. The regulation of cytokines or chemokines in gingival tissues is a complex process that involves many cells of the immune system,Citation20,65-67 or structural tissue cells.Citation68,69 To further enhance this model, additional elements could be introduced into this system, such as neutrophils and B- or T-lymphocytes, following the same protocol as with the Mono-Mac-6 cell lines.
In clinical studies,Citation70 increasing number of T. forsythia, P. gingivalis, P. intermedia, and F. nucleatum correlated with an upregulation of IL-1β, while increasing number of S. oralis, and S. anginosus correlated with up-regulation the TNF-α, which may be in line with our observations in this in vitro experimental system. With regards to in vitro studies, the effects of periodontal bacteria are more representative when part of biofilm communities. Hence, the comparison of the effects of 2 biofilms with slightly altered bacterial composition can be useful for evaluating the relative involvement of different species in tissue cytokine responses.Citation59
In conclusion, we have developed a novel model system to resemble the periodontal pocket, which allows for the simultaneous study of the interaction between oral biofilms, gingival tissues and cells of the immune system, in a biologically relevant environment. To our knowledge, this is the first model that utilized a perfusion bioreactor to study periodontal infection in vitro, and its establishment could also have an impact in the study of other biofilm-related diseases. Moreover, this in vitro system may reduce the need for usage of experimental animal models in periodontal research.
Materials and Methods
Cell culture
The development of the present 3D cell culture model required a combination of gingival epithelial keratinocytes, gingival fibroblasts and monocytic cells. Immortalized human gingival epithelial keratinocytes (HGEK-16) and immortalized human gingival fibroblasts (GFB-16) were established by transducing E6/E7 oncoproteins from human papillomavirus type 16 (HPV16-E6/E7) to primary cells.Citation32 HGEK-16s were maintained in complete epithelial medium (CM-DKSFM) consisting of defined keratinocyte serum free medium (Gibco, 10744-019), supplemented with 100 U/ml penicillin (Sigma, 15140-122), 100 μg/ml streptomycin (Sigma, 15140-122), 2 mM L-glutamine (Sigma, G7513), and 0.25 μg/ml fungizone (Sigma, 15290-018). GFB-16s were maintained in complete fibroblasts medium (CM-DF12) Dulbecco's modified Eagle medium consisting of nutrient mixture F-12 media (Sigma-Aldrich, D8900-10L), supplemented with 1.2 g/ml of Na-Bicarbonate (Sigma-Aldrich, S5761), 10 % PANSera FBS special designed for ES cells (FBS, Bioswisstec AG, 2602), 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine, and 0.05 μg/ml fungizone.
The human myelomonocytic cell line Mono-Mac-6 obtained from the German Collection of Microorganisms and Cell Cultures (Mascheroder, Braunschweig, Germany) was maintained in complete myelomonocytic medium (CM-RPMI) consisting of RPMI-glutamax (Gibco, 72400-021), supplemented with 10% fetal bovine serum, 1% non-essential amino acids (Gibco, 11140-035), 1% sodium pyruvate (Gibco, 11360-039) and 9 μg/ml bovine insulin (Sigma, 8418) 100 U/ml penicillin, 100 μg/ml streptomycin, and 0.05 μg/ml fungizone.
In vitro biofilm formation
An 11 species biofilm was established based on a previously developed Zürich 10 species subgingival biofilm model.Citation43,71 The following strains were used included in this model: Prevotella intermedia ATCC 25611T (OMZ 278), Aggregatibacter actinomycetemcomitans JP2 (OMZ 295), Campylobacter rectus (OMZ 398), Veillonella dispar ATCC 17748T (OMZ 493), Fusobacterium nucleatum subsp. nucleatum (OMZ 598), Streptococcus oralis SK248 (OMZ 607), Treponema denticola ATCC 35405T (OMZ 661), Actinomyces oris (OMZ 745), Streptococcus anginosus ATCC 9895 (OMZ 871), Tannerella forsythia (OMZ 1047) and Porphyromonas gingivalis W50 (OMZ 308). Briefly, 200 μl of bacterial cell suspensions containing equal densities (OD550 = 1.0) of each strain were mixed with 1.6 ml of growth medium consisting 60% saliva,Citation72 10% human serum, 30%mFUM,Citation73 and 0.5% hemin to initiate biofilm formation on hydroxyapatite discs (diameter 13 mm) (Clarkson Chromatography Products, custom made), pre-coated for 4 h with saliva, which was diluted 1:2 in a mixture of 0.9% NaCl and distilled water. A ring that is slightly larger than hydroxyapatite disc was sheathed around the veneer of the disc. The diameter of inner core at one end of the ring is smaller than 13 mm, therefore able to seal the hydroxyapatite disc inside. This end of the ring is around 1 mm longer than the surface of the hydroxyapatite disc, creating a gap between the biofilm surface and the bottom of the well. The discs were incubated for 64 h in anaerobic conditions, with medium renewed at 16 h and 40 h. During the first medium renewal, additional 40 μl of T. denticola (OD550 = 1.0) were further added to each well. The discs were dip-washed in saline at 16 h, 20 h, 24 h, 40 h, 44 h, 48 h and 64 h.
