Abstract
Bacterial eukaryotic-like serine threonine kinases (eSTKs) and serine threonine phosphatases (eSTPs) have emerged as important signaling elements that are indispensable for pathogenesis. Differing considerably from their histidine kinase counterparts, few eSTK genes are encoded within the average bacterial genome, and their targets are pleiotropic in nature instead of exclusive. The growing list of important eSTK/P substrates includes proteins involved in translation, cell division, peptidoglycan synthesis, antibiotic tolerance, resistance to innate immunity and control of virulence factors. Recently it has come to light that eSTK/Ps also directly modulate transcriptional machinery in many microbial pathogens. This novel form of regulation is now emerging as an additional means by which bacteria can alter their transcriptomes in response to host-specific environmental stimuli. Here we focus on the ability of eSTKs and eSTPs in Gram-positive bacterial pathogens to directly modulate transcription, the known mechanistic outcomes of these modifications, and their roles as an added layer of complexity in controlling targeted RNA synthesis to enhance virulence potential.
Abbreviations
| TCS | = | two-component signaling |
| OCS | = | one-component signaling |
| eSTK | = | eukaryotic-like serine-threonine kinase |
| eSTP | = | eukaryotic-like serine-threonine phosphatase |
| PASTA | = | penicillin-binding protein and Ser/Thr kinase associated |
| REC | = | receiver |
| wHTH | = | winged helix-turn-helix |
| PTM | = | posttranslational modification |
| PPM | = | protein phosphatase metal binding |
| ROS | = | reactive oxygen species |
Introduction
Eukaryotic-like serine-threonine kinases (eSTKs) and phosphatases (eSTPs) in bacterial pathogens
To allow their continued survival pathogenic microorganisms have evolved multiple and sophisticated mechanisms to adapt to environmental changes within the host. These mechanisms probe the extracellular environment for variations in, for example, nutrient concentrations, oxygen tension and other perceivable dangers to enable them to elicit the appropriate physiological alterations in response. Extracellular signals are commonly relayed into the cells by means of reversible phosphorylation through protein kinases and phosphatases, and in doing so modulate their target activities. The most well described prokaryotic signaling cascades are the two-component signaling (TCS) systems.Citation1,2 The typical TCS system makes use of a membrane bound sensory histidine kinase capable of detecting extracellular signals and when activated, autophosphorylates a conserved histidine residue within the ATPase catalytic domain of the protein. In the most basic and common system, the phosphoryl group is then transferred onto a conserved aspartate contained within the RECiever (or REC domain) of a single cognate effector, often a response regulator transcription factor.Citation2 This event then triggers the activation of the C-terminal effector domain portion of the protein, the majority of which are DNA-binding domains, to result in subsequent interaction and regulation of specific promoters to facilitate the microbe's timely environmental adaptation.Citation3
Although TCS has traditionally enjoyed the ‘lion's share’ of attention in the bacterial signaling field and were once thought the only means by which microbes could sense their environment, it has more recently come to light that eSTKs and eSTPs also play important roles in prokaryotic adaptation. Unlike their TCS counterparts, eSTK/Ps are usually present at far fewer numbers per bacterial genome, with only one cognate pair found in many Gram-positive species. eSTKs were initially identified in the soil-dwelling, Gram-negative microbe Myxococcus xanthus for their involvement in the normal development of this organism.Citation4 However, most subsequent studies have focused on these signaling cascades as being indispensable for virulence in a variety of human pathogens.Citation5-15 The ‘genomic age’ has revealed a considerable expansion of the eSTK protein family and the presence of their cognate eSTPs in most prokaryotic Phylums, which include important Gram-negative human pathogens such as Pseudomonas aeruginosa,9,Citation16 and many Gram-positives such as Enterococcus faecalis,Citation17 Staphylococcus aureus,12,14,Citation18-20 Mycobacterium tuberculosis,Citation6,21,22 Streptococcus pneumoniae,Citation7,23,24 Streptococcus agalactiaeCitation8 and Streptococcus pyogenes.10,Citation25 This discovery led to the realization that unlike TCS histidine kinases, which almost exclusively target a single response regulator partner, eSTK substrates are instead quite pleiotropic in nature. This feature enables a single eSTK/P pair to regulate a large variety of proteins crucial for important cellular functions, including transcription and translation, cell division, dormancy, antibiotic resistance, bacterial persistence and expression of virulence factors such as exotoxinsCitation20 (). The diversity of eSTK substrates is highlighted by several proteomic studies. Some examples include one from M. tuberculosis where 301 protein substrates were identified for the unusually high 11 eSTKs found in this organism,Citation22,26 another from Listeria monocytogenes where 62 interacting partners were identified for its 2 eSTKs,Citation27 and a study conducted in S. agalactiae where 10 potential substrates were discovered for its single known eSTK.Citation28 However, at present many identified posttranslational modifications (PTMs) associated with regulatory roles in virulence and other important bacterial functions have only been verified by in vitro experimentation, and thus require more difficult genetic and biochemical assays to demonstrate their relevance in vivo. lists the eSTK transcription modulating targets described in this review and highlights these issues. For a more comprehensive list of eSTKs and their substrates see reference Citation29.
Table 1. Gram-positive eSTK-regulated transcription factors involved in virulence and infectionFootnotec
Figure 1. Schematic diagram showing the functional roles of eukaryotic serine-threonine kinases (eSTKs) and phosphatases (eSTPs). eSTKs contain an N-terminal cyclic AMP (cAMP)-dependent Protein Kinase domain (PKA; colored blue) and usually a C-terminal sensory domain. In non-mycobacterial Gram-positive bacteria the sensory domain is often a PASTA domain (purple half-rings). PASTA domains have been shown to bind free muropeptides (yellow balls) and cell wall-targeting antibiotics (e.g. β-lactams, orange diamonds). Once activated, eSTKs will dimerize and their intracellular kinase domains phosphorylate pleiotropic targets to produce specific cellular responses. eSTPs can, in turn, remove the phosphate(s) from an eSTK or substrates. Targets include metabolic enzymes, cell wall building and cell division regulatory enzymes such as DivIVA or FtsZ, translational machinery, and transcription factors (TFs) including sigma factors (σ) and possibly RNA polymerase (RNAP) holoenzyme subunits.
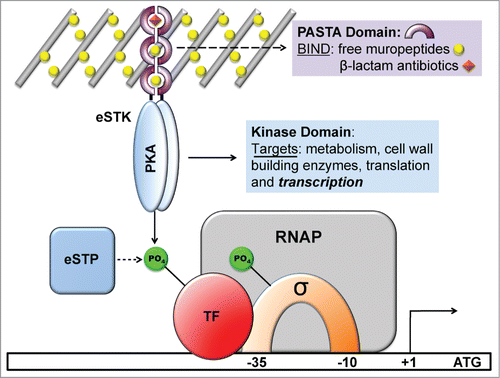
eSTK structure and function
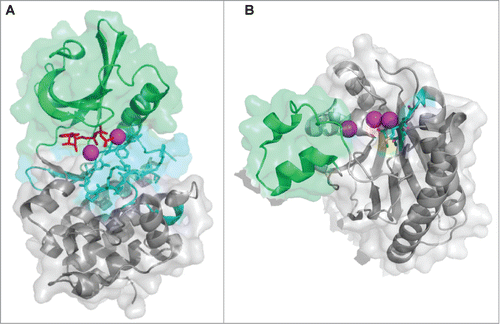
The prokaryotic eSTK family was initially defined as cyclic AMP (cAMP)-dependent protein kinase (PKA) domains responsible for substrate recognition and phosphotransferase activity.Citation30 Analysis of PknB in M. tuberculosis, the first structurally characterized bacterial eSTK PKA domain, reveals a typical bi-lobed organization strikingly similar to its eukaryotic versionsCitation32-34 (). The structure possesses an N-terminal subdomain consisting primarily of β-strands followed by a C-terminal region almost entirely composed of α-helices, while the ATP binding pocket and substrate interaction site are sandwiched in between (). This arrangement is also referred to as a Hanks-type domain VIb fold, which is defined by a conserved lysine present in the ATPase active site and the motif “DXXPXN,” where X is any amino acid.Citation30 This architecture appears to be well conserved across the prokaryotic eSTK family, which suggests that a common mechanism of action is shared among these enzymes.Citation21 However, extensive examination of the available bacterial genomes suggest that these receptor kinases can be further divided into several subclasses of related functions, and therefore might exhibit functional diversity similar to their eukaryotic counterparts.Citation31
Figure 2. Atomic structures of eSTKs and eSTPs. (A) 3.0 Å atomic structure cartoon/surface of the PknB eSTK from M. tuberculosis (PDB code 1MRU; ref. Citation21). The two distinct lobes of the structure can easily be seen: (i) the N-terminal PKA subdomain consisting primarily of β-strands (colored green), and (ii) the a C-terminal subdomain almost entirely composed of α-helices (colored gray). Residues that make up the catalytic active pocket are colored in cyan. These residues include the metal (magnesium) binding domain, conserved Hank's-type ATPase active site motif “DXXPXN” and substrate interaction domain. The phosphothiophosphoric acid-adenylate ester (or AGS) substrate is colored red and magnesium ions colored magenta. (B) Cartoon/surface representation of the 2.65 Å atomic structure of the eSTP from S. agalactiae (PDB code 2PK0; ref.Citation48). The Flap domain, thought to be involved with substrate interactions, is colored green. The conserved “DMGG” catalytic pocket motif that coordinates 2 of the 3 magnesium ions (colored magenta) is shown in cyan. In the DMGG motif the sulfur atoms are colored yellow, nitrogens blue and oxygens red.
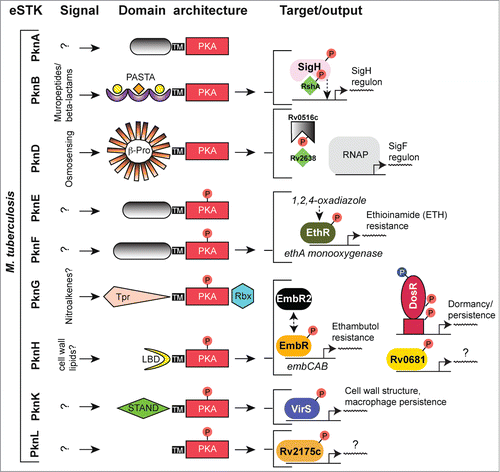
Although some eSTKs, especially those found in Gram-negative species, are predicted to be soluble cytoplasmic proteins,Citation35,36 in Gram-positives most of these kinases possess a single transmembrane region that tethers the PKA module to a C-terminal extracellular sensory domain. This C-terminal sensory feature can take on many forms and is best exemplified by the unusually high number and diversity found in M. tuberculosisCitation22,35,37 (11 total: PknA-B, and PknD-L). The multiple M. tuberculosis sensory domains include a β-propeller extension in PknD, a C-terminal Tpr and N-terminal rubredoxin-like sequence in PknG, a penicillin-binding protein and Ser/Thr kinase associated (PASTA) structure in PknB, and then finally several unidentified C-terminal domains in the remaining M. tuberculosis eSTK sequences (see Fig. 6 and ref. 38 for a review of eSTK domain architecture in M. tuberculosis). Although historically the actual ligands that induce these diverse sensory domains have been limited to the PASTA repeats commonly found in other more canonical Gram-positives (see below), a number of recent studies have now shed further light on this subject. Convincing evidence has now shown that the beta-propeller extension in PknD is sensing osmolarity,Citation39 which can change considerably, for example, during airborne transmission, when replicating within infected macrophages and within granulomas. In response to PknD osmotic stimulation, transcriptional machinery was activated in a kinase-dependent manner.Citation39 Although the actual inducing ligand still remains at large, in the example of PknG experimentation has suggested that nitrated fatty acids (nitroalkenes) could be the natural interacting molecules.167 These compounds are metabolically produced during inflammation and could possibly serve as a means by which the bacteria can sense phagosome-lysosome fusion during macrophage intracellular replication.
Outside of the Mycobacteria, the most commonly studied eSTK extracellular domains have been PASTA repeats,Citation40 which Mycobacteria also possess a homolog of (PknB) (). In contrast to the extensive conservation of the N-terminal catalytic core, PASTA domains show considerable disparity between homologs,Citation41 suggesting that these repeats have evolved to accommodate variations in ligand structure. In support, some studies have indicated a surprising ability of PASTA domains to discriminate between different ligands, with apparently only slight changes in amino acid composition.Citation42 However, given that the eSTK PASTA domains are homologous to known canonical penicillin-binding proteins (PBPs) whose function is the last catalytic step in peptidoglycan biosynthesis,Citation40 it is unlikely that they have evolved for the primary purpose of detecting β-lactam-type antibiotics for which they are named. Nevertheless, PASTA domains are able to bind this class of antimicrobials with variable affinities.Citation43
Thus far, the only known natural ligands of eSTKs described have been DAP-type hydrolysed muropeptide peptidoglycan fragments.Citation42,44 As these findings have to date only been shown with the Bacillus subtilis PASTA-containing eSTK (PrkC),Citation42,44 at the moment we can only infer that other homologs found within pathogenic bacteria have similar receptor specificity. Based on these seminal studies, the activation of eSTKs is thought to initiate by the binding of these muropeptide ligands, resulting in dimerization and subsequent autophosphorylation of the cytoplasmic N-terminal kinase domain. This action then results in phosphorylation of downstream target proteins, which eventually results in the modulation of transcriptional activity (; reviewed in ref.Citation29). Intriguingly, recent work has demonstrated that PASTA-containing eSTKs might perform their functions in a specific area of the cell.Citation45 Veening and colleagues have shown that in S. pneumoniae the PASTA-harboring eSTK (StkP) co-localizes to the bacterial divisosome with 2 of its known regulatory targets that control the cell division process, namely DivIVA and FtsA. Expanding on these studies, Grangeasse and colleagues have proposed that StkP, DivIVA and another protein called GpsB form a triad complex at the divisosome to enable the classic ellipsoid, or ‘cocci’ shape for which the pneumococcal cell is named.168 Collectively, these studies physically place PASTA-containing eSTKs at co-peripheral and septal cell-wall synthesis sites, and suggests that their function is, at least in part, to coordinate bacterial growth by modifying multiple substrates in this process with intricate timing.Citation45
eSTP structure and function
Differing from histidine kinases that often possess inherent phosphatase activity,Citation1 eSTKs apparently lack this enzymatic function. This absent feature has presumably necessitated the evolution of an eSTP phosphatase counterpart to enable the option to revert the cell's physiology back to ‘normality’ following an eSTK phosphorylation-driven stress response. Interestingly, genomes with multiple eSTKs contain cognate eSTP phosphatases in far fewer numbers. For example, the M. tuberculosis genome possesses 11 eSTKs, yet only has a single predicted eSTP.Citation22
The eSTPs belong to the protein phosphatase metal (PPM) binding family of enzymes, which are further subdivided into the PP2C class based on the presence of 11 conserved domains.Citation46 Although classical PP2C phosphatases found in plants and other eukaryotes employ a bi-nuclear magnesium or manganese active site, structural analysis has revealed up to 3 metal ions bound within the catalytic core of the bacterial versionsCitation47-49 (). The conserved eSTP active site is surrounded by a central β-sandwich and flanked by a pair of α-helices, which strikingly parallels the architecture of human versionsCitation50,51 (). A protruding so-called flexible ‘flap’ domain is the major divergent structural feature from eukaryotic versions whose function is thought to involve substrate specificity and modulation of catalytic activityCitation49,52 (shown in green in ). While their precise regulatory roles and consequences remain less understood than their kinase counterparts, available data show that eSTPs regulate the same vital eSTK processes in many pathogenic bacteria including cell division, virulence, antibiotic susceptibility and expression of virulence factors.Citation10-12,19,25,29,53 An interesting recent report by Pancholi and colleagues has expanded the role of eSTPs by demonstrating that the S. pyogenes version is somehow secreted, and can then enter the host cell and nuclear envelope to affect apoptotic signaling cascades.Citation54 Although important in their own right in the regulation of pathogen virulence traits, the scope of this review will focus more on eSTK regulation of transcription.
