Abstract
Translation initiation in eukaryotes requires the involvement of multiple initiation factors (eIFs) that facilitate the binding of the 40 S ribosomal subunit to an mRNA and assemble the 80 S ribosome at the correct initiation codon. eIF4F, composed of eIF4E, eIF4A, and eIF4G, binds to the 5′-cap structure of an mRNA and prepares an mRNA for recruitment of a 40 S subunit. eIF4B promotes the ATP-dependent RNA helicase activity of eIF4A and eIF4F needed to unwind secondary structure present in a 5′-leader that would otherwise impede scanning of the 40 S subunit during initiation. The poly(A) binding protein (PABP), which binds the poly(A) tail, interacts with eIF4G and eIF4B to promote circularization of an mRNA and stimulates translation by promoting 40 S subunit recruitment. Thus, these factors serve essential functions in the early steps of protein synthesis. Their assembly and function requires multiple interactions that are competitive in nature and determine the nature of interactions between the termini of an mRNA. In this review, the domain organization and partner protein interactions are presented for the factors in plants which share similarities with those in animals and yeast but differ in several important respects. The functional consequences of their interactions on factor activity are also discussed.
Introduction
Protein synthesis involves 3 phases: initiation, elongation, and termination. Because the 18 S rRNA of the eukaryotic 40 S ribosomal subunit lacks the region corresponding to the Shine-Dalgarno sequence of the bacterial 16 S rRNA that enables the latter to bind near an initiation codon in a prokaryotic mRNA, the recruitment of the 40 S subunit depends on the action of multiple initiation factors. These factors facilitate 40 S subunit binding at the 5′-cap structure (m7GpppN) of an mRNA and assist in the 5′ to 3′ directional scanning of the 5′-leader to identify the correct initiation codon. Recruitment of the 40 S subunit requires the functions of the cap-binding complex, a group of initiation factors that include eIF4E, which binds to the 5′-cap structure; eIF4A, a DEAD/H-box, ATP-dependent, RNA helicase that uses energy from ATP hydrolysis to unwind secondary structure in the 5′-leader that could inhibit 40 S subunit scanning; and eIF4G, a modular scaffolding protein.Citation1-4 Together, these 3 factors are known as eIF4F. eIF4G stimulates the ATPase activity of eIF4ACitation5 as does eIF4B which is also associated the cap-binding complex.Citation6 Other factors associated with the cap-binding complex include the poly(A) binding protein (PABP), which is bound to the poly(A) tail, and eIF3, which through its interaction with eIF4G and the 40 S subunit, recruits the latter to the mRNA. Thus, the interactions among these factors are essential for their assembly at the 5′-cap and for the translation of most cellular mRNAs. In this review, the interactions among eIF4E, eIF4G, eIF4A, eIF4B, and PABP and the functional consequences of their interactions in plants are examined in detail and are compared with those in animals and yeast.
The Scaffolding Protein eIF4G Organizes the Assembly of the Cap-Binding Factors
Plants express a novel eIF4G isoform not found in other eukaryotes
The scaffolding function of eIF4G is central to the cap-binding complex in that, through its interactions with most of the other components, it maintains the integrity of the complex. eIF4G is the largest subunit of eIF4F and 2 similar isoforms are expressed in most eukaryotes. For example, in animals and yeast, the 2 eIF4G proteins are expressed from distinct genes but are similar in mass and sequenceCitation7,8 whereas the 2 isoforms in plants, referred to as eIF4G and eIFiso4G, differ substantially in mass and sequence.Citation9 eIFiso4G in wheat is only 30% identical to eIF4G from the same speciesCitation9 compared with 53% identity between the 2 isoforms in yeast and 46% identity between the 2 isoforms in humans.Citation7,8 At 86 kDa, eIFiso4G is substantially smaller than eIF4G in plants or other eukaryotes but, like plant eIF4G, it contains 2 HEAT domains that are composed of antiparallel α-helical hairpins known as HEAT repeatsCitation10,11 and these account for most of the shared similarity between the 2 plant isoforms (). The eIF4E interaction domain lies N-proximal to the first of these HEAT domains and its position as well as its sequence is conserved among animal, plant, and yeast eIF4G proteins. Together, eIF4E and eIF4G comprise plant eIF4F while eIFiso4G and eIFiso4E (the isoform of eIF4E in plants) comprise eIFiso4F.
Figure 1. Partner protein interactions of eIF4G in higher eukaryotes. (A) The RNA and protein binding domains in wheat eIF4G (TaeIF4G), eIFiso4G (TaeIFiso4G), human eIF4G (HseIF4G), and yeast eIF4G (SceIF4G) are shown. Interaction domains for partner proteins are indicated by color with a key included. Domains for yeast eIF4G are defined as described previously.Citation37 (B) Below the N-terminal PABP binding site of wheat eIF4G is a sequence logo for the region corresponding to the PABP interaction site in human eIF4G. For this analysis, a sequence alignment from 6 monocot and 7 dicot eIF4G homologs was generated using EBI MUSCLE. The aligned sequences were submitted to WebLogo Berkeley to generate a sequence logo, in which the most conserved one, 2, or sometimes 3 residues at each position are shown as the consensus. Below the protein sequence logo are the sequences of the PABP interaction sites for human eIF4G1 and eIF4G2 as well as the corresponding sequence for wheat eIF4G. In each case, residues conserved with the consensus sequence are colored as in the sequence logo. The logo was constructed from eIF4G homologs from Triticum aestivum (wheat, JN091779), Brachypodium distachyon (Bd1g25002), Oryza sativa (Os07 g36940), Setaria italica (Si028648), Panicum virgatum (Pv00019592 and Pv00063738), Phaseolus vulgaris (010G043700), Ricinus communis (29709), Brassica rapa Chiifu-401 (Bra014505), Thellungiella halophila (Thhalv10005736), Capsella rubella (Cr10016570), Arabidopsis thaliana (AEE80028), and Arabidopsis lyrata (939058).