Development of the 3D organotypic culture system in bioreactor
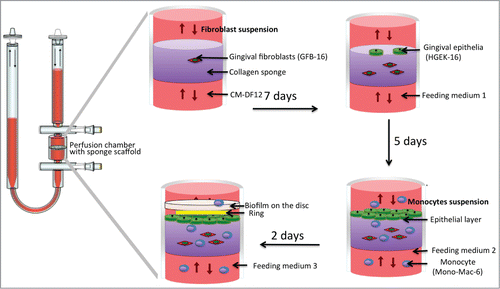
The 3D organotypic culture was established using 3D collagen sponge (porcine collagen, type I) scaffolds (Optimaix, Matricel GmbH, O3D304030) cut into a disc shape with 8 mm in diameter and 3 mm thick in a perfusion bioreactor (UCUP, Cellec Biotek AG, www.cellecbiotek.com, UCUP001) based on a previously developed bioreactor system.Citation74,75 Briefly, the scaffold was embedded within a pair of provided adaptors and placed inside the bioreactor chamber, in order to constrain the cell/medium suspension passing through its pores, avoiding thus convectional flow (). The flow rate was controlled by the PHD Ultra Syringe Pump (included in the U-CUP bioreactor package).
The protocol for the development of the 3D co-culture in the bioreactor is schematically shown in . Initially, 2 × 106 GFB-16 cells suspended in CM-DF12 were perfused through each Ø 8 × 3 mm 3D collagen sponge scaffold, at a superficial velocity of 1000 μm/sec for 15 h, followed by 100 um/sec, as previously has been demonstrated to maximize the seeding efficiency of porous scaffold materials and allow subsequent tissue development.Citation75,76 After 7 days, the bioreactor chambers were opened under sterile hood and 1 × 106 HGEK-16 cells were statically seeded on the top of same 3D collagen sponge in order to form an epithelial layer on its surface. Cells were incubated in feeding medium 1 (CM-DKSFM: CM-DF12 = 1:1) for 3 h to allow cell attachment before initiating again the perfusion at a velocity of 100 μm/sec. After 5 days, 2 x 10Citation6 Mono-Mac-6 cells were injected into the bioreactor system and continued the perfusion at a velocity of 100 um/sec, in feeding medium 2 (CM-DMEM: CM-DF12: CM-RPMI = 1:1:1). After 2 days, the sponge was washed 3 times with feeding medium 3 (antibiotic free CM-DMEM: CM-DF12: CM-RPMI = 1:1:1) to remove any remaining antibiotic, prior to the addition of the biofilm in the system (description in the next section). The removed medium was also retained in order to collect the Mono-Mac-6 cells by centrifugation, and thereafter added back to the system with feeding medium 3. The cultures were fed every 2 d during this process, before being brought in co-culture with the biofilm.
In order to mimic the conditions of periodontal infection, hydroxyapatite discs with or without (control) the biofilms were introduced into the bioreactor chambers. Briefly, the bioreactor chambers were opened under sterile conditions in tissue culture, and the hydroxyapatite discs were placed by press-fit within the provided adaptors, facing the epithelial surface of the generated 3D organotypic tissue, or the collagen sponge alone (control). The chambers were closed again and perfusion flow continued at a flow rate of 135 μm/sec in feeding medium 3 for additional 24 h. The culture temperature was 37°C and the atmospheric conditions 2% O2 and 5% CO2.
