Abstract
Cell adhesion and migration are regulated through the concerted action of cytoskeletal dynamics and adhesion proteins, the activity of which is governed by RhoGTPases. Specific RhoGTPase signaling requires spatio-temporal activation and coordination of subsequent protein-protein and protein-lipid interactions. The nature, location and duration of these interactions are dependent on polarized extracellular triggers, such as cell-cell contact, and intracellular modifying events, such as phosphorylation. RhoA, RhoB, and RhoC are highly homologous GTPases that, however, succeed in generating specific intracellular responses. Here, we discuss the key features that contribute to this specificity. These not only include the well-studied switch regions, the conformation of which is nucleotide-dependent, but also additional regions and seemingly small differences in primary sequence that also contribute to specific interactions. These differences translate into differential surface charge distribution, local exposure of amino acid side-chains and isoform-specific post-translational modifications. The available evidence supports the notion that multiple regions in RhoA/B/C cooperate to provide specificity in binding to regulators and effectors. These specific interactions are highly regulated in time and space. We therefore subsequently discuss current approaches means to visualize and analyze localized GTPase activation using biosensors that allow imaging of isoform-specific, localized regulation.
Abbreviations
| FRET | = | Förster resonance energy transfer |
| GAP | = | GTPase activating protein |
| GDP | = | guanine nucleotide diphosphate |
| GEF | = | guanine nucleotide exchange factor |
| GFP | = | green fluorescent protein |
| GTP | = | guanine nucleotide triphosphate |
| RBD | = | RhoGTPase binding domain |
Introduction
The family of RhoGTPases, part of the superfamily of Ras-like GTP-binding proteins, was first described 3 decades ago.Citation1 There are approximately 20 RhoGTPases described in mammals, a small number of which such as RhoA, Rac1 and Cdc42 have been studied in great detail.Citation2,3 Rho family members show high amino acid sequence and structural homology, but cellular responses can be GTPase-specific, even for the isoforms RhoA, RhoB and RhoC that show 88% sequence homology ().Citation4 RhoGTPases are best known for their regulation of cytoskeletal dynamics and, as a consequence, of cell adhesion and migration.Citation5,6 RhoA controls actin stress fiber formation and acto-myosin contraction at the rear of the cell, Rac1 regulates formation of membrane ruffles and Cdc42 controls formation of filopodial extensions at the leading edge. RhoB expression is regulated by growth factors such as EGF, inflammatory cytokines such as TNFα and stress stimuli including UV radiation.Citation7 RhoB is localized to endosomes as well as to the plasma membrane and has been implicated in cytoskeletal dynamics and the regulation of TNFα signaling in primary human endothelial cells. RhoC is strongly associated with cell motility. MiRNA-mediated regulation of RhoC expression is implicated in metastasis and cancer.Citation8,9 Recent studies using RhoGTPase biosensors, however, indicate a more complex functional relationship between these proteins (see below). It is well accepted that GTPase-specific responses (e.g. cell contraction by RhoA versus protrusion by Rac1) are the result of tightly controlled spatio-temporal activation and signaling.Citation10 Detailed studies based on structural and mutational analysis have identified a series of regions and amino acid residues within RhoGTPases that are required for their specific localization, activation and downstream signaling. These data imply that, within a particular cellular context, RhoGTPases find a way to selectively activate subsets of effector proteins at selected locations within the cell.
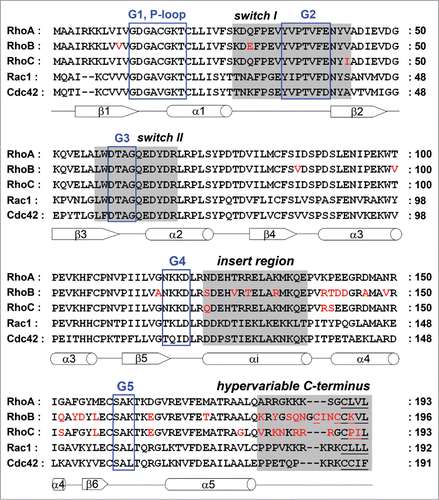
Figure 1. Alignment of the RhoA, RhoB, RhoC, Rac1 and Cdc42 amino acid sequences. G motifs are marked with a blue box. Switch I, switch II, the insert region and the hypervariable C-terminus are highlighted in gray, sites for lipid modification are underlined. Sequence differences in RhoB and RhoC compared to RhoA are shown in red. Secondary structures (α-helices as cylinders, β-strands as arrows) are referring to RhoA and indicated below the amino acid sequence.
Most RhoGTPases are molecular switches cycling between an inactive guanine nucleotide diphosphate (GDP)-bound state and an active guanine nucleotide triphosphate (GTP)-bound state.Citation2,3,11 In the active conformation, GTPases specifically interact with effector proteins to initiate downstream signaling. RhoGTPases show a high affinity for guanine nucleotides, and a slow intrinsic GDP/GTP exchange rate and GTP hydrolysis. Because this does not allow fast on/off rates, cells use a series of regulatory proteins to control spatio-temporal signaling by RhoGTPases. These include Rho guanine nucleotide exchange factors (RhoGEFs, reviewed in refs. 3, 13), which stimulate the release of bound GDP and form a complex with the nucleotide-free GTPase. Since the concentration of GTP in the cell is higher than that of GDP, this favors GTP binding and GTPase activation. Another large group of regulators are the RhoGTPase activating proteins (RhoGAPs, reviewed in refs. 3, 13), which accelerate the slow intrinsic GTP hydrolysis which leads to RhoGTPase inactivation. Finally, most RhoGTPases bind to chaperones, called guanine nucleotide dissociation inhibitors (RhoGDIs), which are cytosolic proteins that lack enzymatic activity. GDIs retain RhoGTPases in the inactive conformation, sequester them from cellular membranes and protect the GTPase from effector binding and proteolytic degradation.Citation12
Until recently, analysis of RhoGTPase signaling mostly relied on biochemical assays or image analysis in combination with ectopic expression of active or inactive mutants. Over the past ∼10 years however, this field has seen a growing set of tools in the form of isoform-specific antibodies as well as biosensors, which allow researchers to study agonist-induced, localized signaling by endogenous RhoGTPases in living cells. In this overview, we will focus on the structure-function relation of a subset of RhoGTPases, limiting the discussion to the closely related family members RhoA, RhoB and RhoC. We will discuss the GTPase-specific interaction with regulators and effectors and discuss the use of GTPase-based biosensors in studies on localized activation and signaling. We will address the main question on how structural differences may allow specificity between such highly homologous GTPases. An important conclusion that emerges from this overview is that the molecular basis of GTPase-specific output lies in the fact that several regions within the GTPase structure, including the hypervariable portion outside the core G (guanine nucleotide binding) domain, provide key contributions to signaling specificity.
The Structural Core of RhoGTPases
The G domain
RhoGTPase are monomeric proteins of around 20 kDa. Their structure comprises the core G domain, a hallmark of RhoGTPases and other members of the Ras-like GTPase superfamilyCitation,2,3,11 next to a short ‘insert region’ and the C-terminal short hypervariable region (). Guanine nucleotide binding is mediated through the G domain which contains a 6-stranded mixed β-sheet surrounded by 5 α-helices and is extended by the helical insert region which is characteristic for the family of RhoGTPases (). The G domain is characterized by 5 conserved sequence motifs G1-G5. The G1 motif (also known as P-loop) coordinates the β-phosphate of the bound nucleotide and the Mg2+ ion which is required for nucleotide binding. RhoA, RhoB and RhoC have identical sequences in this region (). Introducing the steric mutation Gly14Val in RhoA/B/C leads to a constitutively active, ‘GTP-locked’ protein whereas the mutation Thr19Asn in RhoA/B/C results in a low nucleotide affinity which in turn leads to a high affinity binding to GEF proteins. This renders this mutant dominant negative. Both mutants have been commonly used in biochemical and cell biological experiments as tools to identify specific interaction partners and explore the biological activities of the GTPases. In contrast to the G1 motif, low sequence homology among all RhoGTPases, including RhoA, RhoB and RhoC, has been observed for the G4 motif and the G5 motif which both mediate the interaction with the guanine base ().