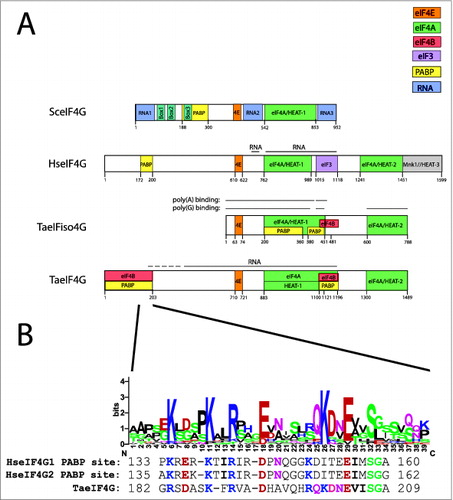
Although all eIF4G proteins contain one or more HEAT domains, they differ in number. Animal eIF4G proteins contain 3 HEAT domains, yeast eIF4G proteins contain one, while, as mentioned, plant eIF4G and eIFiso4G contain 2. The HEAT-1l/MIF4G domain, which contains 5 HEAT repeats in a helical stack, is present in animal, plant, and yeast eIF4GCitation10 whereas the HEAT-2/MA3 domain, also composed of 5 HEAT repeats,Citation11 is found in animal and plant eIF4G.Citation12,13 The HEAT-3 domain, composed of 3 and one half HEAT repeats, is present only in animal eIF4G and functions to bind Mnk eIF4E kinases 1/2.Citation11
eIF4A binds the first 2 HEAT domains of animal eIF4G as well as the region between these domains but each HEAT domain contacts separate surfaces of eIF4A in order to contribute to its stable binding.Citation14 Binding to HEAT-1 promotes eIF4A helicase activity while HEAT-2 serves a modulatory role.Citation6,Citation15-20 The binding of eIF4A to the HEAT domains positions the 2 eIF4A domains containing the ATP- and RNA-binding sites in a partially closed state that facilitates interaction with substrates and the release of reaction products.Citation19,21,22 Binding of eIF4E to human eIF4G also stimulates eIF4A helicase activity by overcoming the autoinhibitory function of the eIF4E-binding site in eIF4G and the ability of eIF4E to do so is independent of its cap-binding function.Citation23 As yeast eIF4G lacks the HEAT-2 domain, eIF4A must bind through the HEAT-1 domain alone which eIF4B may facilitate.Citation24 As the HEAT-1 and HEAT-2 domains of plant eIF4G and eIFiso4G bind eIF4A, they are more similar to the animal homologs than to yeast.Citation12,13 However, like yeast eIF4G, plant eIF4G and eIFiso4G lack the HEAT-3 domain.
Although eIF4A interacts with the eIFiso4G HEAT-1 domain, its binding is abolished if the N-terminal 30–40 residues of HEAT-1 are deleted.Citation12 Moreover, the N-terminal region of the HEAT-1 domain is sufficient to support interaction with eIF4A, suggesting that this region is essential for interaction with eIF4A while the remainder of the HEAT-1 domain serves to stabilize eIF4A binding.Citation12 A 7 amino acid sequence, i.e., ILNKLTP, within this region is important for eIF4A binding as mutations within the sequence abolish eIF4A binding and the asparagine, lysine, and threonine residues in this highly conserved element make direct contacts with specific residues in eIF4A.Citation16,19,25 Although eIF4A binds the HEAT-1 and HEAT-2 domains of plant eIFiso4G and eIF4G, they differ in that the eIFiso4G HEAT-1 domain is sufficient to bind eIF4A whereas a short region immediately C-proximal to the HEAT-1 domain of eIF4G is needed in addition to the HEAT-1 domain,Citation12,13 consistent with observations made with animal eIF4G in which the HEAT-1 domain alone bound eIF4A with lower affinity than did a longer region of eIF4G.Citation26
In animal eIF4G, eIF3 binds to a 90 amino acid region just C-proximal to the HEAT-1 domain through its c, d, and e subunitsCitation27,28 which was reported to bind in a cooperative manner with eIF4ACitation29 although this was not confirmed in a subsequent study.Citation28 Whether eIF3 binds to the corresponding region in plant eIF4G or eIFiso4G remains to be determined.
eIF1 and eIF5 bind yeast eIF4G within an arginine and serine rich domain that lies just N-proximal to the HEAT-1 domain and their interaction with eIF4G is competitive.Citation30,31 eIF1 interacts with eIF3 and is involved in AUG selection whereas eIF5 interacts with eIF2 and eIF3 and promotes their binding to a 40 S subunit as well as their function.Citation32 Whether these factors affect eIF4A binding to the HEAT-1 domain of eIF4G is unknown and whether these factors interact directly with plant and animal eIF4G remains to be determined.
eIF4G and eIFiso4G bind PABP and eIF4B in distinct ways
An interaction domain for PABP lies N-proximal to the eIF4E interaction domain in eIF4G and the position of this interaction domain is conserved in animals, plants, and yeast ().Citation13,17,Citation33-36 The N-terminal PABP interaction domain in wheat eIF4G is contained within the first 200 amino acids.Citation13 In human eIF4G1 and eIF4GII, the 2 isoforms present in this species, the PABP interaction domain lies within a discrete 28–29 amino acid region in the N-terminal portion of each protein.Citation17 To identify the positions that might be conserved among plant eIF4G orthologs in the corresponding region, a graphical logo showing the conservation of sequence at each residue was generated from a sequence alignment of the corresponding region from eIF4G orthologs of 6 monocot and 7 dicot species (). Several residues were observed to be nearly invariant throughout the region including several basic and acidic residues. Comparing the PABP interaction domain of human eIF4G1 and eIF4GII as well as the corresponding region from wheat eIF4G shows that these exhibit comparable similarity to the consensus sequence (). Deletion of this region in wheat eIF4G results in loss of PABP binding,Citation13 suggesting that this region exhibiting conservation with animal eIF4G homologs is important for interaction with PABP. Although specific residues within this region required for PABP binding have not been identified for either animal or plant eIF4G, the conservation suggests that some of these residues may be functionally important.
The presence of an N-terminal RNA-binding domain (referred to as RNA1) in yeast eIF4G1 can functionally substitute for the adjacent PABP-binding domain as can a region between RNA1 and the PABP-binding domain which contains 2 elements, Box1 and Box2 ().Citation37 RNA1 and Box1 assist in the direct binding of PABP to eIF4G1 while RNA1, Box1, Box2, and the PABP-binding domain all contribute to the formation of the eIF4G:PABP:mRNA complex.Citation37 These data suggest that the interaction of PABP with eIF4G is only one of several interactions that stabilizes eIF4G binding to an mRNA, at least in yeast. These findings support the earlier observation that the interaction between eIF4G and PABP in yeast is RNA-dependentCitation33 as the RNA-binding domain RNA1 assists the interaction between PABP and eIF4G.Citation37 Whether functionally analogous sequence elements to RNA1, Box1, and Box2 identified in yeast eIF4G1 are present in plant or animal eIF4G is unknown although no obvious sequences exhibiting conservation with these yeast elements are apparent.
Although only one PABP interaction domain has been identified in animal and yeast eIF4G proteins, a second binding domain is present in plant eIF4G which lies C-proximal to its HEAT-1 domain ()Citation13 which represents a significant difference among these homologs. The observation that the interaction between eIF4G and PABP is RNA-dependent in yeastCitation33 but not in plantsCitation13,34,38 might indicate that the PABP-eIF4G interaction mediated by 2 domains obviates the need for RNA to stabilize the interaction. That RNA is not required for PABP binding to eIF4G in animalsCitation17 as well as in plants suggests that the interaction between eIF4G and PABP in higher eukaryotes may differ fundamentally from that in yeast. However, whether a second PABP binding site is present in animal eIF4G is a question worth re-examining in light of the findings with plant eIF4G.