Biofilm harvesting and quantification by real-time quantitative polymerase chain reaction (qPCR)
For the evaluation of the bacterial composition of the biofilm after 24 h of co-cultured, the discs were removed from the bioreactor and vigorously vortexed for 3 min with 1 ml 0.9 % NaCl, and sonicated at 25 W for 5 seconds. The bacterial suspensions were stored at 20°C before being processed for real-time quantitative PCR (qPCR) analysis. The DNA was extracted from bacterial suspensions using the GenElute™ bacterial genomic DNA kit (Sigma-Aldrich, NA2110) following the gram-positive lysis protocol of manufacturer's guideline with expanded lysis steps from 30 min to 1 h (lysozyme/mutanolysin step), and from 10 min to 30 min (proteinase K step). All primers were designed using online NCBI/ Primer-BLAST tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast) targeted 16S rRNA gene (). The qPCR runs were performed using SYBR® Green PCR Master Mix (Life Technologies, 4309155) in a StepOnePlus™ Real-Time PCR Systems (Applied Biosystems) at 95°C for 10 min, 40 cycles of 95°C for 15 s and 60°C for 1 min. Standard curves were generated using DNA extracted from planktonic cultures of each of the 11 species used in biofilm, and the abundances were calculated using the theoretical genome weight of each organism from each strain according to the NCBI database, as previously described.Citation44
Table 1 Primer sequences and properties
Quantification of cytokine secretion
Upon completion of the experiments, the flow culture medium was collected from bioreactors, and centrifuged at 1500 rpm for 5 min before being filtered through 0.2 μm pore size, and stored at −80°C for further uses. The Cytokine Human Ultrasensitive 10-Plex Panel (Novex®, Thermoscientific, LHC6004) was used to quantify cytokines in the collected medium, according to the manufacturer's instructions. The following cytokines were tested in this assay: Interleukin (IL)-1β, IL-2, IL-4, IL-8, IL-5, IL-6, IL-10, Interferon gamma (IFN-γ), Tumor necrosis factor α (TNF-α), and granulocyte-macrophage colony-stimulating factor (GM-CSF) with sensitivities more than 0.05, 0.05, 0.1, 0.05, 0.1, 0.5, 0.05, 0.1, 0.5, and 0.1 pg/ml, respectively. These cytokines were detected with premixed antibody beads, read on a Luminex®200 system, and analyzed by xPONENT® software (Thermoscientific).
Harvesting of organotypic culture and histological analysis by Masson's Trichrome staining
The collagen sponges containing the generated 3D organotypic gingival tissue were removed from the bioreactor once the experiment was completed. They were then fixed with 3% paraformaldehyde in 2% sucrose/PBS buffer for 2 h, washed 3 × 20 min in PBS, dehydrated in 15% sucrose/PBS for 2 h, and kept in 30% sucrose/PBS overnight at 4°C.
Slices of harvested sponges were introduced into aluminum foil molds filled with 5% gelatin and 5% sucrose solution, before snap frozen in cold 2-methylbutane (around −40°C by dry ice). The frozen blocks were sectioned into 16 mm and transferred to Superfrost® Plus slides (Thermoscientific, 10143352). The slides were dried in room temperature overnight. They were stained with Weigert's iron hematoxylin (Sigma-Aldrich, 03979) for 20 sec, washed in tap water for 2 min, and rinsed in distilled water, stained with solution A (0.5 g acid fuchsin (Sigma-Aldrich, 857343), 0.5 g ponceau sylidine (Sigma-Aldrich, 81465), 1 ml glacial acetic acid (Sigma-Aldrich, 537020), and 99 ml distilled water) for 15 min, rinsed with distilled water, stained with solution B (1 g phosphomolybdic acid (Sigma-Aldrich, 79560) in 100 ml distilled water) for 5 min, rinsed with distilled water, stained with solution C (2 g light green SF yellowish (Sigma-Aldrich, 62110) and 2 ml glacial acetic acid in distilled water to a final volume of 100 ml) for 1 min, rinsed with distilled water, and dehydrated with 100% ethanol.
Scanning electron microscopy
For SEM analysis of the 3D culture, slices of harvested collagen sponges were introduced into 0.185 M Sodium cacodylate buffer for one day, dissected and left for 1 day in cacodylate before being immersed in 2.5% glutaraldehyde in cacodylate for 45 min, washed in cacodylate for 1 h, 5 min in H2O, and then stored at 4°C overnight in 50% acetone. The samples were later dehydrated in a graded series of acetone, dried in CO2, and thereafter mounted on aluminum stubs and sputter-coated with 4 nm of platinum. The samples were imaged using a TESCAN VEGA TS 5136 XM scanning electron microscope (Tescan) at 25 KV.