Figure 2. 3D structures of the RhoA, RhoB and RhoC G domain. (A–C) Ribbon representation of (A) RhoA (4–180 aa; Protein Data Bank (PDB) ID: 1FTN),Citation75 (B) RhoB (4–185 aa; PDB ID: 2FV8)Citation76 and (C) RhoC (3–179 aa; PDB ID: 2GCN).Citation77 Structures are shown in the same orientation. Switch I, switch II and the insert region are highlighted in gray, GDP is shown in yellow sticks. Residues marked in cyan differ among RhoA/B/C (underlined labeling in A) and are different compared to Rac1/Cdc42 (labeling in C). (D–F) Zooms of A–C highlighting that RhoA and RhoB contain at position 43 a valine, whereas RhoC has an isoleucine which has slightly bulkier side chain which underlies isoform-specific GEF- and effector binding (see text). (G–I) Electrostatic potential of the solvent accessible surface of the G domains of (G) RhoA, (H) RhoB and (I) RhoC represented in the same orientation as in A–C, lower panels show cognate structures rotated by 180°. Note that the hypervariable C-termini are not included in this representation. Electrostatic potential was calculated using the Adaptive Poisson-Boltzmann Solver (APBS) software, combined with the PDB2PQR server, at a pH of 7.5 and a threshold of ± 5 kTe−1 (red – negative charge; blue – positive charge).Citation78 Figures were prepared with PyMol (PyMol Molecular Graphics System, Schroedinger, LLC).
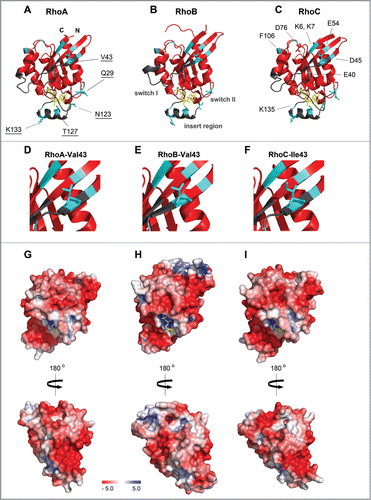
The isoforms RhoA, RhoB and RhoC have an identical sequence in the switch I (residues 27–43 in RhoA) and switch II regions (residues 57–68), except for the positions 29 and 43 which are both located in switch I (). Whereas RhoA and RhoB have a valine at position 43, RhoC encodes a hydrophobic, but bulkier, amino acid, an isoleucine (), which plays a crucial role in RhoGEF and effector binding (see below). Intriguingly, RhoA and RhoC contain at position 29 a glutamine whereas RhoB has a negatively charged glutamate residue at this position (). In contrast, the switch I region among RhoA, Rac1 and Cdc42 shows only low sequence homology (). The switch II region contains a highly conserved glutamine (Gln63 in RhoA/B/C) which is remarkably conserved between all RhoGTPases. This amino acid coordinates the nucleophilic water relative to the GTP γ-phosphate and is thus required for the intrinsic and GAP-catalyzed GTP hydrolysis.Citation2,3,11 Mutating this residue to a leucine or alanine renders a RhoGTPase constitutively active.
The switch I and switch II regions sense whether a GDP or a GTP molecule is bound, and these regions change their conformation accordingly (). In detail, the main chain NH groups of the highly conserved Thr37 (switch I) and Gly62 (switch II) in RhoA form 2 hydrogen bonds with the oxygen of the γ-phosphate in the nucleotide. This phosphate is only present in GTP but not in GDP. Loss of the γ-phosphate following GTP hydrolysis leads to loss of these hydrogen bonds and to the relaxation of the entire switch I and switch II regions into the GDP-bound form. This mechanism, based on these structural changes in the switch regions, is universal to small GTPases and is known as ‘loading-spring’ mechanism.Citation2,11,13 Mainly dependent on these structural differences, regulatory proteins and effectors detect the nucleotide conformation of the RhoGTPase and interact with both switch regions. However, although most described interactions involve the switch regions, there is sufficient evidence for additional portions of small GTPases, such as the insert region or a polybasic C-terminal domain, to contribute or even determine specific interactions with Rho GTPases (see below).
The Rho insert region
A hallmark of all members of the RhoGTPase family is the Rho insert region (residues 123–137 in RhoA) which is located between the G4 and G5 motif and extends the core G domain by about 13 residues ().Citation2,3,11 The insert region is involved in GEF binding,Citation14 but serves primarily in the binding and activation of effector proteins such as the NADPH oxidase,Citation15 IQGAP,Citation16 ROCKCitation17 and mDia.Citation18
The insert region of RhoA and RhoC is identical, except for the Asn123 in RhoA which has, however, a similar side-chain as the Gln123 in RhoC (). The insert region of RhoB differs in 4 amino acids. At position 123, RhoB contains a serine, a polar but smaller residue than the respective asparagine in RhoA and glutamine in RhoC. Furthermore, RhoB harbors a hydrophobic valine instead of a polar threonine in RhoA and RhoC at position 127 and an arginine at position 133 which is also a positively charged residue like the lysine in RhoA and RhoC. More interesting is residue 129 which is a polar threonine in RhoB, but a positively charged arginine in RhoA and RhoC. Whether the insert region may indeed mediate RhoB-specific interactions has not yet been demonstrated. In contrast, it has been shown that this region, which is also remarkably different between RhoA, Rac1 and Cdc42 (), determines isoform-specific binding of these GTPases to several effector proteins (see below).
The C-terminus of RhoGTPases
The C-terminus of RhoGTPases comprises, next to the hypervariable region, the CAAX-box to which a lipid anchor is attached allowing GTPase binding to membranes ().Citation2,19 RhoA, RhoB and RhoC differ in this posttranslational lipid modification which has consequences for their subcellular localization. The lipid anchor regulates the interaction with RhoGDIs and with regions within the plasma membrane (RhoA, RhoC) or endosomal vesicles (RhoB).Citation4,12 RhoA and RhoC are geranylgeranylated, whereas RhoB has both a palmitoyl anchor and a farnesyl or geranylgeranyl group.Citation4
The hypervariable region also functions as protein binding site and determines, at least in part, specific binding to regulatory or effector proteins (see below). In most RhoGTPases, including RhoA and RhoC, this region of around 10 residues harbors a polybasic stretch that binds the inner leaflet of the plasma membrane (). Once more, RhoB shows a special feature since it does not have positively charged amino acids (lysine, arginine) in its C-terminal domain but contains mainly polar residues (serine, glutamine), which may form hydrogen bonds with interaction partners.
In summary, RhoA, RhoB and RhoC have a very similar structure of the G domain and the insert region (). However, the differences in their surface charge distribution in these domains (; Movies S1–S3), are likely important for isoform-specific interactions with GEFs, GAPs, GDIs and effector proteins. Additional charge differences are provided by the hypervariable C-termini (basic in RhoA/C, polar in RhoB; not included in the representation in ). Moreover, the differential targeting of RhoA, RhoB and RhoC as determined by the lipid anchor(s) and/or the hypervariable region will allow localized GTPase activation and signaling, which further contributes to signaling specificity.