Bacterial Transcription Factors
Two-component systems
TCS systems are defined by their ability to accept a phosphate from a cognate histidine kinase partner on a conserved aspartate residue within the REC domain of the protein.1,3,Citation55 Although REC domains can be associated with diversified output domains, they are most often found attached to helix-turn-helix (HTH) motif-containing effectors whose direct role is to bind DNA and modulate RNAP recruitment.Citation56 REC domains can be found N-terminal to HTH effector domains as in the case of XRE, ArsR, HTH-10 and HTH-REC outputs. Alternatively, they can be found C-terminal to the effector module as in the case of TCS OmpR-type regulators having winged HTH DNA-binding motifs,Citation57 and also BetR, AlpA, HlxR, LysR and MerR HTH-containing sequences.Citation56
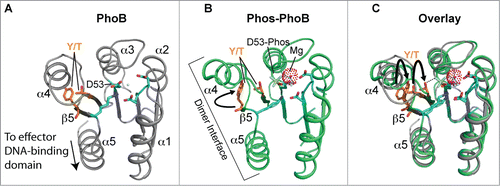
The three-dimensional structure of the canonical REC domain maintains an overall (α/β)5 topology (; refs.Citation58,55). These domains consist of 2 major areas of activity: (i) an acidic catalytic core containing the conserved Asp residue that receives the phosphate from the activated histidine kinase (e.g. Asp53 in PhoB, ), and (ii) a conserved Ser/Thr-Phe/Tyr pair which act as a molecular switch to enable dimerization and activation after the PTM has occurred.55,Citation59 The latter events have been dubbed ‘Y/T-coupling’ and are responsible for driving the post-phosphorylation repositioning of the conserved Ser or Thr residue into the catalytic portion of the protein to help coordinate the newly-formed phospho-aspartate PTM (). This repositioning then allows an invariable Tyr or Phe residue to also move ‘inwards’ to a less solvent-exposed area, which in turn leaves a more statically-favorable α4−β5−α5 dimeric interface for sister REC domains to interact and subsequently transmit the signal to the downstream effector domains (e.g., a HTH-containing motif) via an uncoupling of REC-effector hydrogen bonding.Citation60,61 Once the REC-effector dissociation occurs, the REC C-terminal α5 helix linker is thought to become more flexible. This action then releases the DNA-binding motif to enable recruitment or obstruction of transcription through interactions with sigma (σ) factors and/or RNAP.Citation58 Interestingly, an alternative dimerization site has now been identified within the α1−α5 helical face of the REC structure,Citation62,63,169 and could be important for dimer-to-dimer contacts at promoters, providing additional and more complex options for bacteria to control their transcription.
Taken together, TCS response regulator modulation of transcription continues to show an increasing complexity and likely depends on the precise positioning of the transcription factors on the promoter, which can be influenced by variable environmental conditions.Citation64 Previous genetic and biochemical data, combined with a low-resolution quaternary structure of the TCS response regulator PhoB bound to DNA, σ70 and the RNAP flap domain suggest that this complexity is controlled in part through direct interaction of the TCS regulator's transactivation loop located within the DNA-binding domain of the protein.Citation65,66
Figure 3. Structure and activation of REC domains. (A) Structure of the PhoB 2-component response regulator REC domain from Escherichia coli (PDB 1B00; ref. 158) shown in the inactivated, unphosphorylated state. PhoB exhibits a classical REC (α/β)5 topology, and α4-β5-α5 canonical dimeric interface. The proposed alternative dimer interface is the α1-α5.Citation63,169 The tyrosine and threonine involved in ‘Y/T’ coupling are colored in orange. Notice the tyrosine is protruding into the solvent area of the α4-β5-α5 face to presumably prevent dimerization and activation. Key residues involved in the active pocket phosphorylation events are colored cyan, including the invariant aspartate residue (D53) that is phosphorylated by the cognate histidine kinase. Nitrogen atoms are colored blue and oxygen atoms red. (B) Structure of activated, phosphorylated PhoB (Phospho-PhoB; PDB 1ZES; ref. 62). Upon D53 phosphorylation, the conserved Y/T residues move toward the catalytic pocket (the Y movement is denoted by the arrow) to coordinate the new phosphate group (colored yellow) and allow the dimer to form. The magnesium ion and surrounding electron cloud (Mg) is shown in magenta. (C) Overlay of PhoB and Phospho-PhoB structures from A and B. The black arrows indicate the movements of the Y/T residues to allow dimerization about the α4-β5-α5 interface.
One-component systems
The so-called ‘One-Component’ Signaling (OCS) systems numerically dominate bacterial genomes and are defined by combining both input (sensory) and output (effector) modules into a single functional polypeptide.Citation51 Like TCS effectors, these lone signal transduction proteins are usually transcription factors that contain an HTH motif (approximately 87%).Citation51,67 However, OCS systems differ in that they exhibit greater diversity than their TCS counterparts. Although too numerous to list, OCS modules include more familiar and evolutionarily expansive protein families such as LysR, MarR and TetR-type regulators, as well as more understudied and rare HTH effector motifs. Not being constrained by upstream cognate partners, OCS systems are able to incorporate a wide range of inputs that include metal binding domains and small ligand effectors. For a comprehensive list of these bacterial signaling domains see references 51 and 67. Interestingly, some TCS system regulators have lost their conserved REC domain phospho-accepting aspartate residues and instead have evolved to accommodate more diverse inputs, effectively reverting back to a one-component unit.Citation55 To date, examples of these pseudo-response regulators have been few, however they are important in that most described thus far are required for infection in a variety of pathogens, including RitR from Streptococcus pneumoniae that regulates iron uptake and related responses to reactive oxygen species (ROS),Citation69 the general transcription regulator AmiR from Pseudomonas aeruginosa that regulates transcriptional termination,Citation68 and HP1043 from Helicobacter pylori and ChxR from Chlamydia trachomatis that regulate infection and virulence traits.Citation70,71
In this review we will concentrate on the emerging, alternative form of RNA synthesis control by eSTK phosphorylation of bacterial transcription factors, and the potential mechanistic consequences of these phospho-modifications. Emerging regulatory PTMs including the ability of eSTKs to phosphorylate tyrosines, as well as the new arginine kinases will not be covered in this review as they have recently been described in detail elsewhere.Citation72,73 Here we will focus on eSTK phosphorylation transcription factors on serines, threonines and cysteines to enable timely control of important for virulence and infection traits in pathogenic bacteria (summarized in ). In this regard, the future advancement of the field of prokaryotic PTMs will also be discussed, as a better understanding of these signaling systems will shed light on how eSTK/Ps empower bacteria to cause a range of infections in most important pathogens, and could ultimately offer a realistic focal point of therapeutic strategies in disease prevention.Citation74-77,146
Transcription Factors Regulated by eSTKs and eSTPs
Staphylococcus aureus
As a human pathogen responsible for a wide range of infections, S. aureus is of major clinical significance.Citation78 Underlying this clinical significance is this organism's virulence potential, as well as the increasing difficulties in providing treatment associated with antibiotic resistance. Indeed, the 2 eSTKs in this organism have been strongly implicated in both of these issuesCitation79 (reviewed in ref.Citation14). To date, several S. aureus eSTK transcriptional targets have been identified that control this important pathogen's ability to survive, cause invasive disease and avoid the deleterious effects of antibiotic treatment. These are summarized in .
Figure 4. Overview of eSTK control of transcription in S. aureus. Schematic diagram showing the signaling pathways involved in eSTK control of S. aureus regulatory factors mentioned in this text. Phosphorylation is denoted by the circle with a “P.” Red circles indicate serine or threonine phosphorylation, brown cysteine phosphorylation and blue aspartate phosphorylation. Virulence and other traits that the regulators control are also described, as are regulated operons (if known). The TCS regulators GraR and VraR are phosphorylated by Stk1, as are the OCS regulators CcpA, MgrA and SarA. MgrA is phosphorylated by Stk1 on residues Ser110 and Ser113, and homologous sites yet to be verified experimentally are found in SarA and SarZ (see ). However, aside from being phosphorylated by Stk1, SarA is also phosphorylated by the SA0077 (Stk2) kinase on threonine residues. Stk2 is only present in some S aureus strains harboring methicillin resistance. The question mark labels indicate that the exact site(s) of phosphorylation has not been identified. TM, transmembrane region.
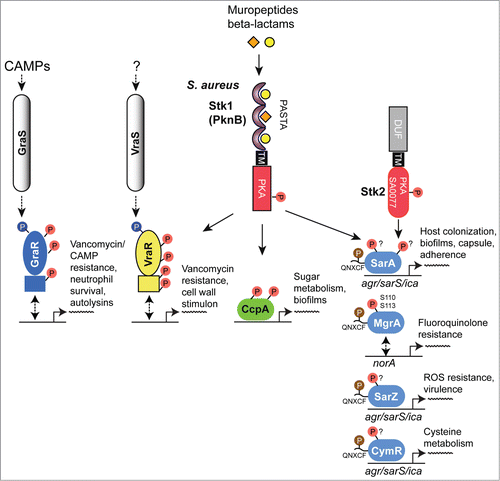
An early observation of eSTK/P-mediated modulation of transcription in S. aureus implicated the global transcription regulator MgrA, a member of the SarA/MgrA family of OCS regulators that contain a dimerization domain and winged HTH motif. However, little is known about how their regulatory effects are modulated in the absence of an obvious sensory input domain.Citation80 What is known is MgrA is responsible for regulating several virulence factors that include α toxin, nuclease, lipase, coagulase and protein A, as well as autolysins and levels of capsular polysaccharide in S. aureus. In addition, MgrA also controls the synthesis of several efflux pumps involved in antimicrobial resistance in this pathogen.Citation81 Work by Hooper and colleagues has shown that addition of S. aureus extracts to purified MgrA protein resulted in an abrogation of its binding to target DNA, which suggested PTMs were responsible. A later publication by the same group identified 2 adjacent serines (Ser110 and Ser113) as being the residues modified by the S. aureus PASTA-containing eSTK Stk1 (also referred to as PknB) and the cognate eSTP RsbU, which reciprocally regulates transcription of the genes encoding 2 drug efflux pumps (norA and norB), and consequently fluoroquinolone resistanceCitation82 (). Interestingly, Ser110 and Ser113 reside within the dimerization domain interface of MgrA (). This positioning is suggestive of a mechanism by which phosphorylation of these residues prevents MgrA dimerization and binding to DNA to allow derepression and therefore subsequent activation of the norA efflux pump under antibiotic imposed stresses.
Figure 5. eSTK phosphorylation sites in S. aureus transcription. Domain architectures and identified phospho-residues (Serine (S), Threonine (T) or Cysteine (C), indicated by the arrows) are shown on the left. Some corresponding published atomic structures, or structural models that we have generated here as part of this review are shown on the right. Important residues are indicated in the structures. MgrA, SarZ, SarA and CymR all belong to the same family of transcription factors referred to here as the SarA/MgrA family and are regulated by phosphorylation of a conserved N-terminal cysteine residue (ref. 88). Only MgrA has been verified as having 2 additional eSTK-regulated sites (Ser110 and Ser113 within the C-terminal dimerization domain; ref. 82). Homologous Ser110/113 sites in SarZ, SarA and CymR regulators were found using 1- and 3-dimensional alignments. DD, dimerization domain; wHTH, winged helix-turn-helix domain; lac-HTH, LacI family HTH domain; Lux-HTH, LuxR family HTH domain; PBD, periplasmic binding domain; REC, receiver domain. Structures that are shown as dimers are depicted with one monomeric unit in color, and the other sister monomeric unit in gray. Phosphorylated residues are shown in red, except the phosphorylated cysteine in the SarZ structure is shown in orange, and the Ser101 and T119 sites in the VraR structure are colored yellow. The SarZ and VraR PDB codes used were 3HSE (ref. 159) and 4GVP (ref. 63), respectively. The GraR DNA-binding domain (DBD) atomic model was generated using a PhoB structure as a molecular template (PDB 1GXP; ref. 160) with Swiss-Model (ref. 161). All atomic figures were created with MacPyMol (ref. 162).
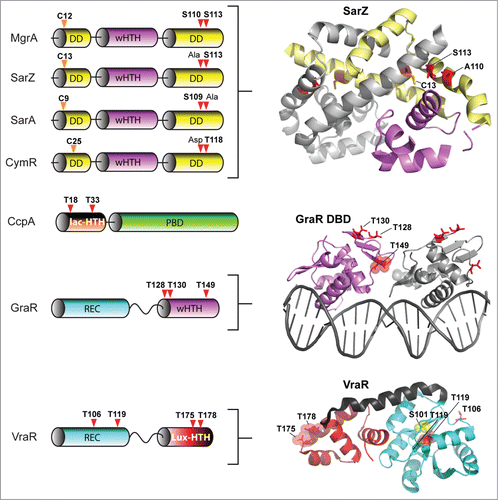
Another S. aureus global transcription regulator SarA,Citation83 a member of the SarA/MgrA family of prokaryotic transcription factors which modulates the expression of over 100 genes in this organism, has also revealed to be directly regulated via eSTK phosphorylation.Citation84 Many of the genes that SarA controls include important virulence determinants such as hla (α toxin) and spa (Spa immune evasion molecule), the master virulence-controlling quorum sensing locus agr (accessory gene regulator), and sarS (staphylococcus accessory regulator), as well as biofilm formation through the regulation of the ica operon to facilitate host colonization.Citation85 This important regulatory element also controls a plethora of other virulence determinants that include toxins (e.g. hemolysins and proteolytic exotoxins), synthesis of the capsule and adherence.Citation80,86 Interestingly, a study by Didier et al. reveals that SarA is phosphorylated by 2 different S. aureus eSTKs. One, Stk1 (see above), controls SarA through threonine-specific phosphorylation, which greatly enhances its in vitro DNA-protein complex formation at promoters of key regulatory determinants agr and sar.Citation84 Intriguingly, the second kinase that phosphorylates SarA, referred to by its gene annotation SA0077 and also called Stk2, only targets serine residue(s) in the protein (). Interestingly, SA0077/Stk2 is strain specific and only found thus far within a subset of methicillin-resistant S. aureus genomes (e.g., N315; ref. 166).
The domain architecture of SA0077 contains 502 amino acids, an N-terminal protein kinase domain, and similar to PASTA-containing versions like Stk1 possesses a single predicted transmembrane segment. However, C-terminal to the transmembrane helix is a predicted helical region of ∼200 amino acids with no homology to known domains or functionally characterized architectures. Contrary to the Stk1 regulatory effect on SarA, phosphorylation by the SA0077 kinase instead produces a decrease in DNA binding to known SarA regulated promoters.Citation84 Although the precise sites of SarA phosphorylation by the SA0077 kinase were not identified, alignment of the available MgrA structure with that of the related SarZ (PDB IDs 2BV6 and 3HSE, respectively; structural alignment = RMS 2.09 Å, 117 to 117 atoms aligned; and data not shown) does show that the related SarA, SarZ and CymR regulators also possess a serine residue at one of the identified MgrA Stk1 phosphorylated positions (Ser110 or Ser113 equivalents; ).Citation82 Collectively these observations allude to a conservation of eSTK recognition in these related transcription factors. These findings are also particularly significant in that Didier et al. demonstrate the potential for multiple eSTK modulations of a single bacterial transcription factor, whose modifications affect the activation of different promoters. These studies add to the rapidly emerging picture of a more complex bacterial transcriptional regulatory network than previously anticipated. Further work is required to determine the precise effects of these phosphoserine PTMs with respect to infection and virulence.