Statistical evaluation
All data present in the experiment derives from triplicate biofilm cultures. For the bacterial determination by qPCR, the values were logarithmically transformed, and then inserted to Prism v.6 software (GraphPad, La Jolla California USA). The statistical differences (P ≤ 0.05) between the groups were calculated by student t-test. For the determination of cytokine concentrations, the statistical differences (P ≤ 0.05) between the groups were calculated by student t-test.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
978721_Supplementary_Materials.zip
Download Zip (3.1 MB)Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Albandar JM. Underestimation of periodontitis in NHANES surveys. J Periodontol 2011; 82:337-41; PMID:21214340; http://dx.doi.org/10.1902/jop.2011.100638
- Brown LJ, Loe H. Prevalence, extent, severity and progression of periodontal disease. Periodontol 2000 1993; 2:57-71; PMID:9673181; http://dx.doi.org/10.1111/j.1600-0757.1993.tb00220.x
- Ohlrich EJ, Cullinan MP, Seymour GJ. The immunopathogenesis of periodontal disease. Aust Dental J 2009; 54 Suppl 1:S2-10; PMID:19737265; http://dx.doi.org/10.1111/j.1834-7819.2009.01139.x
- Kebschull M, Demmer RT, Papapanou PN. "Gum bug, leave my heart alone!"–epidemiologic and mechanistic evidence linking periodontal infections and atherosclerosis. J Dent Res 2010; 89:879-902; PMID:20639510
- Belibasakis GN, Bostanci N. Inflammatory and bone remodeling responses to the cytolethal distending toxins. Cells 2014; 3:236-46; PMID:24709959; http://dx.doi.org/10.3390/cells3020236
- Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontol 2000 2005; 38:135-87; PMID:NOT_FOUND; http://dx.doi.org/10.1111/j.1600-0757.2005.00107.x
- Kuramitsu HK, He X, Lux R, Anderson MH, Shi W. Interspecies interactions within oral microbial communities. Microbiol Mol Biol Rev 2007; 71:653-70; PMID:18063722; http://dx.doi.org/10.1128/MMBR.00024-07
- Rosan B, Lamont RJ. Dental plaque formation. Microbes Infect 2000; 2:1599-607; PMID:11113379
- Jenkinson HF, Lamont RJ. Oral microbial communities in sickness and in health. Trends Microbiol 2005; 13:589-95; PMID:16214341; http://dx.doi.org/10.1016/j.tim.2005.09.006
- Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol 2005; 43:5721-32; PMID:16272510; http://dx.doi.org/10.1128/JCM.43.11.5721-5732.2005
- Sbordone L, Bortolaia C. Oral microbial biofilms and plaque-related diseases: microbial communities and their role in the shift from oral health to disease. Clinl Oral Investig 2003; 7:181-8; PMID:14598129; http://dx.doi.org/10.1007/s00784-003-0236-1
- Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nature Rev Microbiol 2010; 8:481-90; PMID:20514045; http://dx.doi.org/10.1038/nrmicro2337
- Graves D. Cytokines that promote periodontal tissue destruction. J Periodontol 2008; 79:1585-91; PMID:18673014
- Hirschfeld L, Wasserman B. A long-term survey of tooth loss in 600 treated periodontal patients. J Periodontol 1978; 49:225-37; PMID:277674; http://dx.doi.org/10.1902/jop.1978.49.5.225
- Graves DT, Fine D, Teng YT, Van Dyke TE, Hajishengallis G. The use of rodent models to investigate host-bacteria interactions related to periodontal diseases. J Clin Periodontol 2008; 35:89-105; PMID:18199146; http://dx.doi.org/10.1111/j.1600-051X.2007.01172.x
- McMahon KT, Wasfy MO, Yonushonis WP, Minah GE, Falkler WA, Jr. Comparative microbiological and immunological studies of subgingival dental plaque from man and baboons. J Dent Res 1990; 69:55-9; PMID:2303597; http://dx.doi.org/10.1177/00220345900690010901
- Syed SA, Loesche WJ. Bacteriology of human experimental gingivitis: effect of plaque age. Infec Immun 1978; 21:821-9; PMID:711336