eIFiso4G differs from eIF4G in plants in that it lacks most of the region that lies N-terminal of the HEAT-1 domain (). Therefore, the eIFiso4E interaction site lies closer to the eIFiso4G N-terminus than does the eIF4E interaction site in eIF4G and the N-terminal PABP interaction domain present in eIF4G is absent in eIFiso4G. Indeed, as eIFiso4G lacks any sequence analogous to the N-terminal region of eIF4G of plant, animal, or yeast eIF4G proteins, there is no possibility of even functionally analogous sequences for the RNA1, Box1, and Box2 elements of yeast eIF4G1.Citation37 Instead, PABP binds eIFiso4G within the HEAT-1 domain at 2 contiguous sites that overlap extensively with the eIF4A binding site in this same domain ().Citation12 Because the 2 PABP binding sites within the HEAT-1 domain are contiguous, they may be considered as 2 subregions composing a single PABP interaction domain. As the PABP binding site in HEAT-1 lies C-proximal to the eIF4E binding site, eIFiso4G differs substantially from animal and yeast eIF4G where PABP binds to the N-proximal side of the eIF4E binding domain. eIFiso4G differs also from plant eIF4G in that it effectively has one PABP interaction domain in contrast to the 2 present in plant eIF4G ().Citation12
Despite its role in promoting eIF4A function, there has been no report of a direct interaction between eIF4B and eIF4G in animals or yeast although eIF4B is associated with the cap-binding complex.Citation24,26 In contrast, an eIF4B interaction domain was identified in the N-terminal region of plant eIF4G which maps to the same region responsible for binding PABP ().Citation13 Similarly a second eIF4B binding site lies C-proximal to the HEAT-1 domain of plant eIF4G that extensively overlaps with the PABP binding site in this same region ().Citation13 Interestingly, eIF4H, a close homolog of eIF4B, partially displaces eIF4A from animal eIF4G that is bound to a region corresponding to the eIF4B binding site in plant eIF4G.Citation14 As with PABP, eIF4B also binds eIFiso4G at a single site but it differs from PABP in that its binding domain overlaps just the C-terminal end of the HEAT-1 domain ().Citation12 The eIF4B binding domain also overlaps the C-terminal end of the PABP interaction domain in eIFiso4G.
eIF4B and PABP compete for binding plant eIF4G and eIFiso4G
Because eIF4B and PABP bind to 2 similar regions in plant eIF4G and to a similar region in eIFiso4G (), it was possible that they bind either cooperatively or competitively to each region. Assays to determine the nature of their binding revealed that eIF4B and PABP bind competitively to each region of wheat eIF4G and eIFiso4G,Citation12,13 suggesting that the binding of PABP and eIF4B to these sites is mutually exclusive. When PABP and either region of eIF4G were present in equal molar amounts in a competition assay, eIF4B was able to compete with PABP noticeably at substoichiometric molar ratios, suggesting that eIF4B binds wheat eIF4G at each site more strongly than does PABP. Because eIF4B competed with PABP at a lower molar ratio at the HEAT-1-proximal eIF4B/PABP interaction site than at the N-proximal eIF4B/PABP interaction site, it likely binds relatively more strongly than PABP to the HEAT-1-proximal site than it does to the N-terminal site. These findings suggest that the binding of PABP and eIF4B to each of the 2 sites in eIF4G is mutually exclusive but this does not exclude the possibility that PABP and eIF4B could bind simultaneously to eIF4G ().
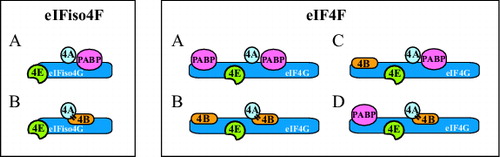
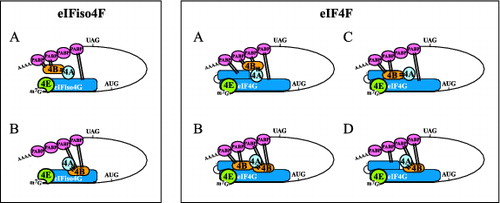
Figure 2. Mutually exclusive binding of PABP and eIF4B to eIFiso4G and eIF4G in plants. For eIFiso4F: (A) binding of PABP or (B) binding of eIF4B is depicted. For eIF4F: (A) binding of PABP to both PABP binding sites, (B) binding of eIF4B to both eIF4B binding sites, (C) binding of eIF4B and PABP to the N- and C-terminal binding sites, respectively, or (D) binding of PABP and eIF4B to the N- and C-terminal binding sites, respectively, is depicted. eIF4B is shown interacting with eIF4A (indicated with bars) but whether eIF4B can bind eIF4A when bound to eIFiso4G or eIF4G is unknown.
The binding sites for PABP and eIF4B overlap extensively in eIFiso4G () and, as with eIF4G, PABP and eIF4B compete for binding eIFiso4G.Citation12 Because eIFiso4G differs from eIF4G in that it lacks the region corresponding to the N-terminal binding site for PABP or eIF4B, the competition between PABP and eIF4B suggests their binding to eIFiso4G is mutually exclusive () and they would not be expected to bind the same molecule of eIFiso4G. This has significant implications for the interactions between the 5′ and 3′ ends of an mRNA as discussed below.
eIF4A does not compete with either eIF4B or PABP for binding plant eIF4G and eIFiso4G
Because the coincident eIF4B and PABP interaction domains in plant eIF4G result in their competitive binding to eIF4G, the observation that the HEAT-1-proximal eIF4B/PABP binding sites partially overlap with the eIF4A binding domain that includes the eIF4G HEAT-1 domain raised the possibility that eIF4B and/or PABP might bind cooperatively or competitively with eIF4A to this region of eIF4G. Binding assays revealed, however, that neither eIF4B or PABP had any effect, either cooperatively or competitively, on eIF4A binding to the HEAT-1 domain,Citation13 suggesting that the eIF4B or PABP binding sites do not overlap sufficiently with the eIF4A interaction domain to prevent eIF4A binding. Similarly, the presence of eIF4A had no detectable effect on the binding of eIF4B or PABP to the HEAT-1-containing region of plant eIF4G. These observations indicate that eIF4A and either eIF4B or PABP can bind plant eIF4G simultaneously but that eIF4B and PABP compete to bind each of the 2 sites in eIF4G. In animals, the binding of eIF4G and eIF4B to eIF4A may be mutually exclusiveCitation26 but this has not been examined for plants.
Because of the extensive overlap between the PABP and eIF4A binding sites in the wheat eIFiso4G HEAT-1 domain, PABP competes with eIF4A for binding to this domain in the absence of the HEAT-2 domain.Citation12 However, when the region of eIFiso4G containing the HEAT-1 and HEAT-2 domains is tested, PABP no longer competes with eIF4A for binding to eIFiso4G.Citation12 Thus, in the absence of the HEAT-2 domain, binding of eIF4A to the eIFiso4G HEAT-1 domain may be weaker and more likely to be displaced by PABP. The presence of the HEAT-2 domain, which also binds eIF4A, likely stabilizes the binding of eIF4A while accommodating binding of PABP to eIFiso4G.
The eIF4B binding site in eIFiso4G overlaps just the C-terminal end of the HEAT-1 domainCitation12 (). Nevertheless, as with PABP, eIF4B competes with eIF4A for binding to the eIFiso4G HEAT-1 domain in the absence of the HEAT-2 domain. However, like PABP, eIF4B does not compete with eIF4A when both the HEAT-1 and HEAT-2 domains of eIFiso4G are present.Citation12 This suggests that, as with PABP, the HEAT-2 domain alleviates the competition between eIF4B and eIF4A, perhaps through stabilizing the binding of eIF4A to eIFiso4G in the presence of eIF4B.