A surprising additional complexity to this story was added by He and colleagues, where they found that the SarA and MgrA regulators in S. aureus act as bona fide functional redox sensors in this pathogen.Citation87,88 Following this result, the same group made the seminal discovery that SarA, MgrA and related regulator SarZ are also controlled through a novel cysteine phosphorylation PTM via an apparently conserved “[Q/N]XCF” motif found within the N-terminus of these proteinsCitation87,88 (). The authors suspect that this novel PTM was missed in previous work due to standard conditions that are normally used in preparation of biological samples for mass spectrometry analysis (e.g. the use of reducing agents such as DTT). This thorough report (ref.Citation88), which includes both in vivo and in vitro evidence as well as structural analysis, demonstrates that Stk1 is responsible for the observed cysteine phosphorylations, and ultimately enables the regulation of several virulence traits, as well as vancomycin resistance in response to oxygen and ROS. Strikingly, cysteine phosphoylation and associated regulatory effects can be readily reversed under reducing conditions. In this same report CymR, an additional SarA/MgrA family member from S. aureus that regulates cysteine metabolism in response to oxygen,Citation89 was also found to be phospho-regulated by Stk1 through a similar N-terminal cysteine residue.Citation88 A collective model for virulence factor regulatory control is thus proposed, whereby cysteine oxidation and phosphorylation act together to control dissociation of SarA/MgrA regulators from target promoters, which results in the activation of virulence factors (e.g., hemolysinCitation88) (). An interesting area of future work will be to extend investigations of this novel PTM to other transcription factors and microbial species. We anticipate that the use of cysteine phosphorylation by pathogenic microbes to control virulence determinants in response to changing oxygen/ROS levels will open up an exciting new area of research in the field of bacterial signal transduction.
Another eSTK/P regulated pathway in S. aureus centers on the LacI/GalR family conserved transcription factor Catabolite control protein A (CcpA) that regulates sugar-related central metabolism in many bacterial species. In this pathway glycolytic intermediates are sensed by the HprK kinase, which, in turn, phosphorylates the intermediate Hpr relay protein on Ser46 to enable complex formation with CcpA and subsequent recognition of Catabolite-responsive elements (CREs) in target promoters.Citation90-92 However, in addition to its well-established role in the regulation of metabolism, CcpA is also responsible for controlling virulence determinants (e.g. biofilm formation) in S. aureus and many other bacterial pathogens.Citation93-96 In S. aureus, in vivo and in vitro studies have recently shown that phosphorylation of CcpA by Stk1 on 2 threonine residues (Thr18 and Thr33; ) controls its function. These residues are located within the CcpA DNA-binding domain and are predicted to hydrogen bond with target DNA.Citation97 In this example, eSTK phosphorylation of CcpA affords the bacteria a simple additional mechanism of regulatory control, whereby protein-DNA interactions can be directly abrogated by the PTMs to allow activation of CcpA-repressed catabolite promoters. Leiba et al. went on to show affected phenotypes through Thr18/33 regulatory control, including formation of biofilms in vitro.Citation97 Interestingly, despite the exceptional conservation of Thr18/33 across multiple bacterial species, CcpA orthologs are unable to accept a phosphate by their native eSTKs, however cross-species phosphorylation is possible.Citation97 Considering the high homology of eSTKs and CcpA orthologs across Gram-positive species, these surprising results suggest that slight changes within amino acid composition might afford a rapid reactivation of ‘dormant‘ eSTK controlled regulatory circuits. How such subtle discrimination by eSTKs between homologous variants of CcpA and other eSTK-regulated substrates is possible when all possess the conserved phosphorylatable residues remains an exciting topic of future research.
In addition to eSTK/P mediated reversible regulation of many OCS systems in S. aureus, several TCS systems are also regulated by phosphoserine-threonine PTMs in this pathogen. One such example comes from the response regulator GraR (for glycopeptide resistance associated), part of the GraSR TCS system associated with expression of the major S. aureus autolysin AtlA, survival within neutrophils, resistance to vancomycin and host cationic antimicrobial peptides (CAMPs), and virulence according to several in vivo infection models.Citation98-100 The GraSR TCS is thought to be ligand-activated by host CAMP interaction with the histidine kinase GraS and the CAMP permease VraG to induce D-alanylation of teichoic acids and MprF-dependent lysylination of phosphatidyl-glycerol (). These modifications then collectively result in an increase in the bacterial surface positive charge and subsequent repulsion of CAMPs.Citation100 On the other hand, besides the noted accumulation of mutations within the GraR response regulator that somehow enable incremental resistance to vancomycin, the precise mechanism of how GraSR conveys resistance to treatment with this antimicrobial still remains largely enigmatic.Citation101 What is known is that GraR is another substrate of the S. aureus PASTA-containing eSTK Stk1.Citation102
By employing in vitro kinase reactions combined with tandem mass spectrometry analyses, Fridman et al. show that GraR phosphorylation by Stk1 occurs on 3 threonine residues, namely Thr128, Thr130 and Thr149Citation102 (). Importantly, experiments reveal that these residues are required for efficient incorporation of D-ala into the bacterial cell wall, which could directly affect the efficiency of vancomycin binding and explain the GraR connection with vancomycin resistance. Additionally, the phosphorylation of these residues enhances the binding of GraR to the vraFG CAMP ABC transporter promoter to control its synthesisCitation102 (). A model of the GraR DNA-binding domain (shown as a dimer bound to DNA in ) places residues Thr128 and Thr130 within the N-terminal antiparallel β-sheet, a structural element known to influence REC-DNA binding domain intra-protein contacts.Citation61 Thus phosphorylation of these sites could potentially hold the 2 domains (REC and DNA-binding domain) apart in an extended conformation to instead allow a shifting of the structural equilibrium in favor of REC dimerization, and subsequent DNA-binding domain interaction with target DNA. Interestingly, Thr149 is predicted to face the other sister HTH motif, and when phosphorylated could thus influence these inter-protein interactions ().
To demonstrate eSTK specificity for the GraR regulator 2 additional Stk1 substrates were used: (1) the related OmpR-type TCS regulator BceR from Bacillus subtilis that confers bacitracin resistance and (2) the NarL family regulator VraR from S. aureus that confers vancomycin resistance and coordinates the cell wall stimulon. Consistent with the interspecies kinase assays described for eSTK phosphorylation of CcpA (above), despite the similarity of these regulators and the conservation of serine-threonine residues at the same positions found in BceR, both were unable to accept a phosphate from the S. aureus eSTK in vitro.Citation102 Thus identification of conserved binding sites only indicates a potential for eSTK regulation, and therefore must be treated with caution. Future directions might include BceR and/or VraR variants from different strains to be tested, as sequences between clinical isolates can vary considerably and could therefore ‘unmask’ substrate specificity though subtle changes in amino acid composition.
In apparent contradiction to the aforementioned results, Molle and colleagues have demonstrated that VraR can indeed be phosphorylated by the S. aureus Stk1 in vitro.Citation103 In these experiments mass spectrometry and mutagenesis were used to identify 4 phospho-accepting VraR residues: Thr106, Thr119, Thr175 and Thr178.Citation103 Similar to parallel studies with CcpA, eSTK phosphorylation negatively impacts the DNA-binding properties of VraR. To explain these results, Thr175 and Thr178 are indeed positioned within the DNA-binding domain where they could inflict charge clashes between conserved protein-DNA or protein-protein contacts (). However, for the modified residues found within the REC domain, Thr106 and Thr119, the potential mechanistic contributions are less obvious. Indeed structures provided by Stock and colleagues are now available for both the inactive and active forms of the S. aureus VraR TCS regulator.Citation63 An interesting feature is the position of Thr119, which is completely buried within the inactive (i.e. unphosphorylated aspartate) VraR structure ( below) and hydrogen bonds to the β5 strand (Ser101) located within the canonical α4−β5−α5 dimeric interface. Upon aspartate phosphorylation and the ensuing REC-DNA binding domain dissociation, Thr119 then becomes solvent-exposed.Citation63 A likely in vivo mechanism one could envision is when VraR is activated by canonical Asp phosphorylation, the S. aureus Stk1 would then have ample access to phosphorylate the now solvent-accessible Thr119 and prevent unwanted reversion back to the inactive state. The S. aureus eSTP could then work to reverse this process under specific required circumstances (i.e., removal of the PTM).
For the second phosphorylated threonine residue in the VraR REC domain (Thr106, ), in the inactive state there are no observed intra- or inter-protomer steric clashes, and its solvent exposure would therefore presumably afford easy accessibility to Stk1 interaction. However in the active (i.e. phosphorylated aspartate) state, this residue moves close to the other, alternative VraR α1−α5 dimerization face of the REC structure.Citation62,63,169 A possible mechanism of activation inhibition was therefore suggested whereby Thr106 modification could interfere with formation of the VraR dimer at the α1−α5 interface by impairing crucial hydrophobic contacts.Citation103
From these observations we can speculate that such simultaneous eSTK control of 2 different VraR dimer interfaces offers a glimpse of how eSTKs and eSTPs might further fine-tune canonical phospho-asparate TCS gene regulation to achieve a more specified phenotypic outcome. Finally, to add further complexity to this system, through elegant biochemical and genetic experiments by Msadek and colleagues it has been shown that in addition to VraR, the S. aureus GraR response regulator also directly interacts with the VraFG transporter promoter (), and the GraS histidine kinase directly binds the transporter itself.Citation100 At present Canova et al. convincingly show that the previously mentioned eSTK-phosphorylated VraR residues are crucial for regulation of antibiotic resistance.Citation103 It will be interesting to understand how these 2 interacting TCS signaling systems (i.e., VraRS and GraRS), both of which are regulated by eSTK/Ps, work together to enable cationic peptide and vancomycin resistance in such an important clinically-relevant pathogen.
Mycobacterium tuberculosis
M. tuberculosis is the causative agent of one of the oldest recorded afflictions of mankind, and is responsible for an annual death toll of millions worldwide. The increasing difficulties associated with its treatment have, in one respect, been attributed to the bacteria's ability to enter a state of dormancy and thus persist within the host. This issue coupled with an alarming increase in antibiotic resistance has now enabled this pathogen to become a source of high morbidity and mortality worldwide.Citation104 A large part of this pathogen's ability to regulate antibiotic resistance and its persistent lifestyle in the host can be attributed to the 11 eSTKs (named PknA through PknL), the single eSTP (called MstP), and a growing list of important eSTK/P regulatory targets (reviewed in refs.Citation6,22). Within this list of regulated proteins are several transcription factors that play crucial roles in the pathogenicity and virulence of M. tuberculosis infection (summarized in ).
Figure 6. Overview of eSTK control of transcription in M. tuberculosis. Schematic diagram showing the signaling pathways involved in eSTK control of M. tuberculosis regulatory factors mentioned in this text. Phosphorylation is denoted by the circle with a “P.” Red circles indicate serine or threonine phosphorylation and blue aspartate phosphorylation. Virulence and other traits that the regulators control is described, as are regulated operons (if known). LBD, lipid-binding domain; β-Pro, β-propeller domain; TM, transmembrane region. Other abbreviations are also described in the text.
The primary point of dormancy regulatory control in M. tuberculosis is mediated by the TCS-like response regulator DosR (also known as DevR and Rv3133c).Citation105 The DosR regulon consists of over 50 genes that are activated in response to hypoxia, nitric oxide and carbon monoxide. These signals are collectively sensed by the 2 cognate histidine kinases DosS and DosT, which through aspartate phosphorylation of DosR control entry of M. tuberculosis into its persistent state.Citation106,107 The discovery that DosR was also regulated by phosphorylation by the eSTK PknH on 2 threonine residues (Thr198 and Thr205) located within its C-terminal helical dimerization interface added an another regulatory complexity to this TCS-like systemCitation108 (). Phosphorylation of the DosR threonines has the effect of enhancing DosR DNA-binding affinity to a level comparable to a similar enhancement through Asp phosphorylation by the cognate histidine kinase DosS. Interestingly, DosR binding to its target DNA is synergistically enhanced by the simultaneous phosphorylation of its threonine residues and the canonical TCS aspartate, as compared to either PTM alone. These data collectively suggest that modulation of the M. tuberculosis hypoxia response is mediated by a convergence of eSTK and TCS regulated pathways, and that phosphorylation by both is required to achieve full activation of the DosR regulon.Citation108 In general, the PknH kinase itself appears to respond to variations of environmental stresses. Treatment of Mycobacteria cells with nitric oxide leads to a two-fold induction of expression the DosR regulon under the control of PknH while exposure to acidic pH and excess heat also reduces PknH expression.Citation108 However, a more recent report has come a step closer to finding the PknH ligand. Av-Gay and colleagues have found that disruption of PknH affects the synthesis of phthiocerol dimycocerosates (PDIMs), a major class of cell wall associated lipids in M. tuberculosis that are required for its virulence.Citation109 Although the actual ligand for PknH remains at large, a crystal structure of the extracellular domain has revealed a large hydrophobic and polar cleft where cell wall related lipids could bind.Citation110 Future work will have to determine the precise inducer of PknH activation and the subsequent downstream effects on regulation of the crucial TCS-like transcription factor DosR.
Of equal importance to mycobacterial persistence is its ability to resist antimicrobial treatments. The transcription factor EmbR is integral to these regulatory functions through the control of 3 genes, embA-C, which mediate resistance to ethambutol, a key frontline antitubercular drug that targets arabinosyltransferases.Citation111 The products of the embCAB operon are involved in the arabinosylation of the cell wall component lipomannan to produce lipoarabinomannan, the former of which induces a potent proinflammatory response and are critical to the structure of the mycobacterial cell envelope.Citation111 In the context of this review, the EmbR regulator is also historically important in that it was the first eSTK substrate to be identified in M. tuberculosis. EmbR is member of the Streptomyces antibiotic regulatory protein (SARP) family of transcription factorsCitation112,113 and maintains a unique structure consisting of an N-terminal OmpR-type DNA-binding domain, followed by a bacterial transcriptional activation (BTA) domain whose fold resembles that of the tetratricopeptid repeat (TPR) architecture of human phosphatases.Citation114 An interesting feature of EmbR is its C-terminal forkhead-associated (FHA) domain, a conserved class of short protein motifs known to facilitate recognition of phospho-threonine-containing peptides, including those belonging to eSTKsCitation112 (). Indeed, several key papers have now shown in great molecular detail how FHA domains can act as intramolecular inhibitors of target protein binding by association with a phospho-threonine group on a different portion of the FHA-containing protein.Citation114-116 To help illustrate this interaction, shows an atomic structure of the EmbR FHA domain (ref. 114) bound to a phospho-threonine peptide.
Figure 7. eSTK phosphorylation sites in M. tuberculosis transcription factors. Annotation is as per . Domain architectures are shown on the left, and 3-dimensional atomic structures and models on the right. The PDB codes for the DosR DNA-binding domain (DBD) and the EmbR-forkhead-associated domain (FHA; shown in blue) in complex with an interacting peptide (in magenta) and phosphate (shown in red) are 3C3W (ref. 163) and 2FF4 (ref. 114), respectively. The RshA anti σ-factor structure was generated with Swiss-Model using an available crystal structure from the anti-sigma factor ChrR (PDB code 2Q1Z; ref. 164) as a molecular template. eSTK phosphorylated residues are shown in red. Abbreviations not previous described are tet-HTH, TetR family helix-turn-helix domain; WHG, tryptophan-histidine-glycine domain presumed to bind a ligand or unknown effector molecule; UnKn ED, unknown effector domain; BTAD, bacterial transcriptional activator domain; wHTH*, wHTH domain missing helix 3; anti-σ: anti-sigma factor; anti-σ ID, anti-sigma factor interacting domain; PD, phosphorylation domain.