Sequence Diversity Determines Binding and Function
RhoGDIs
In contrast to RhoGEFs and RhoGAPs, only 3 RhoGDIs isoforms have been described in mammals.Citation12 RhoGDIs consist of 2 domains. The N-terminal regulatory domain interacts with the switch I and switch II regions of the RhoGTPase. The C-terminal domain, which is required for the membrane extraction of the GTPase, binds to the switch II region, the α3-helix and the lipid anchor. Structural and biochemical studies have identified Thr37, Tyr66, Arg68, Leu69 and Leu72 in RhoA as key residues for GDI-binding.Citation20-22 These amino acids are identical among RhoA, RhoB and RhoC (). However, RhoA and RhoC, but not RhoB, bind to RhoGDI1.Citation12,23 Conversely, RhoB, but not RhoA and RhoC, forms a complex with RhoGDI3. Further studies are required to determine whether these isoform-specific interactions are based on differences in sequence and/or intracellular localization of the Rho proteins.
RhoGEFs
Over 70 different GEFs for RhoGTPases have been described in mammals.Citation3 RhoGEFs bind to both switch regions of the GTPase and thus have overlapping binding sites with GAPs, GDIs and effectors.Citation24 The largest and best-known group of RhoGEFs contains a Dbl Homology (DH) domain, often in combination with a Pleckstrin Homology (PH) domain. Many of these multi-domain proteins show only limited binding specificity which makes spatio-temporal regulation necessary to allow specific signaling. For example, the canonical GEF Dbl as well as the GEFs Vav1, 2, 3 can activate RhoA, but also Rac1 and Cdc42.Citation24 However, there are also examples in which other regions within the GTPase were found to be required for a specific interaction. Leukemia-associated RhoGEF (LARG) and p190RhoGEF specifically activate RhoA, RhoB and RhoC, but not Rac1 and Cdc42, as shown in structural and biochemical studiesCitation25,26 This specificity is mediated through electrostatic interactions and determined by 2 negatively charged residues, Asp45 and Glu54, in the β2- and β3-strand of RhoA/B/C (). These residues are located in between the switch I and II regions, underscoring the notion that residues outside these regions also contribute to signaling (i.e. activation) specificity. In addition, another residue outside of switch II, the Asp76 in the α2-helix of RhoA/B/C, is also crucial for LARG binding (). The residues at position 45, 54 and 76 in Rac1 and Cdc42 are all polar, rather than negatively charged, which may explain the selectivity of the GEFs (). Similarly, the RhoGEF Ect2 (epithelial cell transforming sequence 2) specifically activates RhoA, but not Rac1 and Cdc42, as shown in NIH3T3 fibroblast cells and with purified proteins.Citation27 It was recently described that Ect2 also activates RhoB in human breast and cervical cancer cell lines.Citation28 However, which residues in the GTPase determine the specific binding and whether Ect2 is also an activator for RhoC remains to be investigated.
RhoGEFs that differentiate between the highly related RhoA, RhoB and RhoC isoforms are relatively rare. XPLN, which also belongs to the Dbl-RhoGEF family, shows GEF activity toward RhoA and RhoB, but not RhoC, and promotes assembly of stress fibers and focal adhesions in NIH3T3 fibroblasts.Citation29 Studies with purified proteins and in NIH3T3 fibroblasts indicate that residue 43 defines the isoform distinction, which is one of the very few sequence differences in the N-terminal part of RhoA, RhoB and RhoC ().Citation29,30 Although the structure of the complex of Rho and XPLN has not been resolved yet, it has been suggested that Ile43 in RhoC, which has a slightly bulkier side chain than Val43 in RhoA and RhoB, interferes with the binding and activity of XPLN ().
Another example is the atypical GEF SmgGDS which was found, using purified proteins and studies in HEK293 cells, to activate RhoA and RhoC but not RhoB.Citation31 The C-terminal polybasic region of RhoA and RhoC, in conjunction with the N-terminal portion of the GTPase, is required for the nucleotide exchange by SmgGDS. The lack of exchange activity of SmgGDS toward RhoB may be explained by the different hypervariable region of RhoB which consists mainly of polar residues (see above). This would be reminiscent of the activation of Rac1 by the GEF β–PIX, which is also dependent on the Rac1 hypervariable regionCitation32, albeit that structural evidence for the direct binding of SmgGDS to the C-terminus of a RhoGTPase is currently lacking.
RhoGAPs
Although around 80 different RhoGAPs are known in mammals,Citation3 only a few GAPs have been described to show specificity for the different Rho-like GTPases. In particular, RhoGAPs which are able to differentiate among RhoA, RhoB and RhoC have not yet been identified. Several structural and biochemical studies showed that RhoGTPases interact with RhoGAPs through the P-loop, the switch I and switch II regions.Citation3,33 For example, p190RhoGAP specifically inactivates RhoA, but not Cdc42 and Rac1, as shown with purified proteins and in fibroblasts where it inhibits stress fiber formation.Citation34 Even the GAP domain alone shows this behavior suggesting that certain residues in RhoA determine the isoform specificity. However, the crucial amino acids remain to be identified. In contrast, p190RhoGAP is not able to distinguish between RhoA, RhoB and RhoC, as was shown in several cell types such as fibroblasts, human bladder cancer, breast cancer and melanoma cells.Citation35,36
The RhoGAP ARHGAP21 (also known as ARHGAP10) shows specificity for RhoA and RhoC, but not for Cdc42, to control cancer progression in human PC3 prostate cancer cells.Citation37 In contrast, previous studies using purified proteins and in HeLa cells revealed that ARHGAP21 inactivates predominantly Cdc42 and much less RhoA and Rac1 to regulate cytoskeletal dynamics at the Golgi complex.Citation38 This indicates that the cellular context is a key factor in controlling GTPase specificity of ARHGAP21. Which cellular factors or residues in the RhoGTPase determine the isoform-specific binding to ARHGAP21 have not been investigated in these studies.
Effector proteins
In literature, the role of structural determinants in the binding of activated GTPases to downstream effectors has been most extensively analyzed. For RhoA, RhoB and RhoC, these effectors comprise protein kinases such as ROCK (Rho-associated kinase) and PKN/PRK (Protein kinase C-related kinase), as well as several formins, such as mDia (mammalian diaphanous) or FMNL (formin-like). The studies that have addressed the regions in RhoGTPases that are essential for Rho-effector interactions have usually focused on the so-called effector domain in the N-terminus (amino acids 27–41 in RhoA), which overlaps with the switch I region (). The switch I and II regions show the largest conformational change associated with GTP binding and it is therefore not surprising that these regions are most relevant for effector binding. However, additional regions are also important for isoform-specific downstream signaling.