The competition that exists between PABP and eIF4A or between eIF4B and eIF4A in binding wheat eIFiso4G may be explained by the greater overlap between the binding sites for PABP and eIF4A or for eIF4B and eIF4A in the eIFiso4G HEAT-1 domain compared with the minimal overlap with the eIF4G HEAT-1 region. Although this would appear to be a significant difference between eIFiso4G and eIF4G in plants, the observation that PABP and eIF4A or eIF4B and eIF4A no longer compete when the HEAT-2 domain is also present suggests that intact eIFiso4G and eIF4G would not exhibit differences in their interaction with these partner proteins.
eIF4B, a second bridge between the 5′ and 3′ termini of an mRNA
eIF4B stimulates the RNA-dependent ATP hydrolysis activity and the ATP-dependent RNA helicase activity of animal eIF4A,Citation6 the helicase activity of yeast eIF4ACitation39 and the activity of plant eIF4A in wheat germ extract.Citation40-43 eIF4B increases the affinity of eIF4A for ATP and its helicase processivityCitation6,44,45 in part through coupling ATP hydrolysis to RNA unwinding.Citation46 Together, eIF4B and eIF4G synergistically increase the ATPase activity of eIF4A,Citation26 suggesting a functional interaction between the 2 factors. eIF4B may mediate mRNA binding to ribosomesCitation47-52 or bridge an interaction between eIF4F bound to mRNA and eIF3.Citation53 In yeast, eIF4B promotes eIF4A binding to eIF4G perhaps through a conformational change in the single HEAT domain present in the yeast factor.Citation24 Yeast eIF4B preferentially stimulates translation from mRNAs with moderately stable secondary structure in the 5′ UTR while it stimulates the higher basal level of translation from mRNAs that lack secondary structure only moderately.Citation54 Formation of a 48 S translation initiation complex (i.e., the binding of a 40 S subunit to an mRNA) with animal mRNAs containing a structured 5′ UTR was dependent on eIF4B,Citation55 suggesting that, although the ability of eIF4B to promote eIF4A helicase activity is likely involved in the translation of mRNAs in general, it may be of particular importance for certain types of mRNAs.
Despite its functional conservation among plants, animals, and yeast, only limited conservation is observed for eIF4B from wheat and other species: wheat eIF4B shares just 29% and 24% identity with human and yeast eIF4B, respectively,Citation56 making this factor one of the least conserved of the translation initiation factors. As mentioned above, wheat eIF4B interacts with wheat eIF4G and eIFiso4G and each of these bind to the N-terminal region of eIF4B. For example, eIFiso4G binds at a single site proximal to the N-terminal RNA binding domain of eIF4B ().Citation57
Figure 3. Partner protein interactions of eIF4B in higher eukaryotes. (A) The RNA and protein binding domains in wheat eIF4B (TaeIF4B), human eIF4B (HseIF4B), and yeast eIF4B (SceIF4B) are shown. Interaction domains for partner proteins are indicated by color with a key included. The 7 repeats in yeast eIF4B represent 26 amino acid repeats implicated in promoting translation.Citation59 (B) Below the wheat eIF4B is a sequence logo for the region encompassing the PABP and eIF4A binding site repeats and the RNA binding domain that separates the repeats. A sequence alignment from 16 monocot and 11 dicot eIF4B homologs was generated using EBI MUSCLE. The aligned sequences were submitted to WebLogo Berkeley to generate a sequence logo, in which the most conserved one, 2, or sometimes 3 residues at each position are shown as the consensus. Below the protein sequence logo is the consensus sequence for the region in which residues conserved with the sequence logo are similarly colored. The logo was constructed from eIF4B homologs from Triticum aestivum, Brachypodium distachyon (Bradi5g13500 and Bradi3g47700), Oryza sativa (Os04 g40400 and Os02 g38220), Setaria italica (Si009805 and Si016924), Sorghum bicolor (Sb06 g020170 and Sb04 g024860), Zea mays (GRMZM2G163471, GRMZM2G066815, and GRMZM2G139614), Panicum virgatum (Pavirv00022707 and Pavirv00022708, Pavirv00021798, and Pavirv00059810), Phaseolus vulgaris (l002G259400), Ricinus communis (29792), Brassica rapa Chiifu-401 (Bra026941), Thellungiella halophila (Thhalv10007279 and Thhalv10003946), Capsella rubella (Carubv10008811 and Carubv10016953), Arabidopsis thaliana (AT1G13020 and AT3G26400), and Arabidopsis lyrata (920166 and 484402).
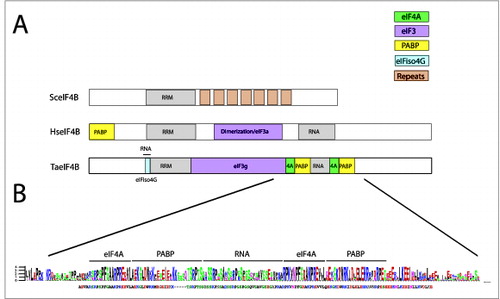
Although a complex between mammalian eIF4B and eIF4A is observed in the presence of RNA and ATP,Citation14,26,58 no direct interaction between the 2 factors has been reported nor has the eIF4A binding site in animal eIF4B been identified. NMR studies with human eIF4H, a factor related to eIF4B that lacks sequence corresponding to the N- and C-terminal ends of eIF4B, revealed binding to the C-terminal domain of eIF4A.Citation14 The C-terminal 72 amino acids of eIF4H were sufficient for this interaction with eIF4A. A direct interaction between eIF4B and eIF4A (as well as PABP), however, has been shown for the plant homologs ().Citation57 eIF4A binds wheat eIF4B at 2 conserved repeats approximately 18 amino acids long that lie on either side of a C-terminal RNA binding domainCitation57 (). PABP also binds immediately C-proximal to each eIF4A-binding site to a conserved 24 amino acid long repeat. As these repeats are conserved, this suggests that plant eIF4B may bind 2 molecules of eIF4A and 2 of PABP.
Despite the fact that eIF4B is poorly conserved, even among plants, the region containing the eIF4A and PABP binding sites represents the most conserved region of the protein in plants. The conservation within this region can be seen with a sequence logo generated from an alignment representing 16 monocot and 11 dicot eIF4B homologs (). The eIF4A binding repeats exhibit the greatest conservation and significant conservation is observed in the PABP binding sites and the RNA binding domain. Interestingly, the C-terminal 72 amino acid region of human eIF4H sufficient for interaction with eIF4A contains the region corresponding to the eIF4A-binding repeat of plant eIF4B and exhibits significant homology with the plant eIF4A-binding site. Repeats with some similarity are also present in yeast eIF4B.Citation59 Whether eIF4B or PABP bind to such elements in animal or yeast eIF4B, however, is unknown. The position of the 2 PABP binding sites in the central region of plant eIF4B differs with the position of the reported PABP binding site in mammalian eIF4B that lies within the N-terminal 80 amino acids of the protein ().Citation60 The lack of conservation in the position of the PABP binding site between mammalian and plant eIF4B is a significant difference between the homologs but may be a result of the poorly conserved nature of eIF4B in eukaryotes. However, the fact that an interaction between eIF4B and PABP has been conserved, at least in higher eukaryotes, suggests that the interaction between these 2 factors is important.