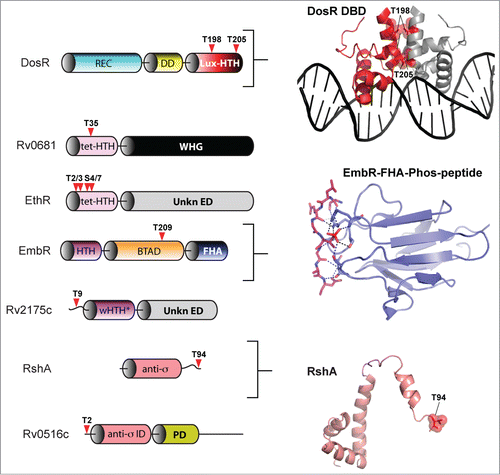
In M. tuberculosis EmbR is also targeted by the PknH kinase, where it is phosphorylated on threonine residues to stimulate interaction with the embCAB promoterCitation111,112 (). These PTMs can then be nullified upon truncation of the FHA domain, or by mutation of highly conserved residues within,Citation112 highlighting its requirement for the EmbR-PknH interaction. Positive regulation of the embCAB operon through EmbR serine-threonine phosphorylation also occurs in vivo, which correlates with an increase in lipoarabinomannan levels and resistance to ethambutol. In an interesting twist to this story, a protein dubbed “EmbR2,” which is 56% identical to EmbR, competes with PknH as a substrate, yet is unable to accept a phosphate. Thus EmbR2 might act as a natural dominant negative regulator of the “real” EmbR to help facilitate an effective inhibition of PknH autophosphorylation.Citation117 In addition to PknH, PknA and PknB are also capable of phosphorylating EmbR in vitro, which can be subsequently dephosphorylated by the cognate eSTP MstP.Citation118 In these studies MstP driven dephosphorylation of EmbR dramatically reduces its DNA binding ability in vitro.Citation118 Although these interactions and their relevance to in vivo regulation of EmbR have yet to be determined, like the SarA factor from S. aureus described above it is interesting to note that several eSTKs might be co-regulating a single transcription factor. Studies showing the precise modified amino acids, their mechanistic and phenotypic outcomes will be of interest in future studies.
In silico predictions have also proven useful for identification of potential phospho-targets of eSTKs. For example this methodology allowed Av-Gay and colleagues to identify targets of PknH, where it was reasoned that the kinase's substrate recognition motif, determined to be “TQLGT,” resembles the core autophosphorylation site of the eSTK itself.Citation119 Forty PknH targets have been identified using this method, including the already characterized EmbR that served as a control, and the putative TetR family transcription factor dubbed “Rv0681.” In EmbR, mutating the predicted phosphorylated BTA domain residue Thr209 produces only a slight decrease in PknH phosphorylation in vitro, suggesting that additional Ser or Thr residues might be modified. Conversely, replacing the single predicted phospho-modified site within Rv0681, Thr35 within the DNA-binding domain, serves to completely abrogate PknH phosphotransfer to this substrate. These findings require verification as relevant in vivo. However, as the recognition sequences for different eSTK homologs are likely to vary,Citation26 in silico and extensive proteomic experiments have thus far proven to be a powerful means from which to identify their pleiotropic substrates.Citation119 In support, a comprehensive study using proteomics to identify eSTK targets and their associated phosphopeptides was able to generate a potential list of eSTK specific recognition motifs. These motifs could then presumably be used to conduct searches for additional eSTK substrates and thus provides a template for many studies to come.Citation26
Another drug that is typically used to treat active tuberculosis is ethionamide (ETH). To be effective, ETH must be activated and its potency is enhanced through chemical modification by the monooxygenase EthA.Citation120 Expression of ethA is, in turn, repressed by the TetR family transcription factor EthR, and thus by association EthR controls levels of ETH resistance in MycobacteriaCitation121 (). In a recent publication, Molle and colleagues show that EthR is an in vitro substrate for the M. tuberculosis eSTK PknF.Citation122 Mass spectrometry identifies 4 phosphorylated residues within the extreme and seemingly unstructured N-terminus of the protein (Thr2, Thr3, Ser4, and Ser7; ). Phosphotransfer by PknF is only absent when either all 4 residues are substituted for, or alternatively can be accomplished by deleting the entire N-terminal sequence.Citation122 Further work demonstrates that in vitro DNA-binding to the EthR target ethA promoter decreases in these serine-threonine mutant EthR versions compared to wild-type.Citation122 Interestingly, the authors go on to show that in the presence of the potent EthR-targeting synthetic anti-mycobacterial antibiotic 1,2,4-oxadiazole, the observed negative effect PknF phosphorylation has on EthR DNA affinity is further enhanced by addition of the drug. From these data the authors proposed a 2-pronged mechanism of PknF mediation of EthR control. First the PTMs inhibit EthR from binding DNA through direct interference with its protein-DNA static interactions. Second, eSTK phosphorylation enhances the effect of 1,2,4-oxadiazole by increasing affinity for its allosteric binding site, which is located in a structurally distinct position from the EthR protein-DNA interface. Collectively, these data indicate a possible M. tuberculosis resistance mechanism for the 1,2,4-oxadiazole class of drugs mediated through eSTK transcription factor phospho-modification. Future studies will include using in vivo techniques and clinical isolates to further test the efficacy of these antibacterial compounds.Citation122
Another M. tuberculosis eSTK targeted transcription factor, Rv2175c, thought to be involved in the control of important cell envelope related functions, was discovered through an initial proteomic-based study in search of novel kinase substrates.Citation123 A follow-up study identifies Thr9 as being the sole residue in Rv2175c (), which is specifically phosphorylated by PknL. Although the exact inducing signal for PknL has yet to be determined, evidence suggests a role similar to that of PknA and PknB in cell wall associated functions, as well as the regulation of glutamine metabolism.Citation124 An available structure of the PknL substrate Rv2175c reveals a surprising OmpR-like TCS winged helix-turn-helix (wHTH) DNA binding motif that is missing its typical third helix, a structural phenomenon usually only observed in phage-derived transcription factors. The wHTH is preceded by a flexible and disordered 16-residue long N-terminus that contains Thr9.Citation125 DNA-binding activity can be completely abrogated by a Thr9 to Asp phospho-mimetic mutation, and yet remains intact after deletion of the first 13 N-terminal residues of the protein containing this residue. With the tertiary structure of the monomer remaining virtually unaffected by phosphorylation,Citation125 it can be concluded that the PTM might affect dimerization, complex-forming interactions with RNAP or other cofactors at the bacterial promoter. Interestingly, the N-terminal regulated extension is unique to M. tuberculosis, not even found in other closely related mycobacterial species,Citation125 which suggests species-specific eSTK control of the Rv2175c regulon. An interesting aspect of these results is that where a plethora of OCS transcription factors similar to Rv2175c have now been identified, it has remained enigmatic as to how they are regulated in the absence of an obvious input domain. This work by Cohen-Gonsaud, Molle and colleagues suggests the exciting prospect that such eSTK control over mycobacterial ‘DNA-binding domain only’ transcription factors might now offer a ubiquitous mechanism for their regulation.
Also contributing to the survival of host-inflicted stresses on M. tuberculosis is the AraC family transcription factor VirS. VirS is homologous to several virulence-regulating transcription factors found in other pathogens (e.g., Shigella) and related mycobacterial species, where it mediates the correct formation of the cell wall structure and protects against low pH conditions.Citation126 The involvement of VirS in these stress responses is regulated through the mycobacterial eSTK PknK, an eSTK required for survival in the early stages of mycobacterial infection and persistence, and likely facilitates adaptation to intracellular environments through translational controlCitation127 (). Along with PknG, PknK is evolutionarily distinct from that of the other M. tuberculosis eSTKs in that it is widely distributed in prokaryotes, both pathogenic and non-pathogenic, and is a soluble protein. At present the signals that PknK responds to are not known, however the presence of a C-terminal STAND domain suggests it might be a signaling hub for many sensory inputs that remain to be elucidated.Citation127 On the other hand some PknK substrates have been revealed. Using VirS protein produced in a heterologous E. coli expression system,Citation128 Kuman et al. were able to show that PknK phosphorylates VirS. The phosphorylated protein exhibited a 2.5 fold higher affinity for the VirS target promoter of mymA that encodes a putative monooxygenase.Citation129 Although the site(s) of phosphorylation have not been elucidated, PknK control of VirS was shown to be relevant in vivo by enabling a regulatory effect over mymA operon transcription.Citation129 The same work by Singh et al. highlights the importance of mymA in controlling the mycobacterial cell wall ultrastructure, persistence within activated macrophages and the specific requirement for infection of the spleen but not the lungs (). These studies further highlight the individual roles mycobacterial eSTKs play in virulence and infection of diverse host tissues.
Although little data is available on the subject, some proteomic studies have implicated eSTKs in the regulation of transcription through direct phosphorylation of core transcriptional machinery. For example, by employing proteomic approaches in the human pathogen S. pneumoniae, the α subunit of RNAP was identified as a possible substrate of the native eSTK (StkP) in this organism.Citation23 Similar techniques have also been used in Listeria monocytogenes to demonstrate interaction between a Listeria eSTK and the α and β subunits of the RNAP holoenzyme.Citation27 Taken together, these findings suggest that eSTKs may use modification of transcription machinery components to influence gene expression on a global scale. However intriguing, data from these experiments have not been verified by additional biochemical and genetic methods. On the other hand, direct regulation of sigma factors by eSTKs and eSTPs and their mechanistic consequences have been demonstrated, especially within M. tuberculosis. One example includes SigH, the alternative sigma factor responsible for regulation of mycobacterial stress responses and required to achieve the full virulence potential of this organism according to a murine model of infection.Citation130
The activity of SigH is post-translationally regulated via interaction with the anti-sigma factor RshA in an effort to control mycobacterial internal redox conditions.Citation130 The SigH-RshA interaction occurs in a redox-dependent manner through 4 conserved cysteine residues within RshA. RshA is itself a substrate of the eSTK PknB and its phosphorylation interferes with the SigH interaction. This PTM might therefore serve to limit the activation of SigH dependent regulons.Citation131 Combing extensive in vivo and in vitro experiments with mass spectrometry analysis, it was determined that RshA can be phosphorylated on just one threonine residue that is relevant to its in vivo activity (Thr94). Intriguingly, Thr94 is located near the RshA C-terminus by helix 4 and is a critical region for sigma factor/anti-sigma factor interactions (). eSTK-mediated phosphorylation of SigH can also be observed on 2 of its threonines (Thr26 and Thr102).Citation131 However, differing from RshA phosphorylation, the SigH PTMs does not produce a pronounced disruption in SigH-RshA complex formation. Possible explanations for this discrepancy could be low phosphorylation stoichiometry, or that SigH phosphorylation may instead be specific for an as-of-yet unidentified non-RshA interacting partner.
M. tuberculosis also possesses many anti-anti-sigma factors that act to sequester and inhibit anti-sigma factors and promote RNAP recruitment under specific environmental circumstances. One of these, the predicted anti-anti-sigma factor Rv0516c, has been identified as a substrate of the eSTK PknD by Alber and colleaguesCitation132 (). The PTM occurs in vitro within the N-terminal sequence on Thr2, and also in vivo when over-expressed with the PknD kinase. Phosphorylation of Rv0516c results in an effective abolition of its interaction with its anti-anti-sigma factor Rv2638, and a subsequent activation of the SigF regulon that responds to osmotic shockCitation39 (). In the same publication by Greenstein et al., the authors expand on these studies by testing a combination of 8 different putative mycobacterium anti-anti-sigma factor substrates for their ability to receive a phosphate from PknA, PknB, PknD, PknE, and PknK eSTKs.Citation132 Results reveal a surprising specificity for these substrates by specific eSTKs. For example, PknA phosphorylates no anti-anti-sigma factors, whereas PnkB and PknE only phosphorylates a single substrate out of the 8 tested. This study shows the potential extensiveness of eSTK regulation of transcription in a single organism, and gives us a glimpse of how eSTKs might similarly control many signaling networks in most bacterial species with surprising discrimination.
Streptococci
The Streptococcus encompasses numerous species of pathogenic Gram-positive bacteria, including but not limited to S. pyogenes (also referred to as Group A streptococci or GAS), S. agalactiae (also called Group B streptococci or GBS) and S. pneumoniae, all of which are capable of causing a diverse array of debilitating diseases. GAS, GBS and S. pneumoniae each possess only a single predicted PASTA domain-containing eSTK/eSTP pair, called Stk1 in GAS/GBS and StkP in S. pneumoniae, all of which are required for infectionCitation7,8,13,25 (reviewed by Burnside and Rajagopal in ref.Citation133). The regulatory effects of eSTK phosphorylation in the streptococci are summarized in .
Figure 8. Overview of eSTK control of transcription in the streptococci. Schematic diagram showing the signaling pathways involved in eSTK control of Group A Streptococci (GAS), Group B Streptococci (GBS) and S. pneumoniae regulatory factors mentioned in this text. In GAS and GBS the PASTA-containing eSTK is called Stk1, and in S. pneumoniae StkP. Stk1 from both GAS and GBS modulates the TCS regulator CovR in these species, whereas WalR has thus far only been shown to be phosphorylated by Stk1 in GAS. Similarly, StkP modulates the TCS regulators RR06 and RitR in S. pneumoniae. The question mark shown above RitR indicates that aside form StkP phosphorylation, regulation from other input signals has not been identified. The phosphorylation PTM is denoted by the circle with a “P.” Red circles indicate serine or threonine phosphorylation, whereas blue indicates aspartate phosphorylation. Virulence and other traits that the regulators control are also described. Transmembrane region (TM). The TCS histidine kinases CovS, WalK and HK06 have 2 TMs each that are not annotated here for purposes of simplicity.
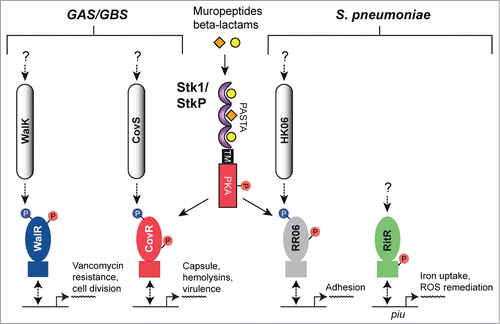
The CovR transcription factor (also referred to as CsrR) is part of the control of virulence (CovRS) TCS system that regulates a large number of genes in both GBS and GAS, including those involved with production of the capsule, major virulence factors, penetration of blood-tissue barriers and avoidance of the immune systemCitation134-136 (). For GBS, strains lacking their eSTK Stk1 were deficient in their ability to survive neutrophil attack and oxidative stress, and the ability to produce their key virulence factor β-hemolysin/cytotolysin (β-H/C).Citation137 This landmark paper by Rajagopal et al. demonstrated for the first time that these processes are regulated by the convergence of the CovRS TCS system and an eSTK, and importantly is the initial example of such co-regulatory control in a pathogen. Subsequent work from the same group has demonstrated that the regulation of β-H/C is mediated through eSTK phosphorylation of Thr65 in CovR, which abolishes binding to the β-H/C promoter and in doing so, alleviates its inhibition on the expression of β-H/C.Citation138 In contrast, phosphorylation of the conserved CovR aspartate residue (Asp53) by the cognate CovS histidine kinase serves to repress β-H/C production. A more recent study has further expanded on these findings to demonstrate how the homologous eSTK and CovRS TCS regulatory pathways in GAS converge to fine-tune global virulence traits through the same site (ref.Citation139 and see below).
