Abstract
The staphylococcal YeaZ is highly conserved in prokaryotic cells and critical for growth of many bacterial pathogens. However, the essentiality for Staphylococcus aureus growth and the biological function of YeaZ behind its essentiality remain undefined. In this study, we created and characterized a defined Pspac-regulated yeaZ expression mutant in S. aureus and demonstrated the indispensability of YeaZ for S. aureus growth. Moreover, we conducted complementation studies, not only confirmed the requirement of YeaZ for S. aureus growth, but also revealed a similarity of essential function between staphylococcal YeaZ and its E. coli homolog. On the other hand, we explored the biological functions of YeaZ and found that YeaZ is involved in the regulation of the transcription of ilv-leu operon that encodes key enzymes responsible for the biosynthesis of the branched-chain amino acids, including isoleucine, leucine, and valine (ILV). qPCR analysis showed that the 6-fold downregulation of YeaZ dramatically elevated approximately 17- to 289-fold RNA levels of ilvD, leuA and ilvA. We further confirmed the transcriptional regulation of the ilv-leu operon by YeaZ using an ilv-promoter-lux reporter system and gel-shift assays and revealed that YeaZ is able to bind the promoter region of ilv. Furthermore, we established that the regulation of ILV biosynthesis isn't associated with YeaZ's essentiality, as the deletion of the ilv-leu operon did not affect the requirement of YeaZ for growth in culture. Our results demonstrate the essentiality of YeaZ for S. aureus growth and suggest that the staphylococcal YeaZ possesses regulatory function.
Background
Staphylococcus aureus is an important pathogen that can cause severe human and animal infections. The prevalence of multi-drug resistant S. aureus, especially methicillin- and vancomycin-resistant S. aureus has caused serious public health concerns.Citation1,2 It is necessary to identify and develop alternative therapeutic strategies to combat S. aureus.
YeaZ is highly conserved within S. aureus, maintaining a 98% to 100% identity at the amino acid sequence level. Furthermore, YeaZ orthologs exist in a variety of pathogens ranging from Escherichia coli to Bacillus anthrax and all homologs possess at least 29% amino acid identity with YeaZ of S. aureus. YeaZ is essential for growth in many bacteria, including E. coli,Citation3 Pseudomonas aeruginosa,Citation4 and Streptococcus pneumoniae.Citation5 In addition, YeaZ plays an important role for bacterial cells to survive in, and exist from, a viable but non-culturable state.Citation6 For example, Salmonella typhimurium YeaZ is a resuscitation-promoting factor,Citation6 which enables bacteria to go from being viable but non-culturable to being culturable.Citation7,8 Moreover, many microbial pathogens, in order to adapt to changing environments, respond to stress by entering a viable but non-culturable stateCitation9 that enables the bacterial cells to resist antibiotics and persist in their environments.Citation10 However, it was unclear whether YeaZ is necessary for S. aureus growth and survival, which is critical to validate whether YeaZ is a potential target for the development of novel broad-spectrum antibacterial agents.
In S. aureus, the yeaZ gene is located on an essential gcp operon that is composed of sa1857, sa1856 (yeaZ), sa1855, and sa1854 (gcp) and are co-transcribed.Citation11 The staphylococcal YeaZ interacts with Gcp, and the interaction between YeaZ and Gcp is likely associated with the essentiality of Gcp.Citation12 Both E. coli and S. typhimurium YeaZ binds to 2 additional essential proteins, YjeE and YgiD,Citation13,14 and the E. coli YeaZ specifically cleaves YgjD (the Gcp homolog).Citation15 However, the staphylococcal YeaZ lacks the ability to cleave Gcp,Citation12 as does S. typhimurium YeaZ.Citation16 These discrepancies suggest that the protease activity of E. coli YeaZ is possibly not involved in its essential mechanism. Structural analysis of YeaZ homologues from E. coli,Citation14 S. typhimurium,Citation15 Thermotoga maritima,Citation17 and Vibrio parahaemolyticusCitation10 indicates that YeaZ belongs to the superfamily of ASKHA (acetate and sugar kinase/Hsc70/actin) Citation18 or HALF (Hsp70/actin-like fold)Citation19 and possesses a classic actin-like nucleotide-binding fold. YeaZ may require interactions with a partner protein and/or cofactors, as S. typhimurium YeaZ has an unusual orientation of the A and B lobes.Citation16 In addition, analysis of crystal packing and protein-protein interfaces suggests YeaZ crystal structure may form two distinct dimers based upon the presence of nucleotides.Citation10 Taken together, the above data indicate that different YeaZ homologs may possess different biological functions.
Recently, we revealed that YeaZ's partner protein Gcp controls the transcription of ilv-leu operon responsible for biosynthesis of the branched-chain amino acids (BCAAs) in S. aureus.Citation11 This led us to hypothesize that YeaZ may also play a role in mediating the transcription of the ilv-leu operon. In S. aureus, the ilv-leu operon consists of ilvDBHC-leuABCD-ilvA 9 genes, which is similar with that of B. subtilis.Citation20-22 Although S. aureus possesses all genes necessary for biosynthesis of BCAAs, the bacterium exhibits an auxotrophic phenotype for BCAAs through an unknown mechanism.Citation23,24 Several mechanisms of regulating the biosynthesis of BCAAs have been revealed and characterized in different organisms.Citation25-27 A global regulator CodY directly regulates the ilv-leu operon in S. aureus.Citation25
In this study, we determined the essentiality of YeaZ for growth in S. aureus and revealed that YeaZ controls the BCAA biosynthesis pathway by transcriptional regulation of the ilv-leu operon. Moreover, we demonstrated that the essentiality of YeaZ is not attributable to its modulation of BCAA biosynthesis. These new findings provide new insights into the biological functions of YeaZ and the regulatory mechanisms of BCAA biosynthesis pathway.