These findings support the notion that plant eIF4B organizes interactions among translation factors to a greater extent than was previously appreciated. Evidence in yeast suggests that the interaction of eIF4B with eIF4G prepares the latter for binding eIF4A.Citation24 As in yeast,Citation61,62 eIF4A is typically lost from preparations of wheat eIF4F (or eIFiso4F),Citation63 suggesting the binding of eIF4A to eIF4G may be transient. Therefore, eIF4B may enhance binding between eIF4A and eIF4G in plants as it does in yeast although no stimulatory effect of eIF4B on eIF4A binding to either eIF4G or eIFiso4G was observed in vitro.Citation12,13
eIF4G, eIFiso4G, and eIF4B bind competitively within the PABP RRM1 domain
PABP contains 4, N-terminal RNA recognition motifs (RRMs) that are tandemly arranged and a C-terminal region that is required for its self-association ().Citation64,65 As each RRM contacts approximately 3 adenosine residues,Citation66 the tandem arrangement allows the protein to bind a continuous stretch of 12 adenosine residues which is the minimum length that PABP requires in order to bind poly(A) RNA.Citation64,67 In animals, the region encompassing RRM1 to RRM2 within PABP is required for interaction with eIF4G whereas in yeast, PABP RRM2 is principally required for eIF4G binding with PABP RRM1 playing a contributing role ().Citation17,68,69 In plants, eIF4G binds PABP within its RRM1 and eIFiso4G binds a region encompassing the C-terminal end of RRM1 and the N-terminal end of RRM2Citation70 (), suggesting that eIF4G proteins interact with PABP within a similar if not quite identical region of PABP across eukaryotes. If the RRM1 of plant PABP is the only domain involved in the interaction with eIF4G, the presence of 2 distinct PABP interaction domains in eIF4GCitation13 suggests that the RRM1 of PABP interacts with both eIF4G sites although presumably with different subsequences as the 2 PABP binding sites in eIF4G do not share any obvious similarity.
Figure 4. Partner protein interactions of PABP in higher eukaryotes. (A) The RNA and protein binding domains in wheat PABP (TaPABP), human PABP (HsPABP), and yeast PABP (ScPABP). are shown. Interaction domains for partner proteins are indicated by color with a key included. (B) Below the wheat PABP is a sequence logo for higher and lower eukaryotic PABP proteins encompassing the eIF4G, eIFiso4G, and eIF4B binding sites of the wheat PABP. For this analysis, a sequence alignment from yeast, animal, and plant PABP homologs was generated using EBI MUSCLE. A sequence logo was generated from the aligned sequences in which the most conserved one or residues at each position are shown as the consensus. The sequences part of RRM1 or RRM2 domains are indicated by the labels above the logo. The RNP2 motif of RRM2 is indicated by the asterisks. The fourth β sheet (β4) of RRM1 and the first β sheet (β1) of RRM2 are indicated by the labels below the logo. The logo was constructed from PABP homologs from Saccharomyces cerevisiae (ScPABP, NM_001179055), Schizosaccharomyces pombe (SpPABP, NM_001018809), Homo sapiens (HsPABP, NM_002568), Xenopus laevis (XlPABP, NM_001086735), Mus musculus (MmPABP, NM_008774), Physcomitrella patens (PpPABP, Pp1s257_72V6), Arabidopsis thaliana (AtPABP, At2g23350), and Triticum aestivum (TaPABP, TAU81318). Below the logo are the sequences from each species used in which invariant residues are colored as in the sequence logo.
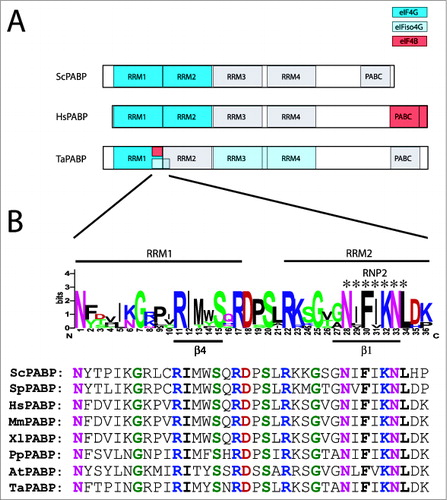
In addition to its principal binding site in PABP RRM1–2, wheat eIFiso4G interacts at a second site within PABP RRM3–4Citation70 (). eIFiso4G binding to the site in RRM1–2 is considerably stronger than its binding to RRM3–4.Citation70 As mentioned above, PABP binds to 2 distinct subregions within the eIFiso4G HEAT-1 domainCitation12 (). The region of PABP encompassing the C-terminal end of RRM1 and the N-terminal end of RRM2 binds the C-terminal subregion within the eIFiso4G HEAT-1 domain whereas PABP RRM3–4 binds the N-terminal subregion of the HEAT-1 domain.Citation12
As with eIF4G and eIFiso4G, plant eIF4B binds within the RRM1 of PABP at a site located within the C-terminal end of this domain (). To identify the residues in this region that might be conserved among eukaryotes, a sequence logo showing the conservation of sequence at each residue was generated from an alignment of the corresponding region from PABP orthologs of yeast, animals, and vascular and non-vascular plants (). The RNP2 motif of RRM2 within this region that makes direct contact with poly(A) RNA is highly conserved as expected. In addition to this motif, several residues are invariant across kingdoms including several present within the linker between RRM1 and RRM2 (). Whether these conserved residues are important for binding eIF4G (as well as eIFiso4G and eIF4B for plant PABP) remains to be determined.
As the binding sites of eIF4B and PABP in plant eIF4G and eIFiso4G overlap causing their binding to eIF4G and eIFiso4G to be mutually exclusive (), it is not surprising that eIF4B and eIFiso4G (and presumably eIF4B and eIF4G although this has not been tested) compete in their binding to PABP.Citation70 These observations indicate that the binding of eIF4B and eIFiso4G to PABP is mutually exclusive as is the binding of eIF4B and PABP to eIFiso4G, suggesting that a molecule of PABP can bind either eIF4B or eIFiso4G but not both simultaneously just as a molecule of eIFiso4G can bind either eIF4B or PABP but not both simultaneously.
eIF4B has been reported to bind mammalian PABP within its C-terminal region () that also interacts with eRF3, Paip1, Paip2.Citation60,Citation71-75 Although the ability of proteins to bind the PABP C-terminal region requires a PAM2 motif that is present among these partner proteins, interestingly, this motif is not present in mammalian eIF4B.