The eSTK-regulated Thr65 residue is located within helix 3 of CovR REC domain. Although an atomic structure of the CovR REC domain is not available, the model shown in predicts Thr65 to reside within a deep cleft, facing the interior of the protein where the phospho-accepting oxygen is hydrogen-bonding to nearby residues. Although the eSTK substrate recognition domain is known to be flexible,Citation21 Thr65 would seem difficult to access within this model static structure. For comparison, in the REC domain of the S. aureus regulator VraR the eSTK-modified Thr119 is completely solvent-inaccessible and facing the interior of the protein ( and data not shown). How eSTKs are able to access such residues is of great interest. One plausible explanation is REC domains exist in a dynamic equilibrium of “on/off” states, and similar to the canonical phosphorylation of the REC aspartate, a serine or threonine modification could shift this equilibrium toward the CovR protein “on” state rather than the “off."Citation140 The predicted Thr65 position in 3-dimensional space is also interesting, where its electron cloud kisses that of the opposing Leu52 residue, the latter of which is located just N-terminal to the CovS-regulated Asp53 (). With this observation in mind we can envision the phosphorylation of Thr65 perturbing the interior of the REC domain, shifting the protein's equilibrium state and altering the position of the canonical phospho-Asp site in such a way as to disfavor CovS interaction and/or Asp53 modification.
Figure 9. Comparison of CovR and VraR Receiver (REC) domain phosphorylation sites. (A) (left panel) Cartoon representation of an atomic model of the CovR REC domain. The model was created with Swiss-Model (ref.Citation161) using the CovR sequence from S. agalactiae and the PhoP structure from M. tuberculosis (PDB code 3R0J; ref.Citation165) as a molecular template. Phosphorylated residues are shown in red. The conserved histidine kinase phosphorylated aspartate (Asp53) is shown in green. Residues that would potentially pose a steric clash with the phosphorylated Thr65 are shown in yellow. (right panel) Surface representation of the left panel rotated 180 degrees. Notice that Thr65 is solvent accessible, but resides in a deep cleft within the interior of the protein. (B) The VraR atomic (REC) structure taken from ref.Citation63 showing the eSTK-regulated Thr119 (colored red) and potential residues that might clash with it in its phosphorylated form (colored yellow). The conserved Asp55 is shown in green.
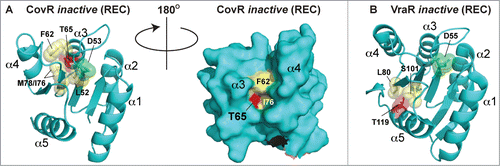
Paralleling the aforementioned findings by Rajagopal and colleagues, Argwal et al. show that the eSTK/eSTP pair from S. pyogenes is also able to reversibly phosphorylate the endogenous CovR GAS homolog. Moreover, in the same work they identify the essential TCS regulator WalR, responsible for controlling crucial pathways involving cell wall metabolism and in vivo accumulation of multidrug resistance,Citation141 as an additional eSTK/P targetCitation10 (for a review on the WalRK TCS see ref. 142). Both CovR and WalR S. pyogenes homologs are phosphorylated on threonine residues as determined by chromatography experiments (). Whereas the WalR modified site(s) have yet to be deciphered, the precise site of eSTK regulation of the GAS CovR is now known to be identical to that of the GBS homolog (i.e. Thr65). These recent and elegant studies by Horstmann et al. (ref. 139) not only demonstrate the relevance of Thr65 to GAS infection and virulence, but also importantly show the potential for eSTK/P cross-species conservation of regulatory control.
A very novel observation stemming from the studies by Pancholi and colleagues was that in infected pharyngeal cells the S. pyogenes eSTP was exported from the pathogen into the host nucleus to initiate pro-apoptotic events and a redirection of the host transcriptome.Citation54 In further support of these findings, Miller et al. have previously identified the S. aureus eSTK PknB as a secreted protein, which was also shown to phosphorylate pro-apoptotic host proteins in vitro.Citation143 eSTK and eSTP modulation of host proteins has also been described in Yersinia sp. Salmonella sp. and several other important bacterial pathogens.Citation144,145 An area of research to expand upon would include determining how eSTK/Ps, especially those containing transmembrane regions and from species that do not possess typical effector secretion systems (e.g. type III), are able export eSTK/Ps out of the bacterial cell, into the host cytoplasm, and in some cases into the nucleus to enable direct rewiring of host transcriptional circuitry.
Despite an available vaccine, S. pneumoniae continues to be one of the leading causes of morbidity and mortality worldwide, especially in young children.Citation146 Understanding how this organism is able to infect such a variety of tissues and cause disease is a crucial step in the progress of producing novel antibiotics and antigens for vaccines. A major focus of preventing pneumococcal disease is deciphering how it adheres to and colonizes the seemingly harsh environment of the nasopharynx, where it is exposed to unwanted oxygen and has to contend with the host immune system.Citation146,147 In another study carried out by Pancholi and colleagues, the pneumococcal TCS response regulator RR06, which controls expression of adhesion elements,Citation148 exhibits reversible regulation by the eSTK/P pair in this pathogen.Citation149 Furthermore, phosphorylation of RR06 on threonine residue(s) results in added stability to its DNA binding (). However the site of modification and in vivo relevance of these modifications still remains at large. An interesting outcome from these studies was that the pneumococcal eSTK/P pair demonstrates control over its virulence properties in a strain-specific manner. These findings hint at how S. pneumoniae and other pathogenic bacteria might use eSTK/P regulation to alter required target specificity through evolving single amino acid changes in their substrates in response to changing host environments. In support, it has recently been reported that point mutations within the GAS homolog of CovR in certain clinical isolates have dramatic effects on expression of CovR-regulated virulence factors, pathogenesis and presumed clinical outcomes.Citation150
During infection streptococci and all other pathogens are reliant on the host to provide their nutritional requirements or perish. For most bacterial pathogens their success is measured in part by host iron availability. Ironically, although iron is the fourth most abundant metal on earth, within the host environment it is surprisingly scarce, where it is found at concentrations far less than required to sustain bacterial growth (<10−9 M on average).Citation151 However when obtained, iron must be kept distant from H2O2 to avoid killing by iron-catalyzed Fenton chemistry.Citation152 In S. pneumoniae this balance is especially important, as it produces up to millimolar quantities of H2O2 as a metabolic byproduct of pyruvate oxidase activity in the presence of oxygen.Citation153
Repressor of iron transport regulator (RitR) is a TCS-like response regulator in S. pneumoniae required for lung infection and responsible for iron uptake and remediation of iron-catalyzed ROS.Citation69,154,155 These actions are carried out in part by RitR-mediated repression of the pneumococcal iron uptake (Piu) transporter, and its simultaneous activation of bacterial ferritin (Dpr) and DNA repair enzymes under high oxygen tension (ref.Citation69 and unpublished data; ). RitR is unusual in that it is a so-called orphan response regulator, which by definition does not possess a cognate histidine kinase partner. Instead RitR is co-transcribed with 6-phosphogluconate dehydrogenase, a key enzyme in the pentose phosphate carbon utilization pathway.Citation69 Moreover, RitR is missing the conserved phosphorylated Asp residue within its REC domain. This posed the interesting question of how this transcription factor is regulated. To answer this question a randomized phage display library was employed to identify a peptide which specifically bound the RitR DNA-binding domain, and whose sequence was identical to the conserved catalytic pocket of the ubiquitous PP2C phosphatase family of proteins that includes eSTPs.Citation24 Titration of purified pneumococcal eSTP to RitR bound to target DNA revealed the formation of a tripartite eSTP-RitR-DNA complex, which can be disrupted by addition of the cognate eSTK partner StkP.Citation24 The pneumococcal eSTK (StkP) and eSTP (PhpP) were then shown to reversibly phosphorylate RitR within its DNA-binding domain to enable gene regulation of the Piu iron-heme transporter.Citation24 Although the exact site(s) of RitR phospho-regulation remain unknown, these studies hint at how bacterial eSTK and eSTPs might be found in complex formation at the site of transcription initiation to direct its control.
Interestingly, several subsequent publications have linked eSTK and eSTP regulation with controlling intracellular iron concentrations and resistance to ROS, including resistance to macrophage and neutrophil oxidative killing in many pathogens (e.g., refs.Citation156,157,25). As eSTK and eSTP regulated functions in the streptococci seem predominantly related to control of virulence factor release and cell envelope related functions, it currently remains unclear how they might also affect responses to a changing redox state, ROS and fluctuating metal ion concentrations. This remains an exciting topic for future investigations.
Future Perspectives
Bacterial eSTKs and eSTPs are ubiquitous signaling proteins found in virtually every important bacterial pathogen, where they are almost invariably required for infection and regulation of virulence. In recent years much progress has been made in understanding how these relays propagate pleiotropic phosphorylation to, for example, modify cell division, avoid host immunity and enable resistance to antimicrobials. One way these global modifications can be accomplished is by directly regulating transcription factors, and even core components of RNA polymerase to then, in turn, initiate a rewiring of transcriptional circuitry with even a single phospho-modification. An overview of possible mechanisms of eSTK/P control of transcription described thus far in the literature is summarized in . With the use of next generation sequencing, exciting possibilities now exist to understand the extent to which eSTK/P phosphorylation affects global transcriptional responses. In this regard, important future directions will include using such technology to determine a pathogen's transcriptome changes through eSTK/P signaling when variable tissues and environmental conditions are encountered during infection. With an ever-accumulating database of clinical isolates to sample, it will also be of future importance to determine how eSTK/Ps and their targets have co-evolved within the host, especially after treatments with antimicrobials. In support, dense genomic sampling of bacterial pathogens has revealed surprising changes in amino acid composition with the potential to accommodate new phosphorylatable sites, which could then enable efficient rewiring of infection and virulence traits. These discoveries combined with newly emerging prokaryotic PTMs such as tyrosine, cysteine and arginine phosphorylations will become increasingly important to more completely understand how the bacterial signaling network is used to successfully cause disease. Finally, a different yet equally important question to be answered is: How do eSTKs and eSTPs discriminate between a diverse array of cellular substrates to enable such precise and coordinated responses? Although specific substrate domain interactions such as FHA modules have been identified and explored, it remains to be determined how eSTK-substrate discrimination occurs with the rest of eSTK/P interacting partners. With the exception of SarA/MgrA family regulators and some sigma factor related proteins reviewed here, most modifications appear to occur on stable portions of the proteins with defined secondary and tertiary structures. Thus it is likely that to enable their substrate specificity, eSTKs and eSTPs might recognize a rigid 3-dimensional motif within their diverse native target substrate folds. For most eSTKs and eSTPs this has yet to be experimentally determined. To begin addressing this problem eSTK/P-substrate co-structural information and accompanying biochemical analyses must be produced. When combined with more randomized combinatorial approaches to substrate interactions, these data should help give us a more complete picture as to how these critical, and until recently underappreciated microbial signaling systems may be further exploited for design of novel antimicrobials.
Figure 10. Possible mechanisms of eSTK/eSTP regulation of transcription. (A) eSTK phosphorylation of a transcription factor can cause either a negative effect (left) or positive effect (right) on the recruitment of transcriptional machinery through direct sigma factor and/or RNA polymerase (RNAP) interactions. (B) eSTK phosphorylation can also directly influence transcription factor binding to target DNA by either obstructing sister monomer interactions, or by directly affecting its physical hydrogen-bonding to target DNA. These influences can result in either a negative (left panel) or positive (right panel) effect on σ-factor/RNAP recruitment. (C) eSTK phosphorylation of both anti-sigma factors (-σ) and anti-anti-sigma factors (−2σ) can result in negative (left) or positive (right) effects on σ-factor/RNAP recruitment.
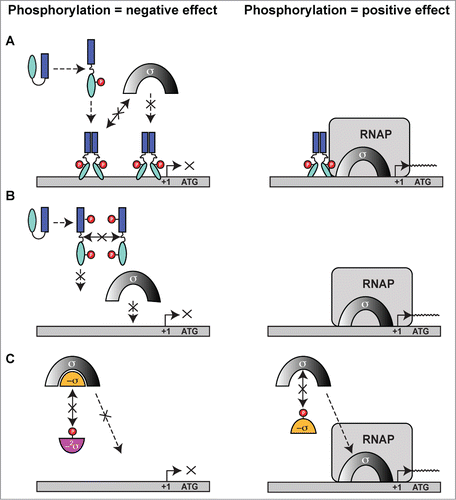
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This manuscript was supported by the Imperial College London Department of Medicine, and the Medical Research Council funded Ph.D. program to the Centre for Molecular Bacteriology and Infection (CMBI).