Results
The conserved protein YeaZ is required for S. aureus growth in vitro; and its essential mechanism is similar to that of E. coli YeaZ
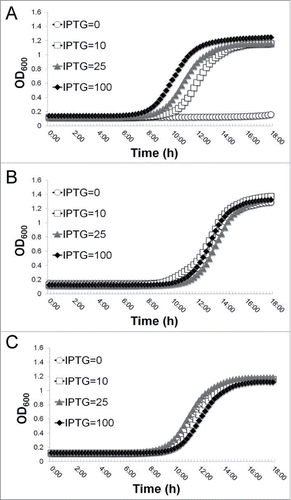
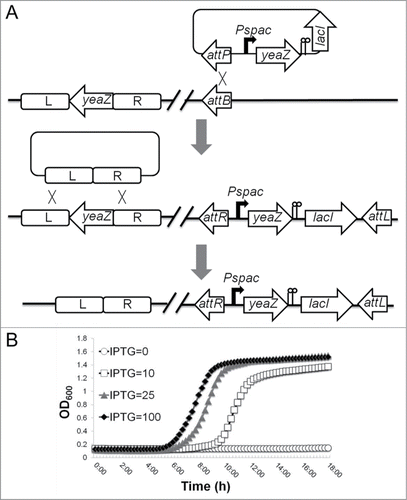
We have revealed that the yeaZ gene is located upstream of the gcp gene in the essential gcp operon.Citation12 To rule out the possible polar effect of yeaZ antisense RNA and further determine the essentiality of YeaZ, we created the defined Pspac-regulated yeaZ expression strains using 2 clinical S. aureus isolates, a hospital-associated MRSA strain, WCUH29, and a community-acquired MRSA strain, 923. The constructs were made by integrating an IPTG-inducible Pspac-driven yeaZ expression cassette into the ϕ11 attB site of chromosomal DNA and then deleting the endogenous yeaZ gene by an allelic gene replacement approach as describedCitation12,28 (). The defined Pspac-regulated yeaZ expression strains showed an IPTG-dependent growth phenotype (). Moreover, we found that without yeaZ expression in trans, the yeaZ deletion mutant was not viable, supporting the indispensability of YeaZ for growth in vitro.
Figure 1. Construction of a defined Pspac-regulated yeaZ expression mutant in S. aureus (A) and the effect of the depletion of YeaZ on growth of S. aureus (B) in tryptic soy broth (TSB) with Tc (5 μg/ml) and different concentrations of IPTG (μM) at 37°C.
To determine the suitability of YeaZ as a potential target for broad-spectrum antibiotics, we investigated whether E. coli YeaZ was able to complement the essentiality of staphylococcal YeaZ. The yeaZ genes were obtained by PCR from S. aureus and E. coli, cloned into a shuttle vector (pYH4-lacI) and reformed into the recombinant plasmids pTL1008 and pTL1108, respectively. The plasmids were electroporated into the defined S. aureus strain with Pspac-regulated yeaZ expression and confirmed using PCR and DNA sequencing. The empty plasmid pYH4-lacI was used as a negative control (). The expression of the staphylococcal YeaZ () in trans reversed the IPTG-dependent growth phenotype of the defined Pspac-regulated yeaZ expression mutant, which further confirmed the requirement of YeaZ for S. aureus growth. Interestingly, the expression of E. coli YeaZ (, EcyeaZ) in trans also complemented the growth defect caused by the depletion of endogenous staphylococcal YeaZ, indicating that both S. aureus and E. coli YeaZ homologs have a similar essential function for bacterial growth.
YeaZ is involved in the regulation of the branched-chain amino acids biosynthesis pathway in S. aureus
Our previous study demonstrated that the YeaZ partner protein, Gcp, affects the branched-chain amino acid biosynthesis pathway.Citation11 The down-regulation of Gcp dramatically elevated the expression of key enzymes that are encoded in the ilv-leu operon and are responsible for the biosynthesis of the branched-chain amino acids isoleucine, leucine, and valine (ILV).Citation11 Because we have demonstrated an interaction between Gcp and YeaZ,Citation12 we hypothesized that YeaZ may coordinate with Gcp to regulate the biosynthesis of branched-chain amino acids. To test this hypothesis, we used qPCR to examine the effect of YeaZ on the transcription of the ilv-leu operon and found that the 6-fold downregulation of YeaZ dramatically induced approximately 17- to 289-fold increases of the transcriptional levels of ilvD, leuA, and ilvA, which are located in the ilv-leu operon (), similar to our findings of Gcp. However, we found no remarkable impact on the transcription of codY and ccpA, which encode a negative (CodY) and a possible positive (CcpA) regulator of the ilv-leu operon, respectively.Citation25-27
Table 1. Gene transcription change due to the depletion of YeaZ
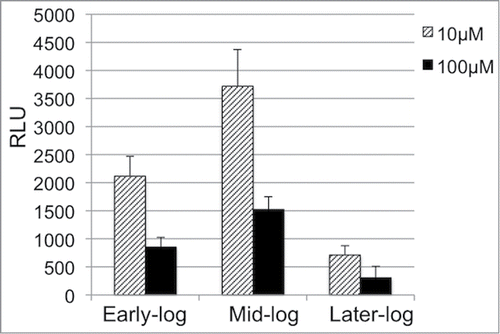
To further confirm the transcriptional regulation, we constructed an ilv promoter-lux reporter fusion in the Pspac-regulated yeaZ expression mutant. This system allows us to effectively down-regulate yeaZ expression and simultaneously to monitor the reporter gene expression by measuring the bioluminescence intensity.Citation29 The Pspac-regulated yeaZ expression mutant carrying a promoterless lux reporter was utilized as a control. The addition of IPTG had no influence on the lux expression in the control strain (data not shown). However, the downregulation of YeaZ from the early log to mid-log phases of growth resulted in a remarkable increase of the bioluminescence intensity (). Taken together, these data suggest that YeaZ transcriptionally represses the expression of the ilv-leu operon.