Functional Consequences of the Interactions Between the Cap-Associated Factors and PABP
PABP and the cap-binding proteins mutually stabilize binding to an mRNA
Given the interactions among the cap-associated initiation factors and PABP, what impact might these interactions have on their function? The 5′-cap and the poly(A) tail synergistically stimulate translation in plants, animals, and yeast which is a consequence of the physical interactions between PABP bound to the poly(A) tail and the cap-associated initiation factors bound at the 5′-cap.Citation76,77 One functional consequence of the physical interaction between plant PABP and eIFiso4G is that PABP increases the binding of eIFiso4F to the 5′-cap by 40-fold and PABP stimulates the ATPase and RNA helicase activity of the eIFiso4F:eIF4A:eIF4B complex.Citation38,78 The interaction of PABP with eIF4G also increases eIF4F binding to the 5′-cap in yeast.Citation79 In mammals, the results have been mixed with depletion of PABP from cell extracts impairing the interaction of eIF4E with the mRNA 5′-capCitation80 but loss of the N-terminal PABP-binding site in eIF4G having no effect on the interaction of eIF4G with eIF4E, at least when tested in vitro.Citation81
Addition of wheat eIF4B to eIFiso4F or the eIFiso4F:PABP complex lowers the activation energy of binding of each to the 5′-cap in plants.Citation82,83 Fluorescence stopped-flow studies of plant PABP:eIFiso4F complex binding to a 5′-cap showed that the PABP-mediated increase in the eIFiso4F affinity for a 5′-cap results from a concentration-independent conformational change and a reduced dissociation rate that enables eIFiso4F to more easily achieve a stable conformation with the 5′-cap.Citation82 This can also explain how binding of PABP increases the ATPase and RNA helicase activity of the cap-associated complex.Citation78 The interaction between PABP and eIF4G stimulates eIF4F binding to an mRNA in mammalian extractsCitation80 and in yeast,Citation84 perhaps by assisting eIF4G to out-compete general RNA-binding proteins that are unable to interact with PABP.Citation85 eIF4B, together with PABP, promotes stable recruitment of eIF4F to an mRNA by accelerating its binding to, and reducing its dissociation from, the 5′-cap.Citation83
The interaction of plant eIFiso4G, eIF4G, or eIF4B with PABP increases binding of the latter to poly(A) RNA by decreasing its dissociation rate.Citation34,86 Moreover, eIF4F (or eIFiso4F) and eIF4B exert a synergistic effect on the poly(A) RNA binding activity of PABP,Citation34 suggesting a functional interaction between all 3 proteins. Therefore, the physical interaction between eIF4G (or eIFiso4G) and PABP mutually stabilizes their association with their respective binding sites while bringing the termini of an mRNA into physical proximity.Citation34,82,86,87 An additional consequence of the physical interaction between PABP and the cap-binding complex is that it serves as a means test to confirm the integrity of an mRNA prior to 40 S subunit recruitment to prevent translation initiation on an mRNA undergoing degradation.Citation88 The interaction between PABP and eIF4G promotes formation of the 48 S pre-initiation complex as well as the assembly of the 80 S ribosome and increases translational efficiency.Citation80,81 That PABP increases 40 S subunit binding to an mRNA demonstrates its stimulatory role in translation initiation,Citation89,90 likely in part through its ability to increase the binding of the cap-associated complex to an mRNA which in turn more efficiently recruits 40 S subunits.
The Mutually Exclusive Binding Between eIF4G and eIF4B to PABP Increases The Number of Interactions Between The Termini of An mRNA
As mentioned, the binding of eIF4B and PABP to eIF4G (or eIFiso4G) in plants is mutually exclusive as is the binding of eIF4B and eIF4G (or eIFiso4G) to PABP. Therefore, because the average length of a poly(A) tail is sufficiently long to support the binding of multiple molecules of PABP, the mutually exclusive nature of eIF4G (or eIFiso4G) and eIF4B binding to PABP suggests they interact with separate molecules of PABP on a poly(A) tail, providing increased stability to the complex through an increase in the number of protein interactions (). Moreover, as PABP and eIF4G (or eIFiso4G) bind to eIF4B at well separated sites and eIF4B can dimerize,Citation91 plant eIF4B likely can bind to PABP and eIF4G (or eIFiso4G) simultaneously, providing yet another means to increase and therefore stabilize the physical interactions between the termini of an mRNA (). Finally, the observation that the combination of eIF4G (or eIFiso4G) and eIF4B synergistically promotes PABP binding to poly(A) RNA in plants and does so in part through promoting multimeric PABP bindingCitation34 suggests that eIF4G (or eIFiso4G) and eIF4B cooperate to interact with PABP and increase the stability of the complex.
Figure 5. Models for possible interactions between PABP and eIFiso4G, eIF4G, and eIF4B during translation initiation in plants. For eIFiso4F: (A) The interaction between the termini of a translating mRNA is facilitated by interactions between PABP to eIFiso4G and eIF4B (indicated with bars). eIFiso4G contains a single binding site for PABP and this interaction is depicted. However, because eIF4A binds eIF4B (which contains 2 eIF4A binding sites), an interaction between eIF4B and 2 molecules of PABP is also depicted. Whether eIF4B can bind 2 molecules of PABP simultaneously and whether eIF4A can bind eIFiso4G and eIF4B simultaneously is unknown. (B) The interaction between the termini of a translating mRNA is facilitated solely by the binding of PABP to eIF4B which in turn is bound to eIFiso4G which contains a single eIF4B binding site. However, because eIF4B contains 2 binding sites for PABP, it is depicted as interacting with 2 molecules of PABP. For eIF4F: (A) The interaction between the termini of a translating mRNA is facilitated through the binding of PABP at the N- and C-terminal PABP binding sites in eIF4G. Because eIF4A binds eIF4B, an interaction between eIF4B and 2 molecules of PABP is also depicted. (B) The interaction between the termini of a translating mRNA is facilitated solely through the binding of PABP to eIF4B which is bound at the N- and C-terminal eIF4B binding sites in eIF4G. Because eIF4B can dimerize, the 2 molecules of eIF4B are shown as a dimer but whether dimerization occurs during binding to eIF4G is unknown. (C) The interaction between the termini of a translating mRNA is facilitated through the binding of 2 molecules of PABP to eIF4B bound at the N-terminal eIF4B binding site in eIF4G and the binding of PABP to the C-terminal PABP binding site in eIF4G. (D) The interaction between the termini of a translating mRNA is facilitated through the binding of PABP to the N-terminal PABP binding site in eIF4G and the binding of 2 molecules of PABP to eIF4B bound at the C-terminal eIF4B binding site in eIF4G.
Regulation of the Interactions Between the Cap-Associated Factors and PABP
The phosphorylation state of PABP determines its type of binding and affinity to poly(A) RNA
Several components of the cap-binding complex and PABP are phosphoproteins and their phosphorylation state can affect the nature and strength of their interaction. For example, PABP is differentially phosphorylated in plants, yeast, and sea urchin.Citation86,Citation92-94 In plants, the phosphorylation state of PABP affects its affinity and type of binding to poly(A) RNA as well as its specificity of its interaction with eIF4G, eIFiso4G, and eIF4B,Citation86 the result of which is to prevent competition between eIFiso4G and eIF4B binding.