Studies based on mutational analysis using chimeras of RhoA and Rac1 demonstrated already in 1995 that these GTPases comprise additional effector-binding regions in the C-terminal portion of the protein.Citation39 Later studies identified the region in the α5-helix (residues 167–179 in RhoA) as being important for effector binding such as shown for the CRIB (Cdc42/Rac1 interactive binding) effectors such as PAK1 (p21 activated kinase 1) or WASP (Wiskott-Aldrich syndrome protein).Citation33,40,41 Similarly, binding of the NADPH oxidase, which belongs to another effector group, to Rac1 is dependent on residues in the α3-helix and the α5-helix and thus regions outside of the classical interaction sites such as the switch regions.Citation42
The hypervariable C-terminus has also been implicated in effector binding. This was initially shown for Rac1, using peptides binding to the p67 subunit of the neutrophil NADPH oxidase.Citation43 Later, a similar finding based on biochemical and structural data has been reported for Rac1 binding to the PRK1 kinase.Citation44 A recent study using purified proteins in scintillation proximity assays analyzing the interactions of RhoA, RhoB and RhoC with the binding domains of the PRK1–3 effector kinases (the HR1 domains) showed that the hypervariable region of RhoB, but not that of RhoA or RhoC, promotes the interaction with the HR1 domain of PRK3, but not of PRK1 or -2.Citation45 The hypervariable region of RhoB allows a distinct, in this case preferential, type of interaction since it harbors primarily polar residues instead of a polybasic stretch as in RhoA and RhoC (). Co-immunoprecipitation experiments indicate, in contrast, that predominantly RhoC, and less RhoA and RhoB, forms a complex with PRK3 in various epithelial cancer cells to promote tumor invasion and metastasis.Citation46
Several studies have implicated the insert region (residues 123–137 in RhoA) in effector binding or regulation. The insert region was found to promote activation of ROCK by RhoA and thus to induce stress fiber formation in NIH3T3 fibroblasts, albeit that the insert region is not involved in ROCK binding.Citation17 A detailed structural and biochemical analysis focusing on the interactions between RhoGTPases and formins, showed that specificity in binding of RhoA and RhoC to mDia1 is determined by the insert helix and Phe106 in the Rho α3-helix (His104 in Rac1/Cdc42) and by Glu40 in the effector domain (Asp38 in Rac1/Cdc42) ().Citation18,47 RhoB, which was not included in this study, also contains the residues Phe106 and Glu40. A previous study indeed showed that RhoB directly interacts with mDia1.Citation48 This interaction was proposed to regulate actin assembly on vesicular membranes and vesicle traffic in human MelJuso melanoma cells.Citation49
The formin FMNL2 specifically binds activated RhoC, but not RhoA or RhoB, to control tumor invasion in MDA-MB435 breast cancer cells.Citation50 Part of this selectivity in effector binding was linked to the RhoC-specific isoleucine at position 43, which is also crucial for specificity in RhoGEF binding (see above, ). Mutating this residue to a valine as in RhoA and RhoB, reduced binding to FNML2, which suggests that this amino acid cooperates with the effector domain in controlling effector specificity. The related FMNL3 was also found to be a specific effector for activated RhoC, but not RhoA, to control cell shape and invasion of human PC3 prostate cancer cells.Citation51 However, the structural basis for this selectivity was not further investigated.
Regulation of RhoGTPases by phosphorylation and ubiquitylation
As an additional layer of complexity determining specificity of signaling, RhoA/B/C are posttranslationally modified in several ways. The best-studied event is the PKA- and PKG-mediated phosphorylation of RhoA on Ser188, which is located in the hypervariable C-terminus (). This phosphorylation serves as a Rho-inactivating signal, as it promotes GDI binding and membrane dissociation and protects RhoA from proteolytic degradation.Citation52 In addition, this phosphorylation interferes with RhoA binding to ROCK,Citation53 further underscoring a role of the C-terminus in effector interactions. It is important to note that this Ser188 is not present in RhoB (Asn191) or RhoC (Arg188) (), which provides the cell with a specific means to regulate RhoA output.
RhoC, but not RhoA, was recently shown to be phosphorylated on Ser73 in the α2-helix by the kinase Akt in SUM149 breast cancer cellsCitation54 although Ser73 is highly conserved among all 3 Rho-isoforms. Intriguingly, phosphorylation of RhoC was required for downstream signaling and invasiveness, which contrasts markedly with the inactivating phosphorylation at Ser188 in RhoA.
Finally, wild type and activated RhoA is ubiquitylated on Lys6 and Lys7 in the β1-strand () by the E3 ubiquitin ligase Smurf1 resulting in local RhoA degradation.Citation55 These residues are also present in RhoB and RhoC and similar regulation at these sites may be expected. A recent study identified activity-independent RhoA ubiquitylation by the FBXL19 ubiquitin E3 ligase as an alternative pathway toward RhoA degradation.Citation56 In this case, poly-ubiquitylation takes place on Lys135 in the insert helix, which is highly conserved among RhoA, RhoB and RhoC (). However, whether FBXL19 can also ubiquitylate RhoB and RhoC remains to be investigated. Although ubiquitylation likely serves to induce proteolytic degradation, there is ample evidence for ubiquitylation to serve as an anchor for protein binding or to drive internalization and lysosomal degradation of membrane-associated proteins.Citation57 Whether ubiquitylation serves such distinct roles in RhoGTPase signaling remains to be established.
Visualization of RhoGTPase Activation and Downstream Signaling
The above overview underscores that signaling specificity involves several different regions within the small RhoGTPase structure, including the well-established effector region, the insert region and the hypervariable C-terminal domain. In addition, regulatory proteins such as GEFs and GAPs contribute to signaling specificity and localized activation and output, although this notion is only partly developed. An important tool in the analysis of GEF-mediated specificity has been the development of GTPase biosensors, which allow visualization of localized GTPase activation. Below, we discuss the available information on biosensors for RhoA/B/C GTPases.
Over the past 2 decades much time has been devoted to developing biosensors to image RhoGTPase activation in living cells. The dominant approach, based on Förster resonance energy transfer (FRET), employs the property that a fluorescent donor molecule can transfer its energy to a nearby energy acceptor when they are in close proximity (∼10 Angstrom). When fluorescent donors and acceptors are used and the emission and excitation spectra of the 2 fluorophores overlap, FRET can be detected.Citation58 By introducing in vivo FRET probes comprising RhoGTPases, researchers can now study the activation of these small proteins with high spatio-temporal resolution.
Current RhoGTPase biosensors display similarities in that 4 specific domains are required. These domains consist of the GTPase of interest, a RhoGTPase-binding domain (RBD) derived from an effector protein and 2 fluorescent proteins, fused to the GTPase and to the RBD in a single chain or bimolecular format. Thus, these probes show where in cells GTPase activation occurs and therefore are essentially “GEF-activity sensors.” One of the first examples of a GTPase biosensor originates from a study of Kraynov et al. who used a bimolecular sensor to detect Rac1 activity in living cells.Citation59 In this study, the sensor consisted of Rac1 fused to a green fluorescent protein (GFP) and a portion of the Rac effector protein Pak1 labeled with Alexa-546 (). By using this approach, increased FRET was observed when the individual molecules came in close proximity, revealing Rac1 activation in the leading edge of migrating fibroblasts.
Figure 3. The structure of RhoGTPase FRET biosensors. Schematic representation of (A) an intermolecular FRET probe and (B and C) intramolecular FRET probes based on (B) Yoshizaki et al.Citation62 and (C) Pertz et al.Citation63 (D–G) The insertion of a fluorescent protein (FP) in the RhoA G domain may lead to mislocalization of the RhoA C-terminus and may impair binding of interaction partners such as RhoGDI. Homology model, calculated using the Phyre2 protein structure prediction server,Citation79 of RhoA-(1–180 aa)-FP-RhoA-C-terminus-(181–193 aa) alone (D) and in complex with RhoGDI (E) as well as of FP-RhoA-(1–193 aa) alone (F) and in complex with RhoGDI (G). Models are based on the structure of RhoA-GDP (PDB ID: 1FTN)Citation75 and GFP (PDB ID: 1H6R)Citation80 as top-scoring prediction events. Position of RhoGDI and GDP (from the complex Rac1-GDP-RhoGDI, PDB ID: 1HH4)Citation81 was obtained through the overlay of Rac1 and RhoA. GDP in yellow sticks; RhoA in green; FP in cyan, GDI in orange. Figures were prepared using PyMol.