Figure 3. Effect of the downregulation of YeaZ on the ilv promoter transcription activity. Relative Bioluminescence intensity of J9230112 during the culture with different concentrations of IPTG (10 μM and 100 μM). The optical density at OD600 nm and bioluminescence intensity of cultures were simultaneously measured at different phases of growth (early-log phase: OD = 0.2; mid-log phase: OD = 0.4; later-log phase: OD = 0.6) at 37°C using a BioTek Synergy plate reader. The experiments were repeated at least 3 times. RLU, relative luminescence units.
YeaZ is capable of binding to the ilv promoter region
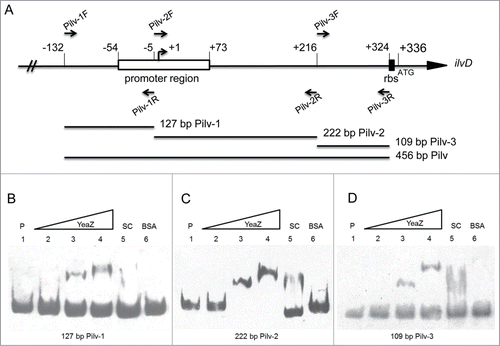
To further characterize the involvement of YeaZ in regulation of the ilv-leu operon, we cloned, expressed, and purified soluble, recombinant staphylococcal YeaZ protein from E. coli using a pGEX-4T-1 expression system, as describedCitation12 (). Then we performed electrophoretic mobility shift assays to determine whether YeaZ is able to bind a DNA fragment of the ilv promoter region. Negative controls included the labeled ilv promoter region without protein and the labeled ilv with BSA protein. The non-labeled ilv promoter region was used as a specific competitor, and an unlabeled unrelated DNA fragment was used as a nonspecific competitor. The gel-shift result showed that the addition of YeaZ to the reaction mixtures retarded the electrophoretic mobility of the Pilv probe (), whereas the addition of a nonspecific DNA competitor had no impact on YeaZ/Pilv complex formation (). The 456 bp Pilv probe covers the −132 bp to +324 bp upstream region of the ilvD (). To further define YeaZ binding region of the ilv promoter region, we conducted gel-shift assays and determined the DNA binding activity of YeaZ using different ilv promoter regions labeled with Biotin. YeaZ strongly bound to the −5 bp to +216 bp region of the ilv promoter (), but weakly bound to the −132 bp to −5 bp and the +212 bp to +324 bp regions (). Taken together, the above results indicate that the essential protein YeaZ has the capacity to bind the promoter region of the ilv-leu operon.
Figure 4. YeaZ binding to the upstream of ilv-leu operon. (A) SDS-PAGE analysis of recombinant staphylococcal YeaZ from E. coli. (B) Gel-shift assay with DIG labeled probe Pilv. P indicates DIG labeled probe without YeaZ; the amount of the labeled Pilv is 200 fmol. The amount of YeaZ from Lane 2 to 5 are 0.5, 1, 2, and 8 μM. SC indicates 100-fold molar excess of specific competitor. The amount of BSA is 15 μM. (C) Gel-shift assay with 100 molar excess competitors. NSC indicates 100-fold molar excess of nonspecific competitor. The amount of YeaZ is 8 μM.
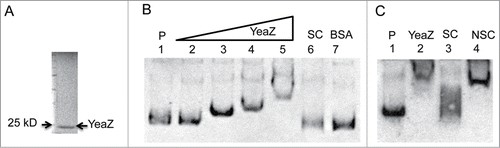
Figure 5. Localization of the YeaZ binding region. (A) Localization of Pilv probes’ primers. Forward primers are listed above the genetic map, and reverse primers are listed below the map. Transcription start site of ilv-leu operon is indicated by +1 and an arrow, and translation start site is indicated by the initiation codon. The ribosome binding site (rbs) and the promoter region of ilvCitation25 are indicated as black and white squares. All the marked positions are indicated as the relative distance to transcription start site. PCR DNA probes for use in gel-shift assay are shown in the lower portion of the figure. (B to D) Gel-shift assays, with each of the different biotinylated PCR probes indicated below the figure. P indicates biotinylated probe without YeaZ; the amount of the biotinylated probes used in each figure are 20 fmol; SC indicates specific competitors each at 300-fold molar excess. All assays were run on 6% acrylamide gels. The amounts of YeaZ in each assay are shown below. (B to D) Lanes 2 to 4 were 0.5, 2, and 8 μM. BSA is added in lane 6 at the amount of 15 μM.