- Reddi D, Bostanci N, Hashim A, Aduse-Opoku J, Curtis MA, Hughes FJ, Belibasakis GN. Porphyromonas gingivalis regulates the RANKL-OPG system in bone marrow stromal cells. Microbes Infect 2008; 10:1459-68; PMID:18789397; http://dx.doi.org/10.1016/j.micinf.2008.08.007
- Reddi D, Brown SJ, Belibasakis GN. Porphyromonas gingivalis induces RANKL in bone marrow stromal cells: involvement of the p38 MAPK. Microb Pathog 2011; 51:415-20; PMID:21939752; http://dx.doi.org/10.1016/j.micpath.2011.09.001
- Hamedi M, Belibasakis GN, Cruchley AT, Rangarajan M, Curtis MA, Bostanci N. Porphyromonas gingivalis culture supernatants differentially regulate interleukin-1beta and interleukin-18 in human monocytic cells. Cytokine 2009; 45:99-104; PMID:19091595; http://dx.doi.org/10.1016/j.cyto.2008.11.005
- Bostanci N, Reddi D, Rangarajan M, Curtis MA, Belibasakis GN. Porphyromonas gingivalis stimulates TACE production by T cells. Oral Microbiology Iimmunol 2009; 24:146-51; PMID:19239642; http://dx.doi.org/10.1111/j.1399-302X.2008.00488.x
- Belibasakis GN, Reddi D, Bostanci N. Porphyromonas gingivalis induces RANKL in T-cells. Inflammation 2011; 34:133-8; PMID:20446027; http://dx.doi.org/10.1007/s10753-010-9216-1
- Jenkinson HF. Beyond the oral microbiome. Environ Microbiol 2011; 13:3077-87; PMID:21906224; http://dx.doi.org/10.1111/j.1462-2920.2011.02573.x
- Radyuk SN, Mericko PA, Popova TG, Grene E, Alibek K. In vitro-generated respiratory mucosa: a new tool to study inhalational anthrax. Biochem Biophys Res Commun 2003; 305:624-32; PMID:12763040; http://dx.doi.org/10.1016/S0006-291X(03)00830-1
- Mussig E, Tomakidi P, Steinberg T. Gingival fibroblasts established on microstructured model surfaces: their influence on epithelial morphogenesis and other tissue-specific cell functions in a co-cultured epithelium: an in-vitro model. J Orofac Orthop 2009; 70:351-62; PMID:19997994
- Bragulla HH, Homberger DG. Structure and functions of keratin proteins in simple, stratified, keratinized and cornified epithelia. J Aanat 2009; 214:516-59; PMID:19422428; http://dx.doi.org/10.1111/j.1469-7580.2009.01066.x
- Karring T, Lang NP, Loe H. The role of gingival connective tissue in determining epithelial differentiation. J Periodontal Res 1975; 10:1-11; PMID:124329; http://dx.doi.org/10.1111/j.1600-0765.1975.tb00001.x
- Dongari-Bagtzoglou A, Kashleva H. Development of a highly reproducible three-dimensional organotypic model of the oral mucosa. Nature protocols 2006; 1:2012-8; PMID:17487190; http://dx.doi.org/10.1038/nprot.2006.323
- Igarashi M, Irwin CR, Locke M, Mackenzie IC. Construction of large area organotypical cultures of oral mucosa and skin. J Oral Pathol Med 2003; 32:422-30; PMID:12846789
- Choe MM, Tomei AA, Swartz MA. Physiological 3D tissue model of the airway wall and mucosa. Nature protocols 2006; 1:357-62; PMID:17406256; http://dx.doi.org/10.1038/nprot.2006.54
- Chai WL, Moharamzadeh K, Brook IM, Emanuelsson L, Palmquist A, van Noort R. Development of a novel model for the investigation of implant-soft tissue interface. J Periodontol 2010; 81:1187-95; PMID:20450401; http://dx.doi.org/10.1902/jop.2010.090648
- Bao K, Akgül B, Bostanci N. Establishment and characterization of immortalized gingival epithelial and fibroblastic cell lines for development of organotypic cultures. Cells, tissues, organs in press; PMID:25471635
- Locke M, Hyland PL, Irwin CR, Mackenzie IC. Modulation of gingival epithelial phenotypes by interactions with regionally defined populations of fibroblasts. J Periodontal Res 2008; 43:279-89; PMID:18447855; http://dx.doi.org/10.1111/j.1600-0765.2007.01028.x