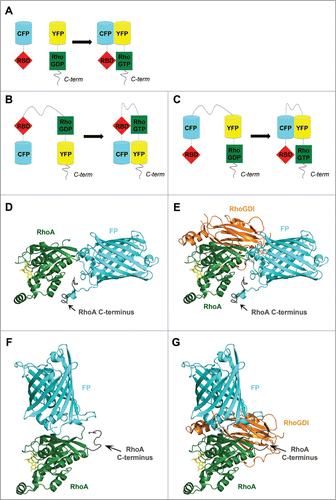
The major disadvantage of a bimolecular biosensor is potentially uneven expression of the 2 constructs. Therefore, Mochizuki et al. introduced the first intramolecular GTPase FRET probe: a Ras and interacting chimeric unit (Raichu-Ras).Citation60 In this approach, the 4 required FRET probe domains were fused to create a single GTPase FRET chain (). The RBD and the GTPase were here placed in the inner core of the molecule, flanked by a yellow-emitting mutant of GFP (YFP) and a cyan emitting mutant of GFP (CFP), respectively. In addition, the C-terminal region of Ras was coupled to the CFP, thereby constitutively targeting the construct to the plasma membrane. A similar strategy was used to make additional Raichu FRET probes for the RhoGTPases Rac1 and Cdc42.Citation61 However, the Ras C-terminal region of the original Raichu FRET probe induces enforced, constitutive membrane targeting of the sensor and prevents regulation by a RhoGDI, thus interfering with proper targeting of the GTPase. Therefore, this region was replaced with the corresponding C-terminal region of the sensor GTPase to create more representative sensors for RhoA, Cdc42 and Rac1.Citation62 In contrast to the design of these GTPase FRET probes, Pertz et al. introduced a RhoA biosensor where the 2 required fluorophores were placed at the central portion of the biosensor, thereby leaving full-length RhoA intact for binding to RhoGDIs and the plasma membrane ().Citation63 The localization of this sensor closely mimics that of endogenous RhoA and as a result, many of the current RhoGTPase biosensors rely on this ‘free C-terminus’ design. In addition to these cellular studies, structural modeling also shows that this is not trivial, since the hypervariable C-terminus may become mislocalized relative to the rest of the GTPase structure (i.e., the G-domain) when a FP is inserted ( compare to ). reflect predictions, based on structural modeling, because such fusion protein structure has not yet been resolved by crystallography or NMR. Since the GTPase C-terminus, in conjunction with more N-terminal regions, contributes to the binding to the GDI and to some of the GEFs, GAPs and effectors, also this region needs to be correctly positioned relative to the rest of the protein in FRET sensors ( compare to ). Finally, additional parameters can be fine-tuned in order to develop biosensors with an optimized FRET efficiency. For example, Fritz et al. used circularly permutated fluorescent protein variants and optimized linker lengths to create a high efficient RhoA biosensor.Citation64
RhoA regulates actin stress fiber formation and acto-myosin contraction at the rear of the cell. However, the use of biosensors has revealed a more complex relationship between the global function of RhoA, i.e. contraction, and its site of activation. Several studies not only observed localized RhoA activation at the contractile rear, but also at the front of randomly migrating cells.Citation63,65-67 More specifically, RhoA activation was detected during the protrusive phase of membrane ruffling where it requires simultaneous Cdc42 activation and where it antagonizes Rac1 activity. Detailed spatio-temporal analyses revealed that RhoA activation is mainly present within a region of 2 μm at the cell edge and highest Rac1 and Cdc42 activation was observed just behind this 2 μm boundary with a delay of 40 seconds.Citation67 Based on these findings, Machacek et al. suggested RhoA to be involved in initial protrusive events, while Rac1 and Cdc42 may be essential in the strengthening and stabilization of these protrusions.
Neither RhoB nor RhoC has been thoroughly studied using biosensors. Currently, there is one published example of a RhoB biosensor, consisting of a CFP-RhoB and YFP-mDia2 construct.Citation68 By expressing these constructs in a murine cell line, an interaction was observed between activated RhoB and mDia2 specifically on endosomes. Zawistowski et al. have recently characterized the first RhoC biosensorCitation69 and compared this FRET probe with an already established RhoA biosensor.Citation63 While comparable RhoA and RhoC activation patterns were observed at the cell edge, temporal differences were shown in the more distal region. Here, RhoC preceded RhoA activation during the formation of cell protrusions, suggestive for a distinct function of these GTPases. Earlier studies had shown that high RhoC activation was observed in the areas around invadapodial structures where it regulates cofilin activity.Citation70,71 This cofilin pathway is essential in the cancer-related processes of tumor cell invasion, migration and metastasis. By restricting cofilin activity to the inner core of invadopodia, RhoC is suggested to be a key player in invadopodia-mediated cancer cell invasion.
FRET imaging in 3 dimensions
Although major progress has been made regarding the use of RhoGTPase biosensors in live cell imaging, the ultimate goal is to use this novel technique in living organisms. The scattering of light in tissues complicates the applicability of this technique,Citation72 but there are already a few examples of RhoGTPase biosensor imaging in 3 dimensional (3D) structures. In a study of Ponik et al., a RhoA biosensor was expressed in breast epithelial cells embedded in a 3D collagen matrix.Citation73 These analyses showed active RhoA at cell-extracellular matrix adhesions, with an inactive RhoA pool at cell-cell contacts. Another example is provided by a study in glioblastoma, the most common and aggressive tumor type in the field of brain cancer.Citation74 Glioblastoma cells that stably expressed a FRET GTPase biosensor (either Rac1, Cdc42 or RhoA) were inoculated into rat brains and different activation patterns of these GTPases were subsequently observed using 2-photon imaging during tumor cell penetration of a specific brain area.
In summary, the introduction of FRET biosensors into the field of GTPase biology has provided us with sophisticated reagents to study the activation of these proteins with high spatio-temporal resolution. Over the past years the design of these FRET sensors has been further optimized, resulting in a biological applicable tool for broad use in live cell and tissue imaging.
Concluding Remarks
An emerging concept from these studies is that specific interactions of small RhoGTPases, inactive or active, with regulators and effectors are not mediated by a single, well-defined part of the protein (i.e., the effector domain). Rather, these interactions may involve several portions of the GTPase at the same time. This is true for interactions with the GDI, some of the GEFs, GAPs as well as a subset of the effectors. Moreover, the classical view of the hypervariable C-terminus driving membrane association through its lipid anchor and a polybasic region is most likely correct but at the same time incomplete. Although some interactions occur in the absence of the GTPase C-terminus, its hypervariable nature is exquisitely fit to contribute to signaling specificity. This portion of the protein acts in concert with the switch I and II regions and perhaps even with the insert region in allowing efficient and selective interactions with upstream regulators and downstream effectors. Thus, the specific outcome of RhoGTPase signaling is the sum of protein-protein and protein-lipid interactions, mediated by a several interacting regions that control selective binding to cellular membranes, localized GEFs and effectors.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
968004_Supplementary_Materials.zip
Download Zip (5.4 MB)Acknowledgments
We thank Dr. Mar Fernandez-Borja for critical reading of the manuscript.
Funding
This work was supported by the Landsteiner Foundation for Blood Transfusion Research (LSBR project no. 903) and a grant from the Netherlands Organization of Scientific Research to PLH (ZonMW MKMD project 40-42600-98-026).