The essentiality of YeaZ for growth is not attributable to its regulation of ilv-leu operon
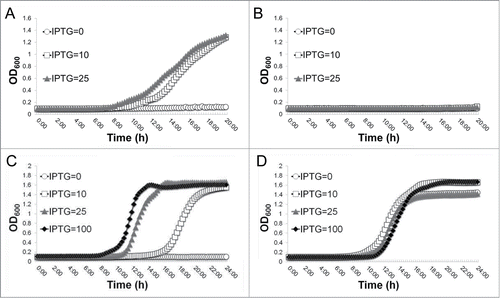
The above results suggested to us that the inhibition of growth when YeaZ is depleted possibly results from the accumulation of these branched-chain amino acids upon the over-expression of the ilv-leu operon. To test this possibility, we examined the impact of ILV on the requirement for YeaZ. Decreasing ILV from the standard concentration of 1.14 mM isoleucine and leucine and 1.28 mM valine to a minimal growth concentration (17.8 nM I and L and 20 nM V) in chemically defined-medium (CDM) had no effect on the requirement for YeaZ, and the growth of the Pspac-regulated yeaZ expression mutant remained IPTG-dependent (). Without limited 8.9 nM ILV, no bacterial growth was observed (). These suggest the accumulation of ILV is not involved in the essential mechanism of YeaZ. To confirm this, we generated knockout mutants of the entire ilv-leu operon in wild-type WCUH29 and in the defined Pspac-regulated yeaZ expression mutant. Then we determined the effect of ilv-leu operon on the importance of YeaZ for growth. The growth of the Pspac-regulated yeaZ expression mutant with the entire ilv-leu operon deleted was IPTG-dependent (), whereas the addition of IPTG had no influence on growth of the ilv-leu operon knockout mutant (). Thus, the direct regulation of the branched-chain amino acid biosynthesis pathway by YeaZ is not associated with the essential mechanism of YeaZ.
Figure 6. Effect of the deletion of the ilv-leu operon on the essentiality of YeaZ for growth. (A) IPTG-dependent growth of Pspac-regulated yeaZ expression mutant in CDM with minimal concentration of ILV (17.8 nM); (B) No bacterial growth in CDM with 8.9 nM ILV. (C) IPTG-dependent growth of the Pspac-regulated yeaZ expression and ilv-leu knockout mutant in CDM; (D) growth curves of the ilv-leu knockout mutant in CDM. Different concentrations of IPTG (μM) were added in the medium to induce the expression of YeaZ. The growth curves are monitored by kinetically measuring the optical density at OD600 nm in the corresponding culture medium in the presence of different concentrations of inducer, IPTG (μM), every 15 min at 37°C using a BioTek Synergy plate reader. These figures are one representative of 3 independent experiments.
Discussion
In this study, we demonstrated that YeaZ is necessary for S. aureus growth in culture and possesses a similar essential function with its E. coli YeaZ homolog. The YeaZ homologs are indispensible for many bacteria, including E. coli,Citation3 P. aeruginosa,Citation4 and S. pneumoniae.Citation5 These results suggest that YeaZ is a potential novel target for the development of a broad-spectrum antibacterial agent. Moreover, we revealed that YeaZ is involved in controlling the biosynthesis of branched-chain amino acids by transcriptional regulation of ilv-leu operon. Previously, we revealed that Gcp indirectly modulates the expression of the ilv-leu operon.Citation11 These data suggest that YeaZ and Gcp may coordinately mediate the ILV biosynthesis pathway.Citation11 Similar with Gcp,Citation11 the essentiality of YeaZ is independent of its repression of the ilv-leu operon, suggesting that up-regulation of ILV-biosynthesis is not a suitable strategy for development of antibacterial agents.
Our gel-shift results indicate that YeaZ exhibits different affinities binding to the different upstream regions of the ilvD gene, which is consistent with other regulators in different bacterial system.Citation30 Moreover, we revealed that the −5 bp to +216 bp upstream region of the ilvD gene is critical for YeaZ binding. These data suggest that YeaZ possesses a regulatory function in S. aureus. Our qPCR assays revealed that YeaZ has a similar effect on the transcriptional level of ilvD, leuA, and ilvA genes within the ilv-leu operon, but has no impact on ilvE, which lies outside of the ilv-leu operon in the S. aureus chromosome and encodes a branched-chain amino acid aminotransferase, a key enzyme responsible for the first step of ILV degradation. Consistent with qPCR analysis, the ilv promoter-lux reporter studies further confirmed the transcriptional regulation of the ilv-leu operon by YeaZ in S. aureus. Moreover, our qPCR showed that YeaZ had no influence on the transcription of codY gene that directly regulates the transcription of ilv-leu operon,Citation27 suggesting that YeaZ may function independently in S. aureus. The identification of YeaZ's repressory effect on the ilv-leu operon in S. aureus is consistent with previous report in E. coli.Citation31
It has been revealed that varied T-box regulation systems are involved in ILV biosynthesis in different bacterial systems.Citation32,33 In S. aureus, a Ile-T-box was found in the up-stream region of ileS, which encodes Ile-tRNA synthetaseCitation33,34; and a weak T-box motif located in the upstream region of ilvDBHC-leuABCD-ilvA operon.Citation35 Thus, we cannot exclude Ile-tRNA dependent regulation of ilv-leu operon in S. aureus.
In B. subtilis it has been revealed that posttranscriptional regulation of the ilv-leu operon by endoribonuclease and exoribonuclease proteins leads to 3 different mRNA transcript lengths of the ilv-leu operon with varied half-life.Citation36 Our data suggest that YeaZ is possibly involved in posttranslational regulation of the ilv-leu operon, since qPCR results exhibited significant variations in the increased transcriptional levels of ilvD, leuA and ilvA genes, which are located in one polycistronic operon, during the depletion of YeaZ. It needs to be defined whether YeaZ affects the activity of endoribonuclease and exoribonuclease in S. aureus.
The observation that the deletion of the ilv-leu operon had no impact on the essentiality of YeaZ for growth, which is consistent with our recent finding that the ilv-leu operon has no impact on the requirement of Gcp,Citation11 indicating the regulation of ILV biosynthesis pathway is one aspect of the biological function of YeaZ, but is not associated with the essential mechanism of YeaZ. This result is consistent with the requirement of YeaZ for bacterial growth in a limited ILV condition. We are currently in the process of determining the full YeaZ regulon and to elucidate the essential mechanism of YeaZ for growth.