References
- Hoch JA, Silhavy TJ, eds. Two-component signal transduction. Washington, D.C.: American Society for Microbiology, 1995
- Stock JB, Stock AM, Mottonen JM. Signal transduction in bacteria. Nature 1990; 344:395-400; PMID:2157156; http://dx.doi.org/10.1038/344395a0
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem 2000; 69:183-215; PMID:10966457; http://dx.doi.org/10.1146/annurev.biochem.69.1.183
- Munoz-Dorado J, Inouye S, Inouye M. A gene encoding a protein serine/threonine kinase is required for normal development of M. xanthus, a gram-negative bacterium. Cell 1991; 67:995-1006; PMID:1835671; http://dx.doi.org/10.1016/0092-8674(91)90372-6
- Galyov EE, Hakansson S, Forsberg A, Wolf-Watz H. A secreted protein kinase of Yersinia pseudotuberculosis is an indispensable virulence determinant. Nature 1993; 361:730-2; PMID:8441468; http://dx.doi.org/10.1038/361730a0
- Molle V, Kremer L. Division and cell envelope regulation by Ser/Thr phosphorylation: Mycobacterium shows the way. Mol Microbiol 2010; 75:1064-77; PMID:20487298; http://dx.doi.org/10.1111/j.1365-2958.2009.07041.x
- Echenique J, Kadioglu A, Romao S, Andrew PW, Trombe MC. Protein serine/threonine kinase StkP positively controls virulence and competence in Streptococcus pneumoniae. Infect Immun 2004; 72:2434-7; PMID:15039376; http://dx.doi.org/10.1128/IAI.72.4.2434-2437.2004
- Rajagopal L, Clancy A, Rubens CE. A eukaryotic type serine/threonine kinase and phosphatase in Streptococcus agalactiae reversibly phosphorylates an inorganic pyrophosphatase and affects growth, cell segregation, and virulence. J Biol Chem 2003; 278:14429-41; PMID:12562757; http://dx.doi.org/10.1074/jbc.M212747200
- Wang J, Li C, Yang H, Mushegian A, Jin S. A novel serine/threonine protein kinase homologue of Pseudomonas aeruginosa is specifically inducible within the host infection site and is required for full virulence in neutropenic mice. J Bacteriol 1998; 180:6764-8; PMID:9852028
- Agarwal S, Pancholi P, Pancholi V. Role of serine/threonine phosphatase (SP-STP) in Streptococcus pyogenes physiology and virulence. J Biol Chem 2011; 286:41368-80; PMID:21917918; http://dx.doi.org/10.1074/jbc.M111.286690
- Martinez MA, Das K, Saikolappan S, Materon LA, Dhandayuthapani S. A serine/threonine phosphatase encoded by MG_207 of Mycoplasma genitalium is critical for its virulence. BMC Microbiol 2013; 13:44; PMID:23432936; http://dx.doi.org/10.1186/1471-2180-13-44
- Cameron DR, Ward DV, Kostoulias X, Howden BP, Moellering RC, Jr., Eliopoulos GM, Peleg AY. Serine/threonine phosphatase Stp1 contributes to reduced susceptibility to vancomycin and virulence in Staphylococcus aureus. J Infect Dis 2012; 205:1677-87; PMID:22492855; http://dx.doi.org/10.1093/infdis/jis252
- Bugrysheva J, Froehlich BJ, Freiberg JA, Scott JR. Serine/threonine protein kinase Stk is required for virulence, stress response, and penicillin tolerance in Streptococcus pyogenes. Infect Immun 2011; 79:4201-9; PMID:21788381; http://dx.doi.org/10.1128/IAI.05360-11
- Ohlsen K, Donat S. The impact of serine/threonine phosphorylation in Staphylococcus aureus. Int J Med Microbiol 2010; 300:137-41; PMID:19783479; http://dx.doi.org/10.1016/j.ijmm.2009.08.016
- Chawla Y, Upadhyay SK, Khan S, Nagarajan SN, Forti F, Nandicoori VK. Protein Kinase B (PknB) of Mycobacterium tuberculosis is essential for growth of the pathogen in vitro as well as for survival within the host. J Biol Chem 2014; 289:13858-75; PMID:24706757; http://dx.doi.org/10.1074/jbc.M114.563536
- Mougous JD, Gifford CA, Ramsdell TL, Mekalanos JJ. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat Cell Biol 2007; 9:797-803; PMID:17558395; http://dx.doi.org/10.1038/ncb1605
- Kristich CJ, Wells CL, Dunny GM. A eukaryotic-type Ser/Thr kinase in Enterococcus faecalis mediates antimicrobial resistance and intestinal persistence. Proc Natl Acad Sci U S A 2007; 104:3508-13; PMID:17360674; http://dx.doi.org/10.1073/pnas.0608742104
- Debarbouille M, Dramsi S, Dussurget O, Nahori MA, Vaganay E, Jouvion G, Cozzone A, Msadek T, Duclos B. Characterization of a serine/threonine kinase involved in virulence of Staphylococcus aureus. J Bacteriol 2009; 191:4070-81; PMID:19395491; http://dx.doi.org/10.1128/JB.01813-08
- Beltramini AM, Mukhopadhyay CD, Pancholi V. Modulation of cell wall structure and antimicrobial susceptibility by a Staphylococcus aureus eukaryote-like serine/threonine kinase and phosphatase. Infect Immun 2009; 77:1406-16; PMID:19188361; http://dx.doi.org/10.1128/IAI.01499-08
- Burnside K, Lembo A, de Los Reyes M, Iliuk A, Binhtran NT, Connelly JE, Lin WJ, Schmidt BZ, Richardson AR, Fang FC, et al. Regulation of hemolysin expression and virulence of Staphylococcus aureus by a serine/threonine kinase and phosphatase. PLoS One 2010; 5:e11071; PMID:20552019; http://dx.doi.org/10.1371/journal.pone.0011071
- Young TA, Delagoutte B, Endrizzi JA, Falick AM, Alber T. Structure of Mycobacterium tuberculosis PknB supports a universal activation mechanism for Ser/Thr protein kinases. Nat Struct Biol 2003; 10:168-74; PMID:12548283; http://dx.doi.org/10.1038/nsb897
- Av-Gay Y, Everett M. The eukaryotic-like Ser/Thr protein kinases of Mycobacterium tuberculosis. Trends Microbiol 2000; 8:238-44; PMID:10785641; http://dx.doi.org/10.1016/S0966-842X(00)01734-0
- Novakova L, Saskova L, Pallova P, Janecek J, Novotna J, Ulrych A, Echenique J, Trombe MC, Branny P. Characterization of a eukaryotic type serine/threonine protein kinase and protein phosphatase of Streptococcus pneumoniae and identification of kinase substrates. FEBS J 2005; 272:1243-54; PMID:15720398; http://dx.doi.org/10.1111/j.1742-4658.2005.04560.x
- Ulijasz AT, Falk SP, Weisblum B. Phosphorylation of the RitR DNA-binding domain by a Ser-Thr phosphokinase: implications for global gene regulation in the streptococci. Mol Microbiol 2009; 71:382-90; PMID:19040630; http://dx.doi.org/10.1111/j.1365-2958.2008.06532.x
- Jin H, Pancholi V. Identification and biochemical characterization of a eukaryotic-type serine/threonine kinase and its cognate phosphatase in Streptococcus pyogenes: their biological functions and substrate identification. J Mol Biol 2006; 357:1351-72; PMID:16487973; http://dx.doi.org/10.1016/j.jmb.2006.01.020
- Prisic S, Dankwa S, Schwartz D, Chou MF, Locasale JW, Kang CM, Bemis G, Church GM, Steen H, Husson RN. Extensive phosphorylation with overlapping specificity by Mycobacterium tuberculosis serine/threonine protein kinases. Proc Natl Acad Sci U S A 2010; 107:7521-6; PMID:20368441; http://dx.doi.org/10.1073/pnas.0913482107
- Lima A, Duran R, Schujman GE, Marchissio MJ, Portela MM, Obal G, Pritsch O, de Mendoza D, Cervenansky C. Serine/threonine protein kinase PrkA of the human pathogen Listeria monocytogenes: biochemical characterization and identification of interacting partners through proteomic approaches. J Proteomics 2011; 74:1720-34; PMID:21406257; http://dx.doi.org/10.1016/j.jprot.2011.03.005
- Silvestroni A, Jewell KA, Lin WJ, Connelly JE, Ivancic MM, Tao WA, Rajagopal L. Identification of serine/threonine kinase substrates in the human pathogen group B streptococcus. J Proteome Res 2009; 8:2563-74; PMID:19309132; http://dx.doi.org/10.1021/pr900069n
- Pereira SF, Goss L, Dworkin J. Eukaryote-like serine/threonine kinases and phosphatases in bacteria. Microbiol Mol Biol Rev 2011; 75:192-212; PMID:21372323; http://dx.doi.org/10.1128/MMBR.00042-10
- Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J 1995; 9:576-96; PMID:7768349
- Tyagi N, Anamika K, Srinivasan N. A framework for classification of prokaryotic protein kinases. PLoS One 2010; 5:e10608; PMID:20520783; http://dx.doi.org/10.1371/journal.pone.0010608
- Ortiz-Lombardia M, Pompeo F, Boitel B, Alzari PM. Crystal structure of the catalytic domain of the PknB serine/threonine kinase from Mycobacterium tuberculosis. J Biol Chem 2003; 278:13094-100; PMID:12551895; http://dx.doi.org/10.1074/jbc.M300660200
- Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell 2002; 109:275-82; PMID:12015977; http://dx.doi.org/10.1016/S0092-8674(02)00741-9
- Madhusudan, Akamine P, Xuong NH, Taylor SS. Crystal structure of a transition state mimic of the catalytic subunit of cAMP-dependent protein kinase. Nat Struct Biol 2002; 9:273-7; PMID:11896404; http://dx.doi.org/10.1038/nsb780
- Krupa A, Srinivasan N. Diversity in domain architectures of Ser/Thr kinases and their homologues in prokaryotes. BMC Genomics 2005; 6:129; PMID:16171520; http://dx.doi.org/10.1186/1471-2164-6-129
- Germain E, Castro-Roa D, Zenkin N, Gerdes K. Molecular mechanism of bacterial persistence by HipA. Mol Cell 2013; 52:248-54; PMID:24095282; http://dx.doi.org/10.1016/j.molcel.2013.08.045
- Barthe P, Mukamolova GV, Roumestand C, Cohen-Gonsaud M. The structure of PknB extracellular PASTA domain from mycobacterium tuberculosis suggests a ligand-dependent kinase activation. Structure 2010; 18:606-15; PMID:20462494; http://dx.doi.org/10.1016/j.str.2010.02.013
- Chakraborti PK, Matange N, Nandicoori VK, Singh Y, Tyagi JS, Visweswariah SS. Signalling mechanisms in Mycobacteria. Tuberculosis (Edinb) 2011; 91:432-40; PMID:21570916; http://dx.doi.org/10.1016/j.tube.2011.04.005
- Hatzios SK, Baer CE, Rustad TR, Siegrist MS, Pang JM, Ortega C, Alber T, Grundner C, Sherman DR, Bertozzi CR. Osmosensory signaling in Mycobacterium tuberculosis mediated by a eukaryotic-like Ser/Thr protein kinase. Proc Natl Acad Sci U S A 2013; 110:E5069-77; PMID:24309377; http://dx.doi.org/10.1073/pnas.1321205110
- Yeats C, Finn RD, Bateman A. The PASTA domain: a beta-lactam-binding domain. Trends Biochem Sci 2002; 27:438; PMID:12217513; http://dx.doi.org/10.1016/S0968-0004(02)02164-3
- Jones G, Dyson P. Evolution of transmembrane protein kinases implicated in coordinating remodeling of gram-positive peptidoglycan: inside versus outside. J Bacteriol 2006; 188:7470-6; PMID:16936012; http://dx.doi.org/10.1128/JB.00800-06
- Squeglia F, Marchetti R, Ruggiero A, Lanzetta R, Marasco D, Dworkin J, Petoukhov M, Molinaro A, Berisio R, Silipo A. Chemical basis of peptidoglycan discrimination by PrkC, a key kinase involved in bacterial resuscitation from dormancy. J Am Chem Soc 2011; 133:20676-9; PMID:22111897; http://dx.doi.org/10.1021/ja208080r
- Maurer P, Todorova K, Sauerbier J, Hakenbeck R. Mutations in Streptococcus pneumoniae penicillin-binding protein 2x: importance of the C-terminal penicillin-binding protein and serine/threonine kinase-associated domains for beta-lactam binding. Microb Drug Resist 2012; 18:314-21; PMID:22455550; http://dx.doi.org/10.1089/mdr.2012.0022
- Shah IM, Laaberki MH, Popham DL, Dworkin J. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell 2008; 135:486-96; PMID:18984160; http://dx.doi.org/10.1016/j.cell.2008.08.039
- Beilharz K, Novakova L, Fadda D, Branny P, Massidda O, Veening JW. Control of cell division in Streptococcus pneumoniae by the conserved Ser/Thr protein kinase StkP. Proc Natl Acad Sci U S A 2012; 109:E905-13; PMID:22431591; http://dx.doi.org/10.1073/pnas.1119172109
- Cohen P. Classification of protein-serine/threonine phosphatases: identification and quantitation in cell extracts. Methods Enzymol 1991; 201:389-98; PMID:1658556; http://dx.doi.org/10.1016/0076-6879(91)01035-Z
- Wehenkel A, Bellinzoni M, Schaeffer F, Villarino A, Alzari PM. Structural and binding studies of the three-metal center in two mycobacterial PPM Ser/Thr protein phosphatases. J Mol Biol 2007; 374:890-8; PMID:17961594; http://dx.doi.org/10.1016/j.jmb.2007.09.076
- Rantanen MK, Lehtio L, Rajagopal L, Rubens CE, Goldman A. Structure of Streptococcus agalactiae serine/threonine phosphatase. The subdomain conformation is coupled to the binding of a third metal ion. FEBS J 2007; 274:3128-37; PMID:17521332; http://dx.doi.org/10.1111/j.1742-4658.2007.05845.x
- Schlicker C, Fokina O, Kloft N, Grune T, Becker S, Sheldrick GM, Forchhammer K. Structural analysis of the PP2C phosphatase tPphA from Thermosynechococcus elongatus: a flexible flap subdomain controls access to the catalytic site. J Mol Biol 2008; 376:570-81; PMID:18164312; http://dx.doi.org/10.1016/j.jmb.2007.11.097
- Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell 2009; 139:468-84; PMID:19879837; http://dx.doi.org/10.1016/j.cell.2009.10.006
- Ulrich LE, Koonin EV, Zhulin IB. One-component systems dominate signal transduction in prokaryotes. Trends Microbiol 2005; 13:52-6; PMID:15680762; http://dx.doi.org/10.1016/j.tim.2004.12.006
- Das AK, Helps NR, Cohen PT, Barford D. Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 A resolution. EMBO J 1996; 15:6798-809; PMID:9003755
- Passalacqua KD, Satola SW, Crispell EK, Read TD. A mutation in the PP2C phosphatase gene in a Staphylococcus aureus USA300 clinical isolate with reduced susceptibility to vancomycin and daptomycin. Antimicrob Agents Chemother 2012; 56:5212-23; PMID:22850507; http://dx.doi.org/10.1128/AAC.05770-11
- Agarwal S, Jin H, Pancholi P, Pancholi V. Serine/threonine phosphatase (SP-STP), secreted from Streptococcus pyogenes, is a pro-apoptotic protein. J Biol Chem 2012; 287:9147-67; PMID:22262847; http://dx.doi.org/10.1074/jbc.M111.316554
- Bourret RB. Receiver domain structure and function in response regulator proteins. Curr Opin Microbiol 2010; 13:142-9; PMID:20211578; http://dx.doi.org/10.1016/j.mib.2010.01.015
- Galperin MY. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol 2006; 188:4169-82; PMID:16740923; http://dx.doi.org/10.1128/JB.01887-05
- Martinez-Hackert E, Stock AM. Structural relationships in the OmpR family of winged-helix transcription factors. J Mol Biol 1997; 269:301-12; PMID:9199401; http://dx.doi.org/10.1006/jmbi.1997.1065
- Gao R, Stock AM. Molecular strategies for phosphorylation-mediated regulation of response regulator activity. Curr Opin Microbiol 2010; 13:160-7; PMID:20080056; http://dx.doi.org/10.1016/j.mib.2009.12.009
- Toro-Roman A, Mack TR, Stock AM. Structural analysis and solution studies of the activated regulatory domain of the response regulator ArcA: a symmetric dimer mediated by the alpha4-beta5-alpha5 face. J Mol Biol 2005; 349:11-26; PMID:15876365; http://dx.doi.org/10.1016/j.jmb.2005.03.059