References
- Madaule P, Axel R. A novel ras-related gene family. Cell 1985; 41:31-40; PMID:3888408; http://dx.doi.org/10.1016/0092-8674(85)90058-3
- Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci 2005; 118:843-6; PMID:15731001; http://dx.doi.org/10.1242/jcs.01660
- Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev 2013; 93:269-309; PMID:23303910; http://dx.doi.org/10.1152/physrev.00003.2012
- Ridley AJ. RhoA, RhoB and RhoC have different roles in cancer cell migration. J Microsc 2013; 251:242-9; PMID:23488932; http://dx.doi.org/10.1111/jmi.12025
- Ridley AJ. RhoGTP ases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol 2006; 16:522-9; PMID:16949823; http://dx.doi.org/10.1016/j.tcb.2006.08.006
- Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol 2005; 21:247-69; PMID: 16212495; http://dx.doi.org/10.1146/annurev.cellbio.21.020604.150721
- Kroon J, Tol S, van AS, Elias JA, Fernandez-Borja M. The small GTPase RhoB regulates TNFalpha signaling in endothelial cells. PLoS One 2013; 8:e75031; PMID:24086429; http://dx.doi.org/10.1371/journal.pone.0075031
- Rosenthal DT, Zhang J, Bao L, Zhu L, Wu Z, Toy K, Kleer CG, Merajver SD. RhoC impacts the metastatic potential and abundance of breast cancer stem cells. PLoS One 2012; 7:e40979; PMID:22911725; http://dx.doi.org/10.1371/journal.pone.0040979
- Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 2007; 449:682-8; PMID:17898713; http://dx.doi.org/10.1038/nature06174
- Pertz O. Spatio-temporal Rho GTPase signaling - where are we now? J Cell Sci 2010; 123:1841-50; PMID:20484664; http://dx.doi.org/10.1242/jcs.064345
- Wittinghofer A, Vetter IR. Structure-function relationships of the G domain, a canonical switch motif. Annu Rev Biochem 2011; 80:943-71; PMID:21675921; http://dx.doi.org/10.1146/annurev-biochem-062708-134043
- Garcia-Mata R, Boulter E, Burridge K. The ‘invisible hand’: regulation of RHO GTPases by RHOGDIs. Nat Rev Mol Cell Biol 2011; 12:493-504; PMID:21779026; http://dx.doi.org/10.1038/nrm3153
- Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science 2001; 294:1299-304; PMID:11701921; http://dx.doi.org/10.1126/science.1062023
- Thomas C, Fricke I, Scrima A, Berken A, Wittinghofer A. Structural evidence for a common intermediate in small G protein-GEF reactions. Mol. Cell 2007; 25:141-9; PMID:17218277
- Nisimoto Y, Freeman JL, Motalebi SA, Hirshberg M, Lambeth JD. Rac binding to p67(phox). Structural basis for interactions of the Rac1 effector region and insert region with components of the respiratory burst oxidase. J Biol Chem 1997; 272:18834-41; PMID:9228059; http://dx.doi.org/10.1074/jbc.272.30.18834
- McCallum SJ, Wu WJ, Cerione RA. Identification of a putative effector for Cdc42Hs with high sequence similarity to the RasGAP-related protein IQGAP1 and a Cdc42Hs binding partner with similarity to IQGAP2. J Biol Chem 1996; 271:21732-7; PMID:8702968; http://dx.doi.org/10.1074/jbc.271.45.28168
- Zong H, Kaibuchi K, Quilliam LA. The insert region of RhoA is essential for Rho kinase activation and cellular transformation. Mol Cell Biol 2001; 21:5287-98; PMID:11463812; http://dx.doi.org/10.1128/MCB.21.16.5287-5298.2001
- Lammers M, Meyer S, Kuhlmann D, Wittinghofer A. Specificity of interactions between mDia isoforms and Rho proteins. J Biol Chem 2008; 283:35236-46; PMID:18829452; http://dx.doi.org/10.1074/jbc.M805634200
- ten Klooster JP, Hordijk PL. Targeting and localized signalling by small GTPases. Biol Cell 2007; 99:1-12; PMID:17155934
- Scheffzek K, Stephan I, Jensen ON, Illenberger D, Gierschik P. The Rac-RhoGDI complex and the structural basis for the regulation of Rho proteins by RhoGDI. Nat Struct Biol 2000; 7:122-6; PMID:10655614; http://dx.doi.org/10.1038/72392
- Longenecker K, Read P, Derewenda U, Dauter Z, Liu X, Garrard S, Walker L, Somlyo AV, Nakamoto RK, Somlyo AP, et al. How RhoGDI binds Rho. Acta Crystallogr D Biol Crystallogr 1999; 55:1503-15; PMID:10489445; http://dx.doi.org/10.1107/S090744499900801X
- Hoffman GR, Nassar N, Cerione RA. Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell 2000; 100:345-56; PMID:10676816; http://dx.doi.org/10.1016/S0092-8674(00)80670-4
- Zalcman G, Closson V, Camonis J, Honore N, Rousseau-Merck MF, Tavitian A, Olofsson B. RhoGDI-3 is a new GDP dissociation inhibitor (GDI). Identification of a non-cytosolic GDI protein interacting with the small GTP-binding proteins RhoB and RhoG. J Biol Chem 1996; 271:30366-74; PMID:8939998; http://dx.doi.org/10.1074/jbc.271.48.30366
- Snyder JT, Worthylake DK, Rossman KL, Betts L, Pruitt WM, Siderovski DP, Der CJ, Sondek J. Structural basis for the selective activation of Rho GTPases by Dbl exchange factors. Nat Struct Biol 2002; 9:468-75; PMID:12006984; http://dx.doi.org/10.1038/nsb796
- Jaiswal M, Gremer L, Dvorsky R, Haeusler LC, Cirstea IC, Uhlenbrock K, Ahmadian MR. Mechanistic insights into specificity, activity, and regulatory elements of the regulator of G-protein signaling (RGS)-containing Rho-specific guanine nucleotide exchange factors (GEFs) p115, PDZ-RhoGEF (PRG), and leukemia-associated RhoGEF (LARG). J Biol Chem 2011; 286:18202-12; PMID:21454492; http://dx.doi.org/10.1074/jbc.M111.226431
- Kristelly R, Gao G, Tesmer JJ. Structural determinants of RhoA binding and nucleotide exchange in leukemia-associated Rho guanine-nucleotide exchange factor. J Biol Chem 2004; 279:47352-62; PMID:15331592; http://dx.doi.org/10.1074/jbc.M406056200
- Solski PA, Wilder RS, Rossman KL, Sondek J, Cox AD, Campbell SL, Der CJ. Requirement for C-terminal sequences in regulation of Ect2 guanine nucleotide exchange specificity and transformation. J Biol Chem 2004; 279:25226-33; PMID:15073184; http://dx.doi.org/10.1074/jbc.M313792200
- Srougi MC, Burridge K. The nuclear guanine nucleotide exchange factors Ect2 and Net1 regulate RhoB-mediated cell death after DNA damage. PLoS One 2011; 6:e17108; PMID:21373644; http://dx.doi.org/10.1371/journal.pone.0017108
- Arthur WT, Ellerbroek SM, Der CJ, Burridge K, Wennerberg K. XPLN, a guanine nucleotide exchange factor for RhoA and RhoB, but not RhoC. J Biol Chem 2002; 277:42964-72; PMID:12221096; http://dx.doi.org/10.1074/jbc.M207401200