- Lekic PC, Pender N, McCulloch CA. Is fibroblast heterogeneity relevant to the health, diseases, and treatments of periodontal tissues? Crit Rev Oral Biol Med 1997; 8:253-68; PMID:9260043; http://dx.doi.org/10.1177/10454411970080030201
- Belibasakis GN, Bostanci N, Reddi D. Regulation of protease-activated receptor-2 expression in gingival fibroblasts and Jurkat T cells by Porphyromonas gingivalis. Cell Biology Int 2010; 34:287-92; PMID:19947912; http://dx.doi.org/10.1042/CBI20090290
- Belibasakis GN, Bao K, Bostanci N. Transcriptional profiling of human gingival fibroblasts in response to multi-species in vitro subgingival biofilms. Mol Oral Microbiol 2014; 29:174-83; PMID:24758474; http://dx.doi.org/10.1111/omi.12053
- Belibasakis GN, Johansson A, Wang Y, Chen C, Kalfas S, Lerner UH. The cytolethal distending toxin induces receptor activator of NF-kappaB ligand expression in human gingival fibroblasts and periodontal ligament cells. Infect Immun 2005; 73:342-51; PMID:15618171; http://dx.doi.org/10.1128/IAI.73.1.342-351.2005
- Carterson AJ, Honer zu Bentrup K, Ott CM, Clarke MS, Pierson DL, Vanderburg CR, Buchanan KL, Nickerson CA, Schurr MJ. A549 lung epithelial cells grown as three-dimensional aggregates: alternative tissue culture model for Pseudomonas aeruginosa pathogenesis. Infect Immun 2005; 73:1129-40; PMID:15664956; http://dx.doi.org/10.1128/IAI.73.2.1129-1140.2005
- Timmins NE, Scherberich A, Fruh JA, Heberer M, Martin I, Jakob M. Three-dimensional cell culture and tissue engineering in a T-CUP (tissue culture under perfusion). Tissue Eng 2007; 13:2021-8; PMID:17590148; http://dx.doi.org/10.1089/ten.2006.0158
- Voisard D, Meuwly F, Ruffieux PA, Baer G, Kadouri A. Potential of cell retention techniques for large-scale high-density perfusion culture of suspended mammalian cells. Biotechnol Bioeng 2003; 82:751-65; PMID:12701141; http://dx.doi.org/10.1002/bit.10629
- McCoy RJ, O'Brien FJ. Influence of shear stress in perfusion bioreactor cultures for the development of three-dimensional bone tissue constructs: a review. Tissue Eng Part B, Rev 2010; 16:587-601; PMID:20799909; http://dx.doi.org/10.1089/ten.teb.2010.0370
- Yeatts AB, Fisher JP. Tubular perfusion system for the long-term dynamic culture of human mesenchymal stem cells. Tissue Eng Part C, Methods 2011; 17:337-48; PMID:20929287; http://dx.doi.org/10.1089/ten.tec.2010.0172
- Ammann TW, Belibasakis GN, Thurnheer T. Impact of early colonizers on in vitro subgingival biofilm formation. PloS One 2013; 8:e83090; PMID:24340084; http://dx.doi.org/10.1371/journal.pone.0083090
- Ammann TW, Bostanci N, Belibasakis GN, Thurnheer T. Validation of a quantitative real-time PCR assay and comparison with fluorescence microscopy and selective agar plate counting for species-specific quantification of an in vitro subgingival biofilm model. J Periodontal Res 2013; 48:517-26; PMID:23278531; http://dx.doi.org/10.1111/jre.12034
- Ammann TW, Gmur R, Thurnheer T. Advancement of the 10-species subgingival Zurich Biofilm model by examining different nutritional conditions and defining the structure of the in vitro biofilms. BMC Microbiol 2012; 12:227; PMID:23040057; http://dx.doi.org/10.1186/1471-2180-12-227
- Belibasakis GN, Guggenheim B, Bostanci N. Down-regulation of NLRP3 inflammasome in gingival fibroblasts by subgingival biofilms: involvement of Porphyromonas gingivalis. Innate Iimu 2013; 19:3-9; PMID:22522430; http://dx.doi.org/10.1177/1753425912444767
- Schroeder HE, Listgarten MA. The gingival tissues: the architecture of periodontal protection. Periodontol 2000 1997; 13:91-120; PMID:9567925 ; http://dx.doi.org/10.1111/j.1600-0757.1997.tb00097.x