In conclusion, this study provides direct evidence that the staphylococcal YeaZ is indispensable for bacterial growth in culture and possesses a similarly essential mechanism to E. coli YeaZ. Moreover, our results demonstrated that YeaZ mediates the biosynthesis of the branched-chain amino acids (BCAAs): isoleucine, leucine and valine (ILV) by transcriptional regulation of the ilv-leu operon. Furthermore, we revealed that the essentiality of YeaZ for growth is not attributable to its modulation of the ilv-leu operon.
Materials and Methods
Bacterial strains, plasmids, and growth conditions
The strains and plasmids used in this study are listed in . E. coli strain DH10B was used for plasmid constructions. Luria-Bertani (LB) liquid medium and LB-agar plates were used for the growth and maintenance of E. coli. S. aureus laboratory strain RN4220 was used as an intermediate host strain prior to introducing plasmids into wild type S. aureus strains. S. aureus WCUH29 and 923 are methicillin-resistant clinical isolates (MRSA) that were used for genetic manipulation and growth characterization. Tryptic soy broth (TSB) medium and chemically defined media (CDM) were used for the cultivation of S. aureus. Glucose was used as a carbon source at a concentration of 56 mM (1%, w/v). All amino acids included in CDM were L-amino acids. When necessary, isoleucine, leucine and valine were left out of the medium during preparation resulting in ILV dropout CDM.
Table 2. Strains and plasmids used in this study
Construction of a defined Pspac-regulated yeaZ expression mutant in S. aureus
The construction of a defined Pspac-regulated yeaZ expression mutant strain in WCUH29 was performed as described.Citation28 Briefly, the yeaZ gene was obtained by PCR with primers listed in , cloned downstream of Pspac promoter in the integration vector pLH1 () with tetracycline for selection. The recombinant plasmid was electroporated into RN4220 and was integrated into the chromosome at the Phage 11 attB site as previously described.Citation37 The chromosome segment containing the Pspac-yeaZ cassette was then transferred from RN4220 to WCUH29 by phage transduction as described using tetracycline selectionCitation38 and confirmed by diagnostic PCR. After the Pspac-yeaZ segment was transduced into the WCUH29 chromosome, the endogenous yeaZ gene was then deleted by homogenous recombination using the temperature sensitive plasmid pKOR1 as described ().Citation39 Using the same strategy, we created the ilv-leu operon deletion mutants in both the defined Pspac-regulated yeaZ expression mutant and the parental wild-type WCUH29 strain. The ilv-leu operon is a 10 kb chromosome segment; to increase the efficiency of allelic replacement events, we deleted the gene cassettes ilvDBHC and leuABCDilvA sequentially in-frame.
Table 3. Primers used in this study
RNA isolation and purification.
Overnight cultures of S. aureus were inoculated at 1% in TSB medium and grown to the mid-exponential (∼3 h) phase of growth. Total RNA was purified from the above cultures, as described.Citation38 Briefly, bacterial cells were harvested by centrifugation at 4,000 ×g, and the RNA was isolated using the SV total RNA isolation system (Promega, Z3100). Contaminating DNA was removed with 2 rounds of DNase treatment (TURBO DNA-free kit, Ambion, AM1907), and the RNA yield was determined spectrophotometrically at 260 nm.
Semi-quantitative real-time RT-PCR (qPCR) analysis
In order to determine the effect of YeaZ on the expression of ilv-leu operon, we employed qPCR to compare the RNA levels as described.Citation11,38 The first strand cDNA was synthesized using reverse transcriptase SuperScript III and random primers (Life Technologies, 18080044). For each RNA sample, we performed duplicate reactions of reverse transcription, as well as a control without reverse transcriptase, PCR reactions were set up in duplicate in order to determine the levels of DNA contamination. Real-time sequence-specific detection and relative quantitation were performed by using VeriQuest SYBR Green qPCR Master Mix (Affymetrix, 75600) with the Stratagene Mx3000P Real Time PCR System. Gene-specific primers were designed to yield 100 ∼ 200 bp specific product (). Relative quantification of the product was calculated using the Comparative CT method, as described for the Stratagene Mx3000P system. The housekeeping gene 16S rRNA was used as an endogenous control.Citation11
Construction of ilvD promoter-lux reporter fusion system
To confirm whether YeaZ mediates the transcription of the ilv-leu operon, we constructed an ilv promoter-lux reporter fusion system as described.Citation11,28 The ilv promoter-lux fusion plasmidCitation11 was electroporated into the defined Pspac-regulated yeaZ expression mutant S. aureus as described.Citation40 Both bioluminescence signals and cell growth were monitored at 37°C by measuring the bioluminescent intensity and optical density at 600 nm with a BioTek Synergy Microplate Reader. To eliminate the effect of bacterial growth, the relative light units (RLU) were calculated (light intensity/OD600) from triplicate readings at different times during growth.