Self-interaction of PABP in Xenopus is mediated through its C-terminal domain.Citation65 When phosphorylated, plant PABP binds poly(A) RNA cooperatively while PABP in a hypophosphorylated state does not bind poly(A) RNA cooperatively but does bind with substantially greater affinity.Citation86 Only under saturating conditions does more than one molecule of hypophosphorylated PABP bind to poly(A) RNA if the RNA is long enough to bind multiple PABP molecules. However, the combination of PABP isoforms of opposite phosphorylation states exhibited the greatest cooperative binding.Citation86 As diverse PABP phosphoisoforms are observed in all plant tissues examinedCitation86,95 and the average poly(A) tail of a typical mRNA sufficiently long to bind multiple molecules of PABP,Citation64,67 the bound PABP is likely be a heterogeneous population of phosphorylated states, consistent with what is observed for polysome-bound PABP.Citation86 Differentially phosphorylated PABP species have been reported in sea urchinCitation92 and the in vivo phosphorylation of a Xenopus laevis embryonic poly(A)-binding protein (known as ePABP or PABPc1-like) at a 4 residue cluster is required for oocyte maturation.Citation96 Although the main cytoplasmic human PABP appears to be extensively post-translationally modified,Citation97 no evidence of its phosphorylation has been detected but not all members of the gene family have been examined to date. Whether the phosphorylated state is important for the interaction between the cap-associated factors and PABP in animals remains to be determined.
The phosphorylation state of PABP determines its binding to eIF4G, eIFiso4G, and eIF4B
The PABP phosphorylation state also affects PABP's binding with its partner proteins.Citation86 The interaction of wheat eIF4G with PABP increases its RNA binding activity and does so by promoting its cooperative binding.Citation86 As this effect is specific to the hypophosphorylated form of PABP, this suggests that eIF4G interacts preferentially with hypophosphorylated PABP. In contrast, the phosphorylation state of plant PABP does not appear to affect the interaction with eIFiso4G.Citation86 eIF4B exhibits a third type of specificity and preferentially increases the RNA binding activity of phosphorylated PABP in plants by increasing its affinity to RNA.Citation86
The phosphorylation state of plant eIF4B also affects the PABP-eIF4G interaction. The presence of eIF4B in its phosphorylated form increased the RNA binding activity of phosphorylated PABP while recombinant eIF4B, lacking phosphorylation, had little effect.Citation86 Only when substantially more recombinant eIF4B was present did it increase the PABP binding to poly(A) RNA but this was specific to the hypophosphorylated form of PABP.Citation86 As wheat eIF4B has a greater effect on PABP's binding activity than does eIF4F or eIFiso4F,Citation34 eIF4B may be particularly important in the interaction between cap-associated factors and PABP during plant translation. Whether this is true in animals where the interaction between eIF4B and PABP also occursCitation60 has not been examined.
The observation that the phosphorylation state of plant PABP affects its binding with partner proteins suggests that eIF4G and eIF4B (or eIFiso4G and eIF4B) would interact with independent molecules of PABP (). As eIF4B binds eIF4G (or eIFiso4G) in plants,Citation12,13,57,91 such a possibility would involve the interaction of 4 molecules (i.e., eIF4G, eIF4B, and 2 molecules of PABP). In addition, as PABP cooperatively binds to poly(A) RNA, the interactions among the 4 molecules may yield a more stable complex. The observation that the combinatorial effect of eIF4G and eIF4B (or eIFiso4G and eIF4B) on the multimeric binding of PABP to poly(A) RNA is synergistic supports this notion.Citation34,86 Because the binding sites for eIF4G, eIFiso4G, and eIF4B overlap in PABP, this also dictates that these components of the cap-binding complex interact with independent molecules of PABP, thereby avoiding competition in their binding.
Regulation of the phosphorylation state of eIF4B correlates with the strength of its interaction with PABP and with active translation
Although the specificity and strength of the PABP-eIF4B interaction is determined by their phosphorylation state in plants, is their phosphorylation regulated either developmentally or in response to environmental changes? Whereas the array of PABP phosphorylated isoforms is fully represented in polysomes and does not substantially change during development or following stress in plants, eIF4B is largely present in its most phosphorylated species in actively growing tissues when protein synthetic rates are highest and it is the phosphorylated species of eIF4B that are preferentially recruited into polysomes.Citation86,93,95 In contrast, eIF4B is present mostly as dephosphorylated species during late seed development or following a heat shock when protein synthesis is repressed.Citation93 Thus, the regulation of eIF4B phosphorylation by developmental or stress-related cues could serve to regulate global protein synthesis in plants. In mammalian cells, eIF4B phosphorylation is regulated by growth factors or stress: it is phosphorylated under conditions of active translation (e.g., following insulin stimulation) but is dephosphorylated following serum depletion or during a heat shock.Citation98-100 Whether the interaction between mammalian eIF4B and PABP is regulated by the phosphorylation state of eIF4B has not been examined in animals.
Concluding Remarks and Future Directions
The presence of multiple interaction domains among the components of the cap-binding complex and PABP and the mutually exclusive binding observed for some partner proteins suggests that more than one set of interactions may be possible during initiation in plants. As mentioned, eIF4G and eIF4B or eIFiso4G and eIF4B likely interact with independent molecules of PABP (). Their interaction with PABP may be direct and simultaneous if multiple PABPs of differing phosphorylation states are bound along a poly(A) tail. Alternatively, it is possible that PABP could bind eIF4B while the latter is bound to eIF4G (or eIFiso4G), so that eIF4B acts as a bridge linking eIF4G (or eIFiso4G) with PABP (). PABP may also bind eIF4B while the latter is bound to eIF4A which in turn is bound to eIF4G (or eIFiso4G), so that eIF4B, through eIF4A, bridges eIF4G (or eIFiso4G) with PABP (). Because eIF4B can dimerize,Citation91 it is possible that each monomer of the eIF4B dimer could contact a PABP while eIF4G (or eIFiso4G) binds to a third PABP. Moreover, the presence of 2 conserved PABP binding sites in eIF4B suggests that the latter may bind 2 molecules of PABP. Alternatively, only one molecule of PABP may bind eIF4B at any one time but the presence of 2 PABP interaction sites in eIF4B may increase the likelihood of interaction between PABP and eIF4B. Given the number of possible interactions among these factors, it is unlikely that the interactions are static but rather are likely to be dynamic and alternate between the examples depicted in during the progression of initiation or in the presence of additional factors that will influence the interactions that predominate. These models will require experimental confirmation before a complete understanding of the possibilities of interaction between the termini of an RNA can be fully appreciated.
Although eIF4B can interact with eIF4G (or eIFiso4G) in plants, it also binds eIF4A directly.Citation12,13,57,91 As an RNA helicase, eIF4A may not only be present as a subunit of eIF4F (or eIFiso4F), it may also be bound to RNA independently of its interaction with eIF4G.Citation4 Therefore, it will be important to determine whether eIF4B can interact with PABP when the former is bound to eIF4G or eIF4A or to both and whether there are different functional consequences between them.
eIF4A is not the sole RNA helicase involved in translation. Ded1 is a DExD/H-box RNA helicase in yeast that binds directly to eIF4G1 to form a translationally repressed Ded1-eIF4F-mRNA intermediate which accumulates in stress granules.Citation101 Ded1 binds to the RNA3 domain of yeast eIF4G which is distinctly different than the interaction of eIF4A with the HEAT-1 domain of eIF4G. Whether DDX3, the Ded1 homolog in humans, is involved in regulating protein synthesis has been controversial.Citation102 The Arabidopsis thaliana gene, At5g14610, encodes a likely Ded1 homolog (80.2%/82.9% amino acid identity/similarity) but whether this binds plant eIF4G or eIFiso4G or exhibits a similar function has not been examined.