- Costea DE, Kulasekara K, Neppelberg E, Johannessen AC, Vintermyr OK. Species-specific fibroblasts required for triggering invasiveness of partially transformed oral keratinocytes. Am J Pathol 2006; 168:1889-97; PMID:16723704; http://dx.doi.org/10.2353/ajpath.2006.050843
- Tomakidi P, Fusenig NE, Kohl A, Komposch G. Histomorphological and biochemical differentiation capacity in organotypic co-cultures of primary gingival cells. J Periodontal Rres 1997; 32:388-400; PMID:9210093; http://dx.doi.org/10.1111/j.1600-0765.1997.tb00549.x
- Rouabhia M, Deslauriers N. Production and characterization of an in vitro engineered human oral mucosa. Biochem Cell Biol 2002; 80:189-95; PMID:11989714; http://dx.doi.org/10.1139/o01-237
- Tomakidi P, Breitkreutz D, Fusenig NE, Zoller J, Kohl A, Komposch G. Establishment of oral mucosa phenotype in vitro in correlation to epithelial anchorage. Cell Tissue Res 1998; 292:355-66; PMID:9560478; http://dx.doi.org/10.1007/s004410051066
- Schroeder HE, Munzel-Pedrazzoli S. Morphometric analysis comparing junctional and oral epithelium of normal human gingiva. Helv Odontol Acta 1970; 14:53-66; PMID:5486280
- Schroeder HE. Quantitative parameters of early human gingival inflammation. Arch Oral Biol 1970; 15:383-400; PMID:5270171
- Schroeder HE. Transmigration and infiltration of leucocytes in human junctional epithelium. Helv Odontol Acta 1973; 17:6-18; PMID:4699377
- Pollanen MT, Gursoy UK, Kononen E, Uitto VJ. Fusobacterium nucleatum biofilm induces epithelial migration in an organotypic model of dento-gingival junction. J Periodontol 2012; 83:1329-35; PMID:22248219; http://dx.doi.org/10.1902/jop.2012.110535
- Paino A, Lohermaa E, Sormunen R, Tuominen H, Korhonen J, Pollanen MT, Ihalin R. Interleukin-1beta is internalised by viable Aggregatibacter actinomycetemcomitans biofilm and locates to the outer edges of nucleoids. Cytokine 2012; 60:565-74; PMID:22898394; http://dx.doi.org/10.1016/j.cyto.2012.07.024
- Dabija-Wolter G, Sapkota D, Cimpan MR, Neppelberg E, Bakken V, Costea DE. Limited in-depth invasion of Fusobacterium nucleatum into in vitro reconstructed human gingiva. Arch Oral Biol 2012; 57:344-51; PMID:22024403; http://dx.doi.org/10.1016/j.archoralbio.2011.09.015
- Thurnheer T, Belibasakis GN, Bostanci N. Colonisation of gingival epithelia by subgingival biofilms in vitro: Role of "red complex" bacteria. Arch Oral Biol 2014; 59:977-86; PMID:24949828
- Belibasakis GN, Thurnheer T, Bostanci N. Interleukin-8 responses of multi-layer gingival epithelia to subgingival biofilms: role of the "red complex" species. PloS One 2013; 8:e81581; PMID:24339946; http://dx.doi.org/10.1371/journal.pone.0081581
- Loesche WJ, Gusberti F, Mettraux G, Higgins T, Syed S. Relationship between oxygen tension and subgingival bacterial flora in untreated human periodontal pockets. Infecti Immun 1983; 42:659-67; PMID:6642647
- Shaddox LM, Wiedey J, Calderon NL, Magnusson I, Bimstein E, Bidwell JA, Zapert EF, Aukhil I, Wallet SM. Local inflammatory markers and systemic endotoxin in aggressive periodontitis. J Dent Res 2011; 90:1140-4; PMID:21730256; http://dx.doi.org/10.1177/0022034511413928
- Navarro-Sanchez AB, Faria-Almeida R, Bascones-Martinez A. Effect of non-surgical periodontal therapy on clinical and immunological response and glycaemic control in type 2 diabetic patients with moderate periodontitis. Journal of clinical periodontology 2007; 34:835-43; PMID:17850602; http://dx.doi.org/10.1111/j.1600-051X.2007.01127.x
- Kardesler L, Buduneli N, Cetinkalp S, Lappin D, Kinane DF. Gingival crevicular fluid IL-6, tPA, PAI-2, albumin levels following initial periodontal treatment in chronic periodontitis patients with or without type 2 diabetes. Inflammat Res 2011; 60:143-51