Cloning, expression and purification of recombinant YeaZ protein
The staphylococcal yeaZ gene was obtained by PCR using the primers listed in and cloned into the BamHI and SalI sites of pGEX-4T-1. The recombinant plasmid was transformed into E. coli strain BL21 (DE3). A 500 ml culture of recombinant YeaZ expression strain in LB medium was incubated aerobically at 37°C to an optical density OD600 nm of 0.6. The recombinant protein expression was subsequently induced by the addition of 1 mM IPTG. After 5 hours of incubation at 37°C, cells were harvested and resuspended in the binding buffer (150 mM NaCl, 20 mM NaH2PO4, pH 7.3) containing 1 mg/ml lysozyme and 1 mM PMSF at room temperature for 1 h. The cells were lysed by sonication on ice; after centrifugation at 25,000 × g for 20 min at 4°C, the supernatant was collected and mixed with Glutathione Sepharose 4B beads (GE Healthcare, 17-0756-01). The purification of YeaZ was conducted according to the manufacturer's instructions. Thrombin was used to cut off YeaZ from GST tag and Benzamidine Sepharose 6B beads (GE Healthcare,17-0568-01) were applied to remove thrombin. The purity of the purified recombinant protein was confirmed by SDS-polyacrylamide gel electrophoresis (PAGE) followed by Coomassie bright blue staining; the protein concentration was measured using the BCATM Protein Assay kit (Thermo Scientific, 23227).
Gel mobility shift DNA binding assay
To determine regulatory mechanism of YeaZ in mediating the transcription of the ilv-leu operon, we performed gel-shift assays. The DNA fragment of the upstream region of ilv was obtained by PCR using the primers listed in . The amplified DNA fragment was purified and labeled with Digoxigenin using a DIG Gel Shift Kit 2nd Generation (Roche, 03353591910) or labeled with Biotin according to the manufacturer's protocol. The DNA-binding and electrophoresis were performed, as describedCitation28,41 Briefly, the purified PCR products were labeled with Digoxigenin using terminal transferase (Roche). The labeled DNA fragments were further purified to remove the redundant DIG-ddUTP and salts. The interaction of YeaZ with DNA was conducted in a 20 μl reaction mixture containing 0.2 pmol DIG-labeled DNA, 1 μg of poly-[d(I–C)], 25 mM NaH2PO4 (pH 8.0), 50 mM NaCl, 2 mM MgCl2, 1 mM DTT, 10% glycerol, 0.1 mM EDTA, and different concentrations of YeaZ protein. For biotin-labeled DNA probes, gel-shift assays were carried out with LightShift Chemiluminescent EMSA Kit (Thermo Scientific, 20148). Unlabeled DNA fragments of the promoter region and internal gene region were added into the reaction with 100 to 300-fold excess to labeled probe as a specific competitor and a nonspecific competitor, respectively. BSA was used as a nonspecific protein binding control. The DNA binding reaction was initiated by the addition of YeaZ and incubated at room temperature for 25 min. Samples were then loaded directly onto a 6% native polyacrylamide gel [acrylamide:bisacrylamide (29:1) in 0.5 × TBE buffer]. Electrophoresis was run for 2 h at 4°C with 7 V/cm, and the gels were transferred to Nylon membrane via electro-blotting in 0.5 × TBE at 300 mA for 90 min at 4°C. After cross-linking of DNA fragments using UV, the membranes were hybridized with anti-digoxigenin-AP antibody and exposed to X-ray film for 4 h to achieve the desired signal. Biotin-labeled probes will be detected using a Chemiluminescent Nucleic Acid Detection Kit (Thermo Scientific, 89880).
Characterization of bacteria growth
The S. aureus strains were inoculated in TSB or CDM with appropriate antibiotics and different concentrations of IPTG (0, 10, 25, and 100 μM) at 37°C. When mentioned, the nutritional complete CDM with 1.143 mM L-isoleucine 1.143 mM L-leucine and 1.280 mM L-valine was serially diluted with ILV dropout CDM to generate ILV titrated CDM. The fresh S. aureus overnight cultures were adjusted to OD600 nm = 1.25, and then were 1:10Citation5 inoculated into the media. The bacterial growth was monitored by kinetically measuring optical density at 600 nm with a BioTek Synergy Microplate reader.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Martin Rosenberg and Mr. Jeffrey W. Hall for critically reading this manuscript.
Funding
This project was supported by grant AI057451 from the National Institute of Allergy and Infectious Disease.
References
- Appelbaum PC. The emergence of vancomycin-intermediate and vancomycin-resistant Staphylococcus aureus. Clin Microbiol Infect 2006; 12(Suppl 1):16–23; http://dx.doi.org/10.1111/j.1469-0691.2006.01344.x
- Loomba PS, Taneja J, Mishra B. Methicillin and vancomycin resistant S. aureus in hospitalized patients. J 2010; 2:275-83; PMID:20927290; http://dx.doi.org/10.4103/0974-777X.68535
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2006; 2: 2006.0008; PMID:16738554; http://dx.doi.org/10.1038/msb4100050
- Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A 2006; 103:2833-8; PMID:16477005; http://dx.doi.org/10.1073/pnas.0511100103