- Kern D, Volkman BF, Luginbuhl P, Nohaile MJ, Kustu S, Wemmer DE. Structure of a transiently phosphorylated switch in bacterial signal transduction. Nature 1999; 402:894-8; PMID:10622255; http://dx.doi.org/10.1038/47273
- Buckler DR, Zhou Y, Stock AM. Evidence of intradomain and interdomain flexibility in an OmpR/PhoB homolog from Thermotoga maritima. Structure 2002; 10:153-64; PMID:11839301; http://dx.doi.org/10.1016/S0969-2126(01)00706-7
- Bachhawat P, Swapna GV, Montelione GT, Stock AM. Mechanism of activation for transcription factor PhoB suggested by different modes of dimerization in the inactive and active states. Structure 2005; 13:1353-63; PMID:16154092; http://dx.doi.org/10.1016/j.str.2005.06.006
- Leonard PG, Golemi-Kotra D, Stock AM. Phosphorylation-dependent conformational changes and domain rearrangements in Staphylococcus aureus VraR activation. Proc Natl Acad Sci U S A 2013; 110:8525-30; PMID:23650349; http://dx.doi.org/10.1073/pnas.1302819110
- Browning DF, Busby SJ. The regulation of bacterial transcription initiation. Nat Rev Microbiol 2004; 2:57-65; PMID:15035009; http://dx.doi.org/10.1038/nrmicro787
- Makino K, Amemura M, Kawamoto T, Kimura S, Shinagawa H, Nakata A, Suzuki M. DNA binding of PhoB and its interaction with RNA polymerase. J Mol Biol 1996; 259:15-26; PMID:8648643; http://dx.doi.org/10.1006/jmbi.1996.0298
- Blanco AG, Canals A, Bernues J, Sola M, Coll M. The structure of a transcription activation subcomplex reveals how sigma(70) is recruited to PhoB promoters. EMBO J 2011; 30:3776-85; PMID:21829166; http://dx.doi.org/10.1038/emboj.2011.271
- Ulrich LE, Zhulin IB. The MiST2 database: a comprehensive genomics resource on microbial signal transduction. Nucleic Acids Res 2010; 38:D401-7; PMID:19900966; http://dx.doi.org/10.1093/nar/gkp940
- O'Hara BP, Norman RA, Wan PT, Roe SM, Barrett TE, Drew RE, Pearl LH. Crystal structure and induction mechanism of AmiC-AmiR: a ligand-regulated transcription antitermination complex. EMBO J 1999; 18:5175-86; PMID:10508151; http://dx.doi.org/10.1093/emboj/18.19.5175
- Ulijasz AT, Andes DR, Glasner JD, Weisblum B. Regulation of iron transport in Streptococcus pneumoniae by RitR, an orphan response regulator. J Bacteriol 2004; 186:8123-36; PMID:15547286; http://dx.doi.org/10.1128/JB.186.23.8123-8136.2004
- Koo IC, Walthers D, Hefty PS, Kenney LJ, Stephens RS. ChxR is a transcriptional activator in Chlamydia. Proc Natl Acad Sci U S A 2006; 103:750-5; PMID:16407127; http://dx.doi.org/10.1073/pnas.0509690103
- Schar J, Sickmann A, Beier D. Phosphorylation-independent activity of atypical response regulators of Helicobacter pylori. J Bacteriol 2005; 187:3100-9; PMID:15838037; http://dx.doi.org/10.1128/JB.187.9.3100-3109.2005
- Grangeasse C, Nessler S, Mijakovic I. Bacterial tyrosine kinases: evolution, biological function and structural insights. Philos Trans R Soc Lond B Biol Sci 2012; 367:2640-55; PMID:22889913; http://dx.doi.org/10.1098/rstb.2011.0424
- Elsholz AK, Turgay K, Michalik S, Hessling B, Gronau K, Oertel D, Mader U, Bernhardt J, Becher D, Hecker M, et al. Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis. Proc Natl Acad Sci U S A 2012; 109:7451-6; PMID:22517742; http://dx.doi.org/10.1073/pnas.1117483109
- Giefing C, Meinke AL, Hanner M, Henics T, Bui MD, Gelbmann D, Lundberg U, Senn BM, Schunn M, Habel A, et al. Discovery of a novel class of highly conserved vaccine antigens using genomic scale antigenic fingerprinting of pneumococcus with human antibodies. J Exp Med 2008; 205:117-31; PMID:18166586; http://dx.doi.org/10.1084/jem.20071168
- Danilenko VN, Osolodkin DI, Lakatosh SA, Preobrazhenskaya MN, Shtil AA. Bacterial eukaryotic type serine-threonine protein kinases: from structural biology to targeted anti-infective drug design. Curr Top Med Chem 2011; 11:1352-69; PMID:21513496; http://dx.doi.org/10.2174/156802611795589566
- Kurosu M, Begari E. Bacterial protein kinase inhibitors. Drug Development Research 2010; 71:168-87
- Pensinger DA, Aliota MT, Schaenzer AJ, Bolden KM, Ansari IU, Vincent WJ, Knight B, Reniere ML, Striker R, Sauer JD. Selective pharmacologic inhibition of a PASTA kinase increases listeria monocytogenes susceptibility to beta-lactam antibiotics. Antimicrob Agents Chemother 2014; 58:4486-94; PMID:24867981; http://dx.doi.org/10.1128/AAC.02396-14
- Lowy FD. Staphylococcus aureus infections. N Engl J Med 1998; 339:520-32; PMID:9709046; http://dx.doi.org/10.1056/NEJM199808203390806
- Tamber S, Schwartzman J, Cheung AL. Role of PknB kinase in antibiotic resistance and virulence in community-acquired methicillin-resistant Staphylococcus aureus strain USA300. Infect Immun 2010; 78:3637-46; PMID:20547748; http://dx.doi.org/10.1128/IAI.00296-10
- Cheung AL, Nishina KA, Trotonda MP, Tamber S. The SarA protein family of Staphylococcus aureus. Int J Biochem Cell Biol 2008; 40:355-61; PMID:18083623; http://dx.doi.org/10.1016/j.biocel.2007.10.032
- Truong-Bolduc QC, Ding Y, Hooper DC. Posttranslational modification influences the effects of MgrA on norA expression in Staphylococcus aureus. J Bacteriol 2008; 190:7375-81; PMID:18805983; http://dx.doi.org/10.1128/JB.01068-08
- Truong-Bolduc QC, Hooper DC. Phosphorylation of MgrA and its effect on expression of the NorA and NorB efflux pumps of Staphylococcus aureus. J Bacteriol 2010; 192:2525-34; PMID:20233929; http://dx.doi.org/10.1128/JB.00018-10
- Chien Y, Manna AC, Projan SJ, Cheung AL. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J Biol Chem 1999; 274:37169-76; PMID:10601279; http://dx.doi.org/10.1074/jbc.274.52.37169
- Didier JP, Cozzone AJ, Duclos B. Phosphorylation of the virulence regulator SarA modulates its ability to bind DNA in Staphylococcus aureus. FEMS Microbiol Lett 2010; 306:30-6; PMID:20337713; http://dx.doi.org/10.1111/j.1574-6968.2010.01930.x
- Valle J, Toledo-Arana A, Berasain C, Ghigo JM, Amorena B, Penades JR, Lasa I. SarA and not sigmaB is essential for biofilm development by Staphylococcus aureus. Mol Microbiol 2003; 48:1075-87; PMID:12753197; http://dx.doi.org/10.1046/j.1365-2958.2003.03493.x
- Cheung AL, Bayer AS, Zhang G, Gresham H, Xiong YQ. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol Med Microbiol 2004; 40:1-9; PMID:14734180; http://dx.doi.org/10.1016/S0928-8244(03)00309-2
- Chen PR, Nishida S, Poor CB, Cheng A, Bae T, Kuechenmeister L, Dunman PM, Missiakas D, He C. A new oxidative sensing and regulation pathway mediated by the MgrA homologue SarZ in Staphylococcus aureus. Mol Microbiol 2009; 71:198-211; PMID:19007410; http://dx.doi.org/10.1111/j.1365-2958.2008.06518.x
- Sun F, Ding Y, Ji Q, Liang Z, Deng X, Wong CC, Yi C, Zhang L, Xie S, Alvarez S, et al. Protein cysteine phosphorylation of SarA/MgrA family transcriptional regulators mediates bacterial virulence and antibiotic resistance. Proc Natl Acad Sci U S A 2012; 109:15461-6; PMID:22927394; http://dx.doi.org/10.1073/pnas.1205952109
- Ji Q, Zhang L, Sun F, Deng X, Liang H, Bae T, He C. Staphylococcus aureus CymR is a new thiol-based oxidation-sensing regulator of stress resistance and oxidative response. J Biol Chem 2012; 287:21102-9; PMID:22553203; http://dx.doi.org/10.1074/jbc.M112.359737
- Mijakovic I, Poncet S, Galinier A, Monedero V, Fieulaine S, Janin J, Nessler S, Marquez JA, Scheffzek K, Hasenbein S, et al. Pyrophosphate-producing protein dephosphorylation by HPr kinase/phosphorylase: a relic of early life? Proc Natl Acad Sci U S A 2002; 99:13442-7; PMID:12359880; http://dx.doi.org/10.1073/pnas.212410399
- Deutscher J, Saier MH, Jr. ATP-dependent protein kinase-catalyzed phosphorylation of a seryl residue in HPr, a phosphate carrier protein of the phosphotransferase system in Streptococcus pyogenes. Proc Natl Acad Sci U S A 1983; 80:6790-4; PMID:6359157; http://dx.doi.org/10.1073/pnas.80.22.6790
- Henkin TM. The role of CcpA transcriptional regulator in carbon metabolism in Bacillus subtilis. FEMS Microbiol Lett 1996; 135:9-15; PMID:8598282; http://dx.doi.org/10.1111/j.1574-6968.1996.tb07959.x
- Seidl K, Stucki M, Ruegg M, Goerke C, Wolz C, Harris L, Berger-Bachi B, Bischoff M. Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob Agents Chemother 2006; 50:1183-94; PMID:16569828; http://dx.doi.org/10.1128/AAC.50.4.1183-1194.2006
- Antunes A, Martin-Verstraete I, Dupuy B. CcpA-mediated repression of Clostridium difficile toxin gene expression. Mol Microbiol 2011; 79:882-99; PMID:21299645; http://dx.doi.org/10.1111/j.1365-2958.2010.07495.x
- Chiang C, Bongiorni C, Perego M. Glucose-dependent activation of Bacillus anthracis toxin gene expression and virulence requires the carbon catabolite protein CcpA. J Bacteriol 2011; 193:52-62; PMID:20971911; http://dx.doi.org/10.1128/JB.01656-09
- Iyer R, Baliga NS, Camilli A. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J Bacteriol 2005; 187:8340-9; PMID:16321938; http://dx.doi.org/10.1128/JB.187.24.8340-8349.2005
- Leiba J, Hartmann T, Cluzel ME, Cohen-Gonsaud M, Delolme F, Bischoff M, Molle V. A novel mode of regulation of the Staphylococcus aureus catabolite control protein A (CcpA) mediated by Stk1 protein phosphorylation. J Biol Chem 2012; 287:43607-19; PMID:23132867; http://dx.doi.org/10.1074/jbc.M112.418913
- Falord M, Mader U, Hiron A, Debarbouille M, Msadek T. Investigation of the Staphylococcus aureus GraSR regulon reveals novel links to virulence, stress response and cell wall signal transduction pathways. PLoS One 2011; 6:e21323; PMID:21765893; http://dx.doi.org/10.1371/journal.pone.0021323
- Meehl M, Herbert S, Gotz F, Cheung A. Interaction of the GraRS two-component system with the VraFG ABC transporter to support vancomycin-intermediate resistance in Staphylococcus aureus. Antimicrob Agents Chemother 2007; 51:2679-89; PMID:17502406; http://dx.doi.org/10.1128/AAC.00209-07
- Falord M, Karimova G, Hiron A, Msadek T. GraXSR proteins interact with the VraFG ABC transporter to form a five-component system required for cationic antimicrobial peptide sensing and resistance in Staphylococcus aureus. Antimicrob Agents Chemother 2012; 56:1047-58; PMID:22123691; http://dx.doi.org/10.1128/AAC.05054-11
- Cui L, Neoh HM, Shoji M, Hiramatsu K. Contribution of vraSR and graSR point mutations to vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother 2009; 53:1231-4; PMID:19124662; http://dx.doi.org/10.1128/AAC.01173-08
- Fridman M, Williams GD, Muzamal U, Hunter H, Siu KW, Golemi-Kotra D. Two unique phosphorylation-driven signaling pathways crosstalk in Staphylococcus aureus to modulate the cell-wall charge: Stk1/Stp1 meets GraSR. Biochemistry 2013; 52:7975-86; PMID:24102310; http://dx.doi.org/10.1021/bi401177n
- Canova MJ, Baronian G, Brelle S, Cohen-Gonsaud M, Bischoff M, Molle V. A novel mode of regulation of the Staphylococcus aureus Vancomycin-resistance-associated response regulator VraR mediated by Stk1 protein phosphorylation. Biochem Biophys Res Commun 2014; 447:165-71; PMID:24704444; http://dx.doi.org/10.1016/j.bbrc.2014.03.128
- Gomez JE, McKinney JD. M. tuberculosis persistence, latency, and drug tolerance. Tuberculosis (Edinb) 2004; 84:29-44; PMID:14670344; http://dx.doi.org/10.1016/j.tube.2003.08.003
- Park HD, Guinn KM, Harrell MI, Liao R, Voskuil MI, Tompa M, Schoolnik GK, Sherman DR. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol Microbiol 2003; 48:833-43; PMID:12694625; http://dx.doi.org/10.1046/j.1365-2958.2003.03474.x
- Roberts DM, Liao RP, Wisedchaisri G, Hol WG, Sherman DR. Two sensor kinases contribute to the hypoxic response of Mycobacterium tuberculosis. J Biol Chem 2004; 279:23082-7; PMID:15033981; http://dx.doi.org/10.1074/jbc.M401230200
- Kumar A, Toledo JC, Patel RP, Lancaster JR, Jr., Steyn AJ. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc Natl Acad Sci U S A 2007; 104:11568-73; PMID:17609369; http://dx.doi.org/10.1073/pnas.0705054104
- Chao JD, Papavinasasundaram KG, Zheng X, Chavez-Steenbock A, Wang X, Lee GQ, Av-Gay Y. Convergence of Ser/Thr and two-component signaling to coordinate expression of the dormancy regulon in Mycobacterium tuberculosis. J Biol Chem 2010; 285:29239-46; PMID:20630871; http://dx.doi.org/10.1074/jbc.M110.132894
- Gomez-Velasco A, Bach H, Rana AK, Cox LR, Bhatt A, Besra GS, Av-Gay Y. Disruption of the serine/threonine protein kinase H affects phthiocerol dimycocerosates synthesis in Mycobacterium tuberculosis. Microbiology 2013; 159:726-36; PMID:23412844; http://dx.doi.org/10.1099/mic.0.062067-0
- Cavazos A, Prigozhin DM, Alber T. Structure of the sensor domain of Mycobacterium tuberculosis PknH receptor kinase reveals a conserved binding cleft. J Mol Biol 2012; 422:488-94; PMID:22727744; http://dx.doi.org/10.1016/j.jmb.2012.06.011
- Sharma K, Gupta M, Pathak M, Gupta N, Koul A, Sarangi S, Baweja R, Singh Y. Transcriptional control of the mycobacterial embCAB operon by PknH through a regulatory protein, EmbR, in vivo. J Bacteriol 2006; 188:2936-44; PMID:16585755; http://dx.doi.org/10.1128/JB.188.8.2936-2944.2006
- Molle V, Kremer L, Girard-Blanc C, Besra GS, Cozzone AJ, Prost JF. An FHA phosphoprotein recognition domain mediates protein EmbR phosphorylation by PknH, a Ser/Thr protein kinase from Mycobacterium tuberculosis. Biochemistry 2003; 42:15300-9; PMID:14690440; http://dx.doi.org/10.1021/bi035150b
- Wietzorrek A, Bibb M. A novel family of proteins that regulates antibiotic production in streptomycetes appears to contain an OmpR-like DNA-binding fold. Mol Microbiol 1997; 25:1181-4; PMID:9350875; http://dx.doi.org/10.1046/j.1365-2958.1997.5421903.x
- Alderwick LJ, Molle V, Kremer L, Cozzone AJ, Dafforn TR, Besra GS, Futterer K. Molecular structure of EmbR, a response element of Ser/Thr kinase signaling in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 2006; 103:2558-63; PMID:16477027; http://dx.doi.org/10.1073/pnas.0507766103