- Sloan CM, Quinn CV, Peters JP, Farley J, Goetzinger C, Wernli M, DeMali KA, Ellerbroek SM. Divergence of Rho residue 43 impacts GEF activity. Small GTPases 2012; 3:15-22; PMID:22673745; http://dx.doi.org/10.4161/sgtp.19557
- Hamel B, Monaghan-Benson E, Rojas RJ, Temple BR, Marston DJ, Burridge K, Sondek J. SmgGDS is a guanine nucleotide exchange factor that specifically activates RhoA and RhoC. J Biol Chem 2011; 286:12141-8; PMID:21242305; http://dx.doi.org/10.1074/jbc.M110.191122
- ten Klooster JP, Leeuwen I, Scheres N, Anthony EC, Hordijk PL. Rac1-induced cell migration requires membrane recruitment of the nuclear oncogene SET. EMBO J 2007; 26:336-45; PMID:17245428; http://dx.doi.org/10.1038/sj.emboj.7601518
- Dvorsky R, Ahmadian MR. Always look on the bright site of Rho: structural implications for a conserved intermolecular interface. EMBO Rep 2004; 5:1130-6; PMID:15577926; http://dx.doi.org/10.1038/sj.embor.7400293
- Ridley AJ, Self AJ, Kasmi F, Paterson HF, Hall A, Marshall CJ, Ellis C. rho family GTPase activating proteins p190, bcr and rhoGAP show distinct specificities in vitro and in vivo. EMBO J 1993; 12:5151-60; PMID:8262058
- Thomas S, Overdevest JB, Nitz MD, Williams PD, Owens CR, Sanchez-Carbayo M, Frierson HF, Schwartz MA, Theodorescu D. Src and caveolin-1 reciprocally regulate metastasis via a common downstream signaling pathway in bladder cancer. Cancer Res 2011; 71:832-41; PMID:21148751; http://dx.doi.org/10.1158/0008-5472.CAN-10-0730
- Wang L, Yang L, Luo Y, Zheng Y. A novel strategy for specifically down-regulating individual Rho GTPase activity in tumor cells. J Biol Chem 2003; 278:44617-25; PMID:12939257; http://dx.doi.org/10.1074/jbc.M308929200
- Lazarini M, Traina F, Machado-Neto JA, Barcellos KS, Moreira YB, Brandao MM, Verjovski-Almeida S, Ridley AJ, Saad ST. ARHGAP21 is a RhoGAP for RhoA and RhoC with a role in proliferation and migration of prostate adenocarcinoma cells. Biochim Biophys Acta 2013; 1832:365-74; PMID:23200924
- Dubois T, Paleotti O, Mironov AA, Fraisier V, Stradal TE, De Matteis MA, Franco M, Chavrier P. Golgi-localized GAP for Cdc42 functions downstream of ARF1 to control Arp23 complex and F-actin dynamics. Nat Cell Biol 2005; 7:353-64; PMID:15793564; http://dx.doi.org/10.1038/ncb1244
- Diekmann D, Nobes CD, Burbelo PD, Abo A, Hall A. Rac GTPase interacts with GAPs and target proteins through multiple effector sites. EMBO J 1995; 14:5297-305; PMID:7489719
- Bishop AL, Hall A. Rho GTPases and their effector proteins. Biochem J 2000; 348(Pt 2):241-55; PMID:10816416; http://dx.doi.org/10.1042/0264-6021:3480241
- Hemsath L, Dvorsky R, Fiegen D, Carlier MF, Ahmadian MR. An electrostatic steering mechanism of Cdc42 recognition by Wiskott-Aldrich syndrome proteins. Mol. Cell 2005; 20:313-24; PMID:16246732
- Toporik A, Gorzalczany Y, Hirshberg M, Pick E, Lotan O. Mutational analysis of novel effector domains in Rac1 involved in the activation of nicotinamide adenine dinucleotide phosphate (reduced) oxidase. Biochemistry 1998; 37:7147-56; PMID:9585526; http://dx.doi.org/10.1021/bi9800404
- Joseph G, Gorzalczany Y, Koshkin V, Pick E. Inhibition of NADPH oxidase activation by synthetic peptides mapping within the carboxyl-terminal domain of small GTP-binding proteins. Lack of amino acid sequence specificity and importance of polybasic motif. J Biol Chem 1994; 269:29024-31; PMID:7961867
- Modha R, Campbell LJ, Nietlispach D, Buhecha HR, Owen D, Mott HR. The Rac1 polybasic region is required for interaction with its effector PRK1. J Biol Chem 2008; 283:1492-500; PMID:18006505; http://dx.doi.org/10.1074/jbc.M706760200
- Hutchinson CL, Lowe PN, McLaughlin SH, Mott HR, Owen D. Differential binding of RhoA, RhoB, and RhoC to protein kinase C-related kinase (PRK) isoforms PRK1, PRK2, and PRK3: PRKs have the highest affinity for RhoB. Biochemistry 2013; 52:7999-8011; PMID:24128008; http://dx.doi.org/10.1021/bi401216w
- Unsal-Kacmaz K, Ragunathan S, Rosfjord E, Dann S, Upeslacis E, Grillo M, Hernandez R, Mack F, Klippel A. The interaction of PKN3 with RhoC promotes malignant growth. Mol Oncol 2012; 6:284-98; PMID:22217540; http://dx.doi.org/10.1016/j.molonc.2011.12.001
- Rose R, Weyand M, Lammers M, Ishizaki T, Ahmadian MR, Wittinghofer A. Structural and mechanistic insights into the interaction between Rho and mammalian Dia. Nature 2005; 435:513-8; PMID:15864301; http://dx.doi.org/10.1038/nature03604
- Watanabe N, Kato T, Fujita A, Ishizaki T, Narumiya S. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat Cell Biol 1999; 1:136-43; PMID:10559899; http://dx.doi.org/10.1038/11056
- Fernandez-Borja M, Janssen L, Verwoerd D, Hordijk P, Neefjes J. RhoB regulates endosome transport by promoting actin assembly on endosomal membranes through Dia1. J Cell Sci 2005; 118:2661-70; PMID:15944396; http://dx.doi.org/10.1242/jcs.02384
- Kitzing TM, Wang Y, Pertz O, Copeland JW, Grosse R. Formin-like 2 drives amoeboid invasive cell motility downstream of RhoC. Oncogene 2010; 29:2441-8; PMID:20101212; http://dx.doi.org/10.1038/onc.2009.515
- Vega FM, Fruhwirth G, Ng T, Ridley AJ. RhoA and RhoC have distinct roles in migration and invasion by acting through different targets. J Cell Biol 2011; 193:655-65; PMID:21576392; http://dx.doi.org/10.1083/jcb.201011038
- Rolli-Derkinderen M, Sauzeau V, Boyer L, Lemichez E, Baron C, Henrion D, Loirand G, Pacaud P. Phosphorylation of serine 188 protects RhoA from ubiquitinproteasome-mediated degradation in vascular smooth muscle cells. Circ Res 2005; 96:1152-60; PMID:15890975; http://dx.doi.org/10.1161/01.RES.0000170084.88780.ea
- Nusser N, Gosmanova E, Makarova N, Fujiwara Y, Yang L, Guo F, Luo Y, Zheng Y, Tigyi G. Serine phosphorylation differentially affects RhoA binding to effectors: implications to NGF-induced neurite outgrowth. Cell Signal 2006; 18:704-14; PMID:16109481; http://dx.doi.org/10.1016/j.cellsig.2005.06.010