- Zalacain M, Biswas S, Ingraham KA, Ambrad J, Bryant A, Chalker AF, Iordanescu S, Fan J, Fan F, Lunsford RD, et al. A global approach to identify novel broad-spectrum antibacterial targets among proteins of unknown function. J Mol Microbiol Biotechnol 2003; 6:109-26; PMID:15044829; http://dx.doi.org/10.1159/000076741
- Panutdaporn N, Kawamoto K, Asakura H, Makino SI. Resuscitation of the viable but non-culturable state of Salmonella enterica serovar Oranienburg by recombinant resuscitation-promoting factor derived from Salmonella Typhimurium strain LT2. Int J Food Microbiol 2006; 106:241-7; PMID:16213054; http://dx.doi.org/10.1016/j.ijfoodmicro.2005.06.022
- Shleeva M, Mukamolova GV, Young M, Williams HD, Kaprelyants AS. Formation of 'non-culturable' cells of Mycobacterium smegmatis in stationary phase in response to growth under suboptimal conditions and their Rpf-mediated resuscitation. Microbiology 2004; 150:1687-97; PMID:15184555; http://dx.doi.org/10.1099/mic.0.26893-0
- Ravagnani A, Finan CL, Young M. A novel firmicute protein family related to the actinobacterial resuscitation-promoting factors by non-orthologous domain displacement. BMC Genomics 2005; 6:39; PMID:15774001; http://dx.doi.org/10.1186/1471-2164-6-39
- Oliver JD. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol Revi 2010; 34:415-25
- Aydin I, Saijo-Hamano Y, Namba K, Thomas C, Roujeinikova A. Structural analysis of the essential resuscitation promoting factor YeaZ suggests a mechanism of nucleotide regulation through dimer reorganization. PLOS One 2011; 6:e23245; PMID:21858042; http://dx.doi.org/10.1371/journal.pone.0023245
- Lei T, Yang J, Zheng L, Markowski T, Witthuhn BA, Ji Y. The essentiality of staphylococcal gcp is independent of its repression of branched-chain amino acids biosynthesis. PLOS One 2012; 7:e46836; PMID:23056478; http://dx.doi.org/10.1371/journal.pone.0046836
- Lei T, Liang X, Yang J, Yan M, Zheng L, Walcheck B, Ji Y. The C-terminal domain of the novel essential protein Gcp is critical for interaction with another essential protein YeaZ of Staphylococcus aureus. PLOS One 2011; 6:e20163; PMID:21625506; http://dx.doi.org/10.1371/journal.pone.0020163
- Nichols CE, Lamb HK, Thompson P, El Omari K, Lockyer M, Charles I, Hawkins AR, Stammers DK. Crystal structure of the dimer of two essential Salmonella typhimurium proteins, YgjD & YeaZ and calorimetric evidence for the formation of a ternary YgjD-YeaZ-YjeE complex. Protein Sci 2013; 22:628-40; PMID:23471679; http://dx.doi.org/10.1002/pro.2247
- Jeudy S, Stelter M, Coutard B, Kahn R, Abergel C. Preliminary crystallographic analysis of the Escherichia coli YeaZ protein using the anomalous signal of a gadolinium derivative. Acta Crystallogr Sect F Struct Biol Cryst Commun 2005; 61:848-51; PMID:16511176; http://dx.doi.org/10.1107/S1744309105025856
- Handford JI, Ize B, Buchanan G, Butland GP, Greenblatt J, Emili A, Palmer T. Conserved network of proteins essential for bacterial viability. J Bacteriol 2009; 191:4732-49; PMID:19376873; http://dx.doi.org/10.1128/JB.00136-09
- Nichols CE, Johnson C, Lockyer M, Charles IG, Lamb HK, Hawkins AR, Stammers DK. Structural characterization of Salmonella typhimurium YeaZ, an M22 O-sialoglycoprotein endopeptidase homolog. Proteins 2006; 64:111-23; PMID:16617437; http://dx.doi.org/10.1002/prot.20982
- Xu Q, McMullan D, Jaroszewski L, Krishna SS, Elsliger MA, Yeh AP, Abdubek P, Astakhova T, Axelrod HL, Carlton D, et al. Structure of an essential bacterial protein YeaZ (TM0874) from Thermotoga maritima at 2.5 A resolution. Acta Crystallogr Sect F Struct Biol Cryst Commun 2010; 66:1230-6; PMID:20944216; http://dx.doi.org/10.1107/S1744309109022192
- Buss KA, Cooper DR, Ingram-Smith C, Ferry JG, Sanders DA, Hasson MS. Urkinase: structure of acetate kinase, a member of the ASKHA superfamily of phosphotransferases. J Bacteriol 2001; 183:680-6; PMID:11133963; http://dx.doi.org/10.1128/JB.183.2.680-686.2001
- Hurley JH. The sugar kinase/heat shock protein 70/actin superfamily: implications of conserved structure for mechanism. Annual review of biophysics and biomolecular structure 1996; 25:137-62; PMID:8800467; http://dx.doi.org/10.1146/annurev.bb.25.060196.001033
- Brinsmade SR, Kleijn RJ, Sauer U, Sonenshein AL. Regulation of CodY activity through modulation of intracellular branched-chain amino acid pools. J Bacteriol 2010; 192:6357-68; PMID:20935095; http://dx.doi.org/10.1128/JB.00937-10
- Grandoni JA, Zahler SA, Calvo JM. Transcriptional regulation of the ilv-leu operon of Bacillus subtilis. J Bacteriol 1992; 174:3212-9; PMID:1577690
- Shivers RP, Sonenshein AL. Bacillus subtilis ilvB operon: an intersection of global regulons. Mol Microbiol 2005; 56:1549-59; PMID:15916605; http://dx.doi.org/10.1111/j.1365-2958.2005.04634.x
- Lincoln RA, Leigh JA, Jones NC. The amino acid requirements of Staphylococcus aureus isolated from cases of bovine mastitis. Veterinary Microbiology 1995; 45:275-9; PMID:7571379; http://dx.doi.org/10.1016/0378-1135(95)00041-8