- Barthe P, Roumestand C, Canova MJ, Kremer L, Hurard C, Molle V, Cohen-Gonsaud M. Dynamic and structural characterization of a bacterial FHA protein reveals a new autoinhibition mechanism. Structure 2009; 17:568-78; PMID:19368890; http://dx.doi.org/10.1016/j.str.2009.02.012
- Nott TJ, Kelly G, Stach L, Li J, Westcott S, Patel D, Hunt DM, Howell S, Buxton RS, O'Hare HM, et al. An intramolecular switch regulates phosphoindependent FHA domain interactions in Mycobacterium tuberculosis. Sci Signal 2009; 2:ra12; PMID:19318624; http://dx.doi.org/10.1126/scisignal.2000212
- Molle V, Reynolds RC, Alderwick LJ, Besra GS, Cozzone AJ, Futterer K, Kremer L. EmbR2, a structural homologue of EmbR, inhibits the Mycobacterium tuberculosis kinase/substrate pair PknH/EmbR. Biochem J 2008; 410:309-17; PMID:17999640; http://dx.doi.org/10.1042/BJ20071384
- Sharma K, Gupta M, Krupa A, Srinivasan N, Singh Y. EmbR, a regulatory protein with ATPase activity, is a substrate of multiple serine/threonine kinases and phosphatase in Mycobacterium tuberculosis. FEBS J 2006; 273:2711-21; PMID:16817899; http://dx.doi.org/10.1111/j.1742-4658.2006.05289.x
- Zheng X, Papavinasasundaram KG, Av-Gay Y. Novel substrates of Mycobacterium tuberculosis PknH Ser/Thr kinase. Biochem Biophys Res Commun 2007; 355:162-8; PMID:17286964; http://dx.doi.org/10.1016/j.bbrc.2007.01.122
- DeBarber AE, Mdluli K, Bosman M, Bekker LG, Barry CE, 3rd. Ethionamide activation and sensitivity in multidrug-resistant Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 2000; 97:9677-82; PMID:10944230; http://dx.doi.org/10.1073/pnas.97.17.9677
- Baulard AR, Betts JC, Engohang-Ndong J, Quan S, McAdam RA, Brennan PJ, Locht C, Besra GS. Activation of the pro-drug ethionamide is regulated in mycobacteria. J Biol Chem 2000; 275:28326-31; PMID:10869356
- Leiba J, Carrere-Kremer S, Blondiaux N, Dimala MM, Wohlkonig A, Baulard A, Kremer L, Molle V. The Mycobacterium tuberculosis transcriptional repressor EthR is negatively regulated by Serine/Threonine phosphorylation. Biochem Biophys Res Commun 2014; 446:1132-8; PMID:24667600; http://dx.doi.org/10.1016/j.bbrc.2014.03.074
- Canova MJ, Veyron-Churlet R, Zanella-Cleon I, Cohen-Gonsaud M, Cozzone AJ, Becchi M, Kremer L, Molle V. The Mycobacterium tuberculosis serine/threonine kinase PknL phosphorylates Rv2175c: mass spectrometric profiling of the activation loop phosphory-lation sites and their role in the recruitment of Rv2175c. Proteomics 2008; 8:521-33; PMID:18175374; http://dx.doi.org/10.1002/pmic.200700442
- Lakshminarayan H, Rajaram A, Narayanan S. Involvement of serine threonine protein kinase, PknL, from Mycobacterium tuberculosis H37Rv in starvation response of mycobacteria. J Microbial Biochem Technol 2009; 1:30-6; http://dx.doi.org/10.4172/1948-5948.1000006
- Cohen-Gonsaud M, Barthe P, Canova MJ, Stagier-Simon C, Kremer L, Roumestand C, Molle V. The Mycobacterium tuberculosis Ser/Thr kinase substrate Rv2175c is a DNA-binding protein regulated by phosphorylation. J Biol Chem 2009; 284:19290-300; PMID:19457863; http://dx.doi.org/10.1074/jbc.M109.019653
- Singh A, Jain S, Gupta S, Das T, Tyagi AK. mymA operon of Mycobacterium tuberculosis: its regulation and importance in the cell envelope. FEMS Microbiol Lett 2003; 227:53-63; PMID:14568148; http://dx.doi.org/10.1016/S0378-1097(03)00648-7
- Malhotra V, Arteaga-Cortes LT, Clay G, Clark-Curtiss JE. Mycobacterium tuberculosis protein kinase K confers survival advantage during early infection in mice and regulates growth in culture and during persistent infection: implications for immune modulation. Microbiology 2010; 156:2829-41; PMID:20522497; http://dx.doi.org/10.1099/mic.0.040675-0
- Canova MJ, Kremer L, Molle V. pETPhos: a customized expression vector designed for further characterization of Ser/Thr/Tyr protein kinases and their substrates. Plasmid 2008; 60:149-53; PMID:18597845; http://dx.doi.org/10.1016/j.plasmid.2008.05.002
- Kumar P, Kumar D, Parikh A, Rananaware D, Gupta M, Singh Y, Nandicoori VK. The Mycobacterium tuberculosis protein kinase K modulates activation of transcription from the promoter of mycobacterial monooxygenase operon through phosphorylation of the transcriptional regulator VirS. J Biol Chem 2009; 284:11090-9; PMID:19251699; http://dx.doi.org/10.1074/jbc.M808705200
- Song T, Dove SL, Lee KH, Husson RN. RshA, an anti-sigma factor that regulates the activity of the mycobacterial stress response sigma factor SigH. Mol Microbiol 2003; 50:949-59; PMID:14617153; http://dx.doi.org/10.1046/j.1365-2958.2003.03739.x
- Park ST, Kang CM, Husson RN. Regulation of the SigH stress response regulon by an essential protein kinase in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 2008; 105:13105-10; PMID:18728196; http://dx.doi.org/10.1073/pnas.0801143105
- Greenstein AE, MacGurn JA, Baer CE, Falick AM, Cox JS, Alber T. M. tuberculosis Ser/Thr protein kinase D phosphorylates an anti-anti-sigma factor homolog. PLoS Pathog 2007; 3:e49; PMID:17411339; http://dx.doi.org/10.1371/journal.ppat.0030049
- Burnside K, Rajagopal L. Aspects of eukaryotic-like signaling in Gram-positive cocci: a focus on virulence. Future Microbiol 2011; 6:747-61; PMID:21797690; http://dx.doi.org/10.2217/fmb.11.62
- Whidbey C, Harrell MI, Burnside K, Ngo L, Becraft AK, Iyer LM, Aravind L, Hitti J, Waldorf KM, Rajagopal L. A hemolytic pigment of Group B Streptococcus allows bacterial penetration of human placenta. J Exp Med 2013; 210:1265-81; PMID:23712433; http://dx.doi.org/10.1084/jem.20122753
- Jiang SM, Cieslewicz MJ, Kasper DL, Wessels MR. Regulation of virulence by a two-component system in group B streptococcus. J Bacteriol 2005; 187:1105-13; PMID:15659687; http://dx.doi.org/10.1128/JB.187.3.1105-1113.2005
- Federle MJ, McIver KS, Scott JR. A response regulator that represses transcription of several virulence operons in the group A streptococcus. J Bacteriol 1999; 181:3649-57; PMID:10368137
- Rajagopal L, Vo A, Silvestroni A, Rubens CE. Regulation of cytotoxin expression by converging eukaryotic-type and two-component signalling mechanisms in Streptococcus agalactiae. Mol Microbiol 2006; 62:941-57; PMID:17005013; http://dx.doi.org/10.1111/j.1365-2958.2006.05431.x
- Lin WJ, Walthers D, Connelly JE, Burnside K, Jewell KA, Kenney LJ, Rajagopal L. Threonine phosphorylation prevents promoter DNA binding of the Group B Streptococcus response regulator CovR. Mol Microbiol 2009; 71:1477-95; PMID:19170889; http://dx.doi.org/10.1111/j.1365-2958.2009.06616.x
- Horstmann N, Saldana M, Sahasrabhojane P, Yao H, Su X, Thompson E, Koller A, Shelburne SA, 3rd. Dual-site phosphorylation of the control of virulence regulator impacts group a streptococcal global gene expression and pathogenesis. PLoS Pathog 2014; 10:e1004088; PMID:24788524; http://dx.doi.org/10.1371/journal.ppat.1004088
- Volkman BF, Lipson D, Wemmer DE, Kern D. Two-state allosteric behavior in a single-domain signaling protein. Science 2001; 291:2429-33; PMID:11264542; http://dx.doi.org/10.1126/science.291.5512.2429
- Howden BP, McEvoy CR, Allen DL, Chua K, Gao W, Harrison PF, Bell J, Coombs G, Bennett-Wood V, Porter JL, et al. Evolution of multidrug resistance during Staphylococcus aureus infection involves mutation of the essential two component regulator WalKR. PLoS Pathog 2011; 7:e1002359; PMID:22102812; http://dx.doi.org/10.1371/journal.ppat.1002359
- Dubrac S, Bisicchia P, Devine KM, Msadek T. A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol Microbiol 2008; 70:1307-22; PMID:19019149; http://dx.doi.org/10.1111/j.1365-2958.2008.06483.x
- Miller M, Donat S, Rakette S, Stehle T, Kouwen TR, Diks SH, Dreisbach A, Reilman E, Gronau K, Becher D, et al. Staphylococcal PknB as the first prokaryotic representative of the proline-directed kinases. PLoS One 2010; 5:e9057; PMID:20140229; http://dx.doi.org/10.1371/journal.pone.0009057
- Odendall C, Rolhion N, Forster A, Poh J, Lamont DJ, Liu M, Freemont PS, Catling AD, Holden DW. The Salmonella kinase SteC targets the MAP kinase MEK to regulate the host actin cytoskeleton. Cell Host Microbe 2012; 12:657-68; PMID:23159055; http://dx.doi.org/10.1016/j.chom.2012.09.011
- Canova MJ, Molle V. Bacterial serine/threonine protein kinases in host-pathogen interactions. J Biol Chem 2014; 289:9473-9; PMID:24554701; http://dx.doi.org/10.1074/jbc.R113.529917
- Kadioglu A, Weiser JN, Paton JC, Andrew PW. The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat Rev Microbiol 2008; 6:288-301; PMID:18340341; http://dx.doi.org/10.1038/nrmicro1871
- Moffitt KL, Gierahn TM, Lu YJ, Gouveia P, Alderson M, Flechtner JB, Higgins DE, Malley R. T(H)17-based vaccine design for prevention of Streptococcus pneumoniae colonization. Cell Host Microbe 2011; 9:158-65; PMID:21320698; http://dx.doi.org/10.1016/j.chom.2011.01.007
- Standish AJ, Stroeher UH, Paton JC. The two-component signal transduction system RR06/HK06 regulates expression of cbpA in Streptococcus pneumoniae. Proc Natl Acad Sci U S A 2005; 102:7701-6; PMID:15897461; http://dx.doi.org/10.1073/pnas.0409377102
- Agarwal S, Pancholi P, Pancholi V. Strain-specific regulatory role of eukaryote-like serine/threonine phosphatase in pneumococcal adherence. Infect Immun 2012; 80:1361-72; PMID:22311926; http://dx.doi.org/10.1128/IAI.06311-11
- Horstmann N, Sahasrabhojane P, Suber B, Kumaraswami M, Olsen RJ, Flores A, Musser JM, Brennan RG, Shelburne SA, 3rd. Distinct single amino acid replacements in the control of virulence regulator protein differentially impact streptococcal pathogenesis. PLoS Pathog 2011; 7:e1002311; PMID:22028655; http://dx.doi.org/10.1371/journal.ppat.1002311
- Pishchany G, Skaar EP. Taste for blood: hemoglobin as a nutrient source for pathogens. PLoS Pathog 2012; 8:e1002535; PMID:22412370; http://dx.doi.org/10.1371/journal.ppat.1002535
- Yesilkaya H, Andisi VF, Andrew PW, Bijlsma JJ. Streptococcus pneumoniae and reactive oxygen species: an unusual approach to living with radicals. Trends Microbiol 2013; 21:187-95; PMID:23415028; http://dx.doi.org/10.1016/j.tim.2013.01.004
- Pericone CD, Park S, Imlay JA, Weiser JN. Factors contributing to hydrogen peroxide resistance in Streptococcus pneumoniae include pyruvate oxidase (SpxB) and avoidance of the toxic effects of the fenton reaction. J Bacteriol 2003; 185:6815-25; PMID:14617646; http://dx.doi.org/10.1128/JB.185.23.6815-6825.2003
- Ong CL, Potter AJ, Trappetti C, Walker MJ, Jennings MP, Paton JC, McEwan AG. Interplay between manganese and iron in pneumococcal pathogenesis: role of the orphan response regulator RitR. Infect Immun 2013; 81:421-9; PMID:23184523; http://dx.doi.org/10.1128/IAI.00805-12
- Throup JP, Koretke KK, Bryant AP, Ingraham KA, Chalker AF, Ge Y, Marra A, Wallis NG, Brown JR, Holmes DJ, et al. A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol Microbiol 2000; 35:566-76; PMID:10672179; http://dx.doi.org/10.1046/j.1365-2958.2000.01725.x
- Shakir SM, Bryant KM, Larabee JL, Hamm EE, Lovchik J, Lyons CR, Ballard JD. Regulatory interactions of a virulence-associated serine/threonine phosphatase-kinase pair in Bacillus anthracis. J Bacteriol 2010; 192:400-9; PMID:19915022; http://dx.doi.org/10.1128/JB.01221-09
- Nguyen L, Pieters J. The Trojan horse: survival tactics of pathogenic mycobacteria in macrophages. Trends Cell Biol 2005; 15:269-76; PMID:15866031; http://dx.doi.org/10.1016/j.tcb.2005.03.009
- Sola M, Gomis-Ruth FX, Serrano L, Gonzalez A, Coll M. Three-dimensional crystal structure of the transcription factor PhoB receiver domain. J Mol Biol 1999; 285:675-87; PMID:9878437; http://dx.doi.org/10.1006/jmbi.1998.2326
- Poor CB, Chen PR, Duguid E, Rice PA, He C. Crystal structures of the reduced, sulfenic acid, and mixed disulfide forms of SarZ, a redox active global regulator in Staphylococcus aureus. J Biol Chem 2009; 284:23517-24; PMID:19586910; http://dx.doi.org/10.1074/jbc.M109.015826
- Blanco AG, Sola M, Gomis-Ruth FX, Coll M. Tandem DNA recognition by PhoB, a two-component signal transduction transcriptional activator. Structure 2002; 10:701-13; PMID:12015152; http://dx.doi.org/10.1016/S0969-2126(02)00761-X
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 2003; 31:3381-5; PMID:12824332; http://dx.doi.org/10.1093/nar/gkg520
- DeLano WL. The PyMOL User's Manual. San Carlos, CA: DeLano Scientific, 2002
- Wisedchaisri G, Wu M, Sherman DR, Hol WG. Crystal structures of the response regulator DosR from Mycobacterium tuberculosis suggest a helix rearrangement mechanism for phosphorylation activation. J Mol Biol 2008; 378:227-42; PMID:18353359; http://dx.doi.org/10.1016/j.jmb.2008.02.029
- Campbell EA, Greenwell R, Anthony JR, Wang S, Lim L, Das K, Sofia HJ, Donohue TJ, Darst SA. A conserved structural module regulates transcriptional responses to diverse stress signals in bacteria. Mol Cell 2007; 27:793-805; PMID:17803943; http://dx.doi.org/10.1016/j.molcel.2007.07.009
- Wang S, Engohang-Ndong J, Smith I. Structure of the DNA-binding domain of the response regulator PhoP from Mycobacterium tuberculosis. Biochemistry 2007; 46:14751-61; PMID:18052041; http://dx.doi.org/10.1021/bi700970a
- Chung A, Duclos B. Stp1 and Stk1: the Yin and Yang of vancomycin sensitivity and virulence in vancomycin-intermediate Staphylococcus aureus strains. J Infect Dis 2012; 205:1625-7; PMID:22492848; http://dx.doi.org/10.1093/infdis/jis255
- Gil M, Grana M, Schopfer FJ, Wagner T, Denicola A, Freeman BA, Alzari PM, Batthyany C, Duran R. Inhibition of Mycobacterium tuberculosis PknG by non-catalytic rubredoxin domain specific modification: reaction of an electrophilic nitro-fatty acid with the Fe-S center. Free Radic Biol Med 2013; 65:150-61.
- Fleurie A, Manuse S, Zhao C, Campo N, Cluzel C, Lavergne JP, Freton C, Combet C, Guiral S, Soufi B, et al. Interplay of the serine/threonine-kinase StkP and the paralogs DivIVA and GpsB in pneumococcal cell elongation and division. PLoS Genet 2014; 10:e1004275.
- Trajtenberg F, Albanesi D, Ruetalo N, Botti H, Mechaly AE, Nieves M, Aguilar PS, Cybulski L, Larrieux N, de Mendoza D, et al. Allosteric activation of bacterial response regulators: the role of the cognate histidine kinase beyond phosphorylation. MBio 2014; 5.