- Lehman HL, Van Laere SJ, van Golen CM, Vermeulen PB, Dirix LY, van Golen KL. Regulation of inflammatory breast cancer cell invasion through Akt1PKBalpha phosphorylation of RhoC GTPase. Mol Cancer Res 2012; 10:1306-18; PMID:22896661; http://dx.doi.org/10.1158/1541-7786.MCR-12-0173
- Ozdamar B, Bose R, Barrios-Rodiles M, Wang HR, Zhang Y, Wrana JL. Regulation of the polarity protein Par6 by TGFbeta receptors controls epithelial cell plasticity. Science 2005; 307:1603-9; PMID:15761148; http://dx.doi.org/10.1126/science.1105718
- Wei J, Mialki RK, Dong S, Khoo A, Mallampalli RK, Zhao Y, Zhao J. A new mechanism of RhoA ubiquitination and degradation: roles of SCF(FBXL19) E3 ligase and Erk2. Biochim Biophys Acta 2013; 1833:2757-64; PMID:23871831
- Clague MJ, Liu H, Urbe S. Governance of endocytic trafficking and signaling by reversible ubiquitylation. Dev Cell 2012; 23:457-67; PMID:22975321
- Tsien RY, Miyawaki A. Seeing the machinery of live cells. Science 1998; 280:1954-5; PMID:9669950; http://dx.doi.org/10.1126/science.280.5371.1954
- Kraynov VS, Chamberlain C, Bokoch GM, Schwartz MA, Slabaugh S, Hahn KM. Localized Rac activation dynamics visualized in living cells. Science 2000; 290:333-7; PMID:11030651; http://dx.doi.org/10.1126/science.290.5490.333
- Mochizuki N, Yamashita S, Kurokawa K, Ohba Y, Nagai T, Miyawaki A, Matsuda M. Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature 2001; 411:1065-68; PMID:11429608; http://dx.doi.org/10.1038/35082594
- Itoh RE, Kurokawa K, Ohba Y, Yoshizaki H, Mochizuki N, Matsuda M. Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol Cell Biol 2002; 22:6582-91; PMID:12192056; http://dx.doi.org/10.1128/MCB.22.18.6582-6591.2002
- Yoshizaki H, Ohba Y, Kurokawa K, Itoh RE, Nakamura T, Mochizuki N, Nagashima K, Matsuda M. Activity of Rho-family GTPases during cell division as visualized with FRET-based probes. J Cell Biol 2003; 162:223-32; PMID:12860967; http://dx.doi.org/10.1083/jcb.200212049
- Pertz O, Hodgson L, Klemke RL, Hahn KM. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature 2006; 440:1069-72; PMID:16547516; http://dx.doi.org/10.1038/nature04665
- Fritz RD, Letzelter M, Reimann A, Martin K, Fusco L, Ritsma L, Ponsioen B, Fluri E, Schulte-Merker S, van Rheenen J, et al. A versatile toolkit to produce sensitive FRET biosensors to visualize signaling in time and space. Sci Signal 2013; 6:rs12; PMID:23882122; http://dx.doi.org/10.1126/scisignal.2004135
- El-Sibai M, Pertz O, Pang H, Yip SC, Lorenz M, Symons M, Condeelis JS, Hahn KM, Backer JM. RhoAROCK-mediated switching between Cdc42- and Rac1-dependent protrusion in MTLn3 carcinoma cells. Exp Cell Res 2008; 314:1540-52; PMID:18316075; http://dx.doi.org/10.1016/j.yexcr.2008.01.016
- Kurokawa K, Matsuda M. Localized RhoA activation as a requirement for the induction of membrane ruffling. Mol Biol Cell 2005; 16:4294-303; PMID:15987744
- Machacek M, Hodgson L, Welch C, Elliott H, Pertz O, Nalbant P, Abell A, Johnson GL, Hahn KM, Danuser G. Coordination of Rho GTPase activities during cell protrusion. Nature 2009; 461:99-103; PMID:19693013; http://dx.doi.org/10.1038/nature08242
- Wallar BJ, Deward AD, Resau JH, Alberts AS. RhoB and the mammalian Diaphanous-related formin mDia2 in endosome trafficking. Exp Cell Res 2007; 313:560-71; PMID:17198702; http://dx.doi.org/10.1016/j.yexcr.2006.10.033
- Zawistowski JS, Sabouri-Ghomi M, Danuser G, Hahn KM, Hodgson L. A RhoC biosensor reveals differences in the activation kinetics of RhoA and RhoC in migrating cells. PLoS One 2013; 8:e79877; PMID:24224016; http://dx.doi.org/10.1371/journal.pone.0079877
- Bravo-Cordero JJ, Oser M, Chen X, Eddy R, Hodgson L, Condeelis J. A novel spatiotemporal RhoC activation pathway locally regulates cofilin activity at invadopodia. Curr Biol 2011; 21:635-44; PMID:21474314; http://dx.doi.org/10.1016/j.cub.2011.03.039
- Bravo-Cordero JJ, Sharma VP, Roh-Johnson M, Chen X, Eddy R, Condeelis J, Hodgson L. Spatial regulation of RhoC activity defines protrusion formation in migrating cells. J Cell Sci 2013; 126:3356-69; PMID:23704350; http://dx.doi.org/10.1242/jcs.123547
- Aoki K, Kiyokawa E, Nakamura T, Matsuda M. Visualization of growth signal transduction cascades in living cells with genetically encoded probes based on Forster resonance energy transfer. Philos Trans R Soc Lond B Biol Sci 2008; 363:2143-51; PMID:18343776; http://dx.doi.org/10.1098/rstb.2008.2267
- Ponik SM, Trier SM, Wozniak MA, Eliceiri KW, Keely PJ. RhoA is down-regulated at cell-cell contacts via p190RhoGAP-B in response to tensional homeostasis. Mol Biol Cell 2013; 24:1688-3; PMID:23552690
- Hirata E, Yukinaga H, Kamioka Y, Arakawa Y, Miyamoto S, Okada T, Sahai E, Matsuda M. In vivo fluorescence resonance energy transfer imaging reveals differential activation of Rho-family GTPases in glioblastoma cell invasion. J Cell Sci 2012; 125:858-68; PMID:22399802; http://dx.doi.org/10.1242/jcs.089995
- Wei Y, Zhang Y, Derewenda U, Liu X, Minor W, Nakamoto RK, Somlyo AV, Somlyo AP, Derewenda ZS. Crystal structure of RhoA-GDP and its functional implications. Nat Struct Biol 1997; 4:699-703; PMID:9302995; http://dx.doi.org/10.1038/nsb0997-699
- Soundararajan M, Turnbull A, Fedorov O, Johansson C, Doyle DA. RhoB can adopt a Mg2+ free conformation prior to GEF binding. Proteins 2008; 72:498-505; PMID:18393397; http://dx.doi.org/10.1002/prot.22017
- Dias SM, Cerione RA. X-ray crystal structures reveal two activated states for RhoC. Biochemistry 2007; 46:6547-58; PMID:17497936; http://dx.doi.org/10.1021/bi700035p
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A 2001; 98:10037-41; PMID:11517324
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 2009; 4:363-71; PMID:19247286; http://dx.doi.org/10.1038/nprot.2009.2
- Ostergaard H, Henriksen A, Hansen FG, Winther JR. Shedding light on disulfide bond formation: engineering a redox switch in green fluorescent protein. EMBO J 2001; 20:5853-62; PMID:11689426; http://dx.doi.org/10.1093/emboj/20.21.5853
- Grizot S, Faure J, Fieschi F, Vignais PV, Dagher MC, Pebay-Peyroula E. Crystal structure of the Rac1-RhoGDI complex involved in nadph oxidase activation. Biochemistry 2001; 40:10007-13; PMID:11513578; http://dx.doi.org/10.1021/bi010288k