- Onoue Y, Mori M. Amino acid requirements for the growth and enterotoxin production by Staphylococcus aureus in chemically defined media. Int J Food Microbiol 1997; 36:77-82; PMID:9168317; http://dx.doi.org/10.1016/S0168-1605(97)01250-6
- Majerczyk CD, Dunman PM, Luong TT, Lee CY, Sadykov MR, Somerville GA, Bodi K, Sonenshein AL. Direct targets of CodY in Staphylococcus aureus. J Bacteriol 2010; 192:2861-77; PMID:20363936; http://dx.doi.org/10.1128/JB.00220-10
- Ludwig H, Meinken C, Matin A, Stulke J. Insufficient expression of the ilv-leu operon encoding enzymes of branched-chain amino acid biosynthesis limits growth of a Bacillus subtilis ccpA mutant. J Bacteriol 2002; 184:5174-8; PMID:12193635; http://dx.doi.org/10.1128/JB.184.18.5174-5178.2002
- Tojo S, Satomura T, Morisaki K, Deutscher J, Hirooka K, Fujita Y. Elaborate transcription regulation of the Bacillus subtilis ilv-leu operon involved in the biosynthesis of branched-chain amino acids through global regulators of CcpA, CodY and TnrA. Mol Microbiol 2005; 56:1560-73; PMID:15916606; http://dx.doi.org/10.1111/j.1365-2958.2005.04635.x
- Fan F, Lunsford RD, Sylvester D, Fan J, Celesnik H, Iordanescu S, Rosenberg M, McDevitt D. Regulated ectopic expression and allelic-replacement mutagenesis as a method for gene essentiality testing in Staphylococcus aureus. Plasmid 2001; 46:71-5; PMID:11535039; http://dx.doi.org/10.1006/plas.2001.1526
- Yan M, Yu C, Yang J, Ji Y. Development of shuttle vectors for evaluation of essential regulator regulated gene expression in Staphylococcus aureus. Plasmid 2009; 61:188-92; PMID:19245820; http://dx.doi.org/10.1016/j.plasmid.2009.02.001
- Cue D, Lei MG, Lee CY. Activation of sarX by Rbf is required for biofilm formation and icaADBC expression in Staphylococcus aureus. J Bacteriol 2013; 195:1515-24; PMID:23354746; http://dx.doi.org/10.1128/JB.00012-13
- Hashimoto C, Sakaguchi K, Taniguchi Y, Honda H, Oshima T, Ogasawara N, Kato J. Effects on transcription of mutations in ygjD, yeaZ, and yjeE genes, which are involved in a universal tRNA modification in Escherichia coli. J Bacteriol 2011; 193:6075-9; PMID:21873492; http://dx.doi.org/10.1128/JB.05733-11
- Vitreschak AG, Lyubetskaya EV, Shirshin MA, Gelfand MS, Lyubetsky VA. Attenuation regulation of amino acid biosynthetic operons in proteobacteria: comparative genomics analysis. FEMS Microbiol Lett 2004; 234:357-70; PMID:15135544; http://dx.doi.org/10.1111/j.1574-6968.2004.tb09555.x
- Gutierrez-Preciado A, Henkin TM, Grundy FJ, Yanofsky C, Merino E. Biochemical features and functional implications of the RNA-based T-box regulatory mechanism. Microbiol Mol Biol Rev 2009; 73:36-61; PMID:19258532; http://dx.doi.org/10.1128/MMBR.00026-08
- Grundy FJ, Haldeman MT, Hornblow GM, Ward JM, Chalker AF, Henkin TM. The Staphylococcus aureus ileS gene, encoding isoleucyl-tRNA synthetase, is a member of the T-box family. J Bacteriol 1997; 179:3767-72; PMID:9171428
- Reiss S, Pane-Farre J, Fuchs S, Francois P, Liebeke M, Schrenzel J, Lindequist U, Lalk M, Wolz C, Hecker M, et al. Global analysis of the Staphylococcus aureus response to mupirocin. Antimicrob Agents Chemother 2012; 56:787-804; PMID:22106209; http://dx.doi.org/10.1128/AAC.05363-11
- Mader U, Hennig S, Hecker M, Homuth G. Transcriptional organization and posttranscriptional regulation of the Bacillus subtilis branched-chain amino acid biosynthesis genes. J Bacteriol 2004; 186:2240-52; PMID:15060025; http://dx.doi.org/10.1128/JB.186.8.2240-2252.2004
- Luong TT, Lee CY. Improved single-copy integration vectors for Staphylococcus aureus. J Microbiol Methods 2007; 70:186-90; PMID:17512993; http://dx.doi.org/10.1016/j.mimet.2007.04.007
- Ji Y, Marra A, Rosenberg M, Woodnutt G. Regulated antisense RNA eliminates alpha-toxin virulence in Staphylococcus aureus infection. J Bacteriol 1999; 181:6585-90; PMID:10542157
- Bae T, Schneewind O. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 2006; 55:58-63; PMID:16051359; http://dx.doi.org/10.1016/j.plasmid.2005.05.005
- Zheng L, Yang J, Landwehr C, Fan F, Ji Y. Identification of an essential glycoprotease in Staphylococcus aureus. FEMS Microbiol Lett 2005; 245:279-85; PMID:15837383; http://dx.doi.org/10.1016/j.femsle.2005.03.017
- Liang H, Zhao YT, Zhang JQ, Wang XJ, Fang RX, Jia YT. Identification and functional characterization of small non-coding RNAs in Xanthomonas oryzae pathovar oryzae. BMC genomics 2011; 12:87; PMID: 21276262; http://dx.doi.org/10.1186/1471-2164-12-87
- Kreiswirth BN, Lofdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 1983; 305:709-12; PMID:6226876; http://dx.doi.org/10.1038/305709a0
- Ji Y, Zhang B, Van SF, Horn, Warren P, Woodnutt G, Burnham MKR, Rosenberg M. Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science 2001; 293:2266-9; PMID:11567142
- Boyle-Vavra S, Yin S, Daum RS. The VraS/VraR two-component regulatory system required for oxacillin resistance in community-acquired methicillin-resistant Staphylococcus aureus. FEMS Microbiol Lett 2006; 262:163-71; PMID:16923071; http://dx.doi.org/10.1111/j.1574-6968.2006.00384.x