- Duarte PM, de Oliveira MC, Tambeli CH, Parada CA, Casati MZ, Nociti FH, Jr. Overexpression of interleukin-1beta and interleukin-6 may play an important role in periodontal breakdown in type 2 diabetic patients. J Periodontal Res 2007; 42:377-81; PMID:17559636; http://dx.doi.org/10.1111/j.1600-0765.2006.00961.x
- Bostanci N, Allaker RP, Belibasakis GN, Rangarajan M, Curtis MA, Hughes FJ, McKay IJ. Porphyromonas gingivalis antagonises Campylobacter rectus induced cytokine production by human monocytes. Cytokine 2007; 39:147-56; PMID:17709256; http://dx.doi.org/10.1016/j.cyto.2007.07.002
- Bostanci N, Akgul B, Tsakanika V, Allaker RP, Hughes FJ, McKay IJ. Effects of low-dose doxycycline on cytokine secretion in human monocytes stimulated with Aggregatibacter actinomycetemcomitans. Cytokine 2011; 56:656-61; PMID:21962932; http://dx.doi.org/10.1016/j.cyto.2011.08.039
- Bostanci N, Allaker R, Johansson U, Rangarajan M, Curtis MA, Hughes FJ, McKay IJ. Interleukin-1alpha stimulation in monocytes by periodontal bacteria: antagonistic effects of Porphyromonas gingivalis. Oral Microbiol Immun 2007; 22:52-60; PMID:17241171; http://dx.doi.org/10.1111/j.1399-302X.2007.00322.x
- Belibasakis GN, Johansson A, Wang Y, Chen C, Lagergard T, Kalfas S, Lerner UH. Cytokine responses of human gingival fibroblasts to Actinobacillus actinomycetemcomitans cytolethal distending toxin. Cytokine 2005; 30:56-63; PMID:15804596; http://dx.doi.org/10.1016/j.cyto.2004.11.008
- Belibasakis GN, Guggenheim B. Induction of prostaglandin E(2) and interleukin-6 in gingival fibroblasts by oral biofilms. FEMS Immunol Med Microbiol 2011; 63:381-6; PMID:22092565; http://dx.doi.org/10.1111/j.1574-695X.2011.00863.x
- Teles RP, Gursky LC, Faveri M, Rosa EA, Teles FR, Feres M, Socransky SS, Haffajee AD. Relationships between subgingival microbiota and GCF biomarkers in generalized aggressive periodontitis. J Clin Periodontol 2010; 37:313-23; PMID:20447254; http://dx.doi.org/10.1111/j.1600-051X.2010.01534.x
- Guggenheim B, Gmur R, Galicia JC, Stathopoulou PG, Benakanakere MR, Meier A, Thurnheer T, Kinane DF. In vitro modeling of host-parasite interactions: the 'subgingival' biofilm challenge of primary human epithelial cells. BMC Microbiol 2009; 9:280; PMID:20043840; http://dx.doi.org/10.1186/1471-2180-9-280
- Guggenheim B, Giertsen E, Schupbach P, Shapiro S. Validation of an in vitro Biofilm Model of Supragingival Plaque. J Dent Res 2001; 80:363-70; PMID:11269730; http://dx.doi.org/10.1177/00220345010800011201
- Gmur R, Guggenheim B. Antigenic heterogeneity of Bacteroides intermedius as recognized by monoclonal antibodies. Infect Immun 1983; 42:459-70; PMID:6196291
- Braccini A, Wendt D, Jaquiery C, Jakob M, Heberer M, Kenins L, Wodnar-Filipowicz A, Quarto R, Martin I. Three-dimensional perfusion culture of human bone marrow cells and generation of osteoinductive grafts. Stem Cells 2005; 23:1066-72; PMID:16002780; http://dx.doi.org/10.1634/stemcells.2005-0002
- Wendt D, Marsano A, Jakob M, Heberer M, Martin I. Oscillating perfusion of cell suspensions through three-dimensional scaffolds enhances cell seeding efficiency and uniformity. Biotechnol Bioeng 2003; 84:205-14; PMID:12966577; http://dx.doi.org/10.1002/bit.10759
- Wendt D, Stroebel S, Jakob M, John GT, Martin I. Uniform tissues engineered by seeding and culturing cells in 3D scaffolds under perfusion at defined oxygen tensions. Biorheology 2006; 43:481-8; PMID:16912419
- Belibasakis GN, Ozturk VO, Emingil G, Bostanci N. Soluble triggering receptor expressed on myeloid cells 1 (sTREM-1) in gingival crevicular fluid: association with clinical and microbiologic parameters. J Periodontol 2014; 85:204-10; PMID:23659423; http://dx.doi.org/10.1902/jop.2013.130144