Another RNA helicase, DHX29, a member of the SF2 DEAH/RHA family, promotes translation in animals through its helicase activity.Citation103 DHX29 mainly binds around helix 16 of the 40 S subunit and likely functions to induce conformational changes in the small ribosomal subunit that open and close the mRNA entrance to aid 43S complexes to unwind structured sequences in an mRNA.Citation104 However, DHX29 is not known to be a component of the cap-binding complex. Proteins encoded by the A. thaliana genes, At1g48650 and At3g26560, exhibit limited homology with DHX29 (49.2%/54.9% and 50.7%/56.3% amino acid identity/similarity, respectively) but any potential role in translation has not been examined.
Plant eIF4G contains 2 binding sites each for eIF4B and PABP.Citation13 Whether this results in the binding of one or 2 molecules of each partner protein is also an outstanding question but this represents a significant difference between the eIF4G and eIFiso4G isoforms in plants,Citation12,13 a difference that does not exist between eIF4G isoforms in other eukaryotes. The binding of eIF4B and PABP to eIFiso4G is mutually exclusive but the fact that plant eIF4G contains 2 binding sites each for eIF4B and PABP raises the possibility that eIF4B and PABP could bind simultaneously to eIF4G () although this has not been demonstrated. How such differences between the 2 eIF4G isoforms may affect their function is not known although preferences between plant eIF4G and eIFiso4G in the mRNAs they translate have been reported.Citation9,Citation105-107 Future work will need to address whether PABP and eIF4B can bind the same molecule of eIF4G simultaneously in plants, and if not, whether PABP and eIF4B cycle through eIF4G during translation initiation as has been reported for eIF4A.Citation108
The interaction between PABP and the cap-binding complex raises the question of how important is the interaction to protein synthesis? The observation that many mRNAs naturally lacking a 5′-cap or a poly(A) tail, but which have evolved functional equivalents, still retain a requirement for an interaction between the termini of the mRNA,Citation109-114 suggesting that the interaction between the termini of an mRNA is a common feature among mRNAs and that such an interaction provides a benefit to the synthesis of protein from an mRNA.
Depletion of PABP from mammalian extracts reduced eIF4E binding to a cap, 48 S complex formation, and translation, all of which could be restored by the addition of wild-type PABP but not by a mutant of PABP unable to interact with eIF4G.Citation80 In contrast, analysis in vivo suggested that deletion of the PABP binding site from mammalian eIF4GI had little effect on the latter's ability to support translation.Citation81 However, mammalian eIF4G lacking the binding sites for eIF4A or eIF3 also had a minimal effect on the ability of eIF4G to support translation in the same study.Citation81 As eIF4G lacking the binding sites for eIF4A or eIF3 showed a marked reduction in activity during translation in vitro and eIF4G lacking the PABP binding site was less effective than native eIF4G,Citation81 the siRNA knockdown approach used to suppress endogenous eIF4G expression in vivo may not have been stringent enough to assay fully the contribution that the interaction between PABP and eIF4G has on translation. When hypertonic shock was used to disassemble polysomes and eIF4F in mammalian cells, eIF4G lacking the PABP binding site was incorporated into eIF4F as efficiently as native eIF4G but was less efficiently incorporated into polysomes.Citation81 In Xenopus oocytes, eIF4G lacking the PABP binding site inhibited translation of co-injected mRNA suggesting it had a dominant negative effect.Citation115
Although these studies might suggest that the interaction between PABP and eIF4G is not essential but may contribute to optimal translation in animals, the interaction between PABP and eIF4B was not considered. In the context of a translating mRNA, this second interaction may stabilize the association of PABP with the cap-binding complex even if the PABP binding site in eIF4G is deleted. Conservation of this interaction in animals and plants suggests it serves a function, despite the fact that eIF4B is the least conserved of all the initiation factors. Until all means for the interaction between PABP and components of the cap-binding complex have been removed, it is difficult to estimate the magnitude that the interaction makes to translation. It is also possible that the interaction is more important in one species than another so its contribution will need to be accurately assessed in animals, plants, and yeast individually.
Although the factors providing the necessary specificity for binding mRNA are eIF4E and PABP, the RNA binding domains in plant eIF4G, eIFiso4G, eIF4A, and eIF4B suggest that these likely contribute to stabilizing the complex to an mRNA as observed for animal and yeast eIF4G,Citation37,116,117 while the presence of protein interaction domains in each factor serves to bring together the activities required to promote the binding of 40 S subunits and their subsequent scanning. The interaction of PABP with the cap-associated factors not only increases these activities and stabilizes the binding of the complex to the 5′-cap, but may promote the re-recruitment of 40 S subunits following translation termination, thereby increasing the yield of protein from a given mRNA before its eventual destruction.
The interaction between PABP and eIF4G may be of particular importance under conditions of competitive translation. Thus, in some translation lysates, the abundance of unengaged eIF4F or PABP may mask competitive translation.Citation105,106 Such a possibility is supported by the observation that the degree to which translation is cap-dependent in wheat germ lysate is inversely proportional to the concentration of PABP or eIF4G/eIFiso4G.Citation9 The interaction between PABP and eIF4G during competitive translation in animals may be particularly important in the presence of general ribonucleoproteins (e.g., YB-1) as their presence permits only those proteins with sufficient strength and specificity to compete for binding.Citation85 In the case of YB-1 for example, the interaction of PABP with eIF4G overcomes the inhibition imposed by this ribonucleoprotein on the eIF4E-5′-cap interaction.Citation85 Thus, the combinatorial strength provided by the multiple interactions among PABP, eIF4G, eIF4E, eIF4A, eIF4B, and eIF3 likely come into greatest play when presented with the full array of RNA binding proteins in a cell as well as a diverse population of mRNAs that are competing for their binding.
The numerous interactions among PABP, eIF4G, eIF4E, eIF4A, eIF4B, and eIF3 support the notion of functional redundancy within the complex. Such functional redundancy does not mean that individual interactions are of no importance as each likely contributes to the rate of assembly and overall stability of the complex. Functional redundancy also means that individual interactions can be targets for regulation in order to modulate the rate of assembly and complex stability as a means to regulate global protein synthesis in response to developmental or environmental signals. For example, developmental cues during late embryo development resulting in a translationally quiescent state by seed maturity correlate with changes in the phosphorylation state of eIF4B that would reduce its interaction with PABP.Citation93 Global repression of protein synthesis in wheat following an exposure to elevated temperature similarly results in the rapid dephosphorylation of eIF4BCitation93 and a reduction in its binding to PABP and to eIF4F and eIFiso4F.Citation86 Although exposure to heat-stress does not alter the distribution of plant PABP isoforms, a reduction in the poly(A)-binding activity of PABP and its binding to eIF4F and eIFiso4F is observed which may be a consequence of the reduced eIF4B interaction that would normally increase the poly(A)-binding activity of PABP and its association with eIF4F and eIFiso4F.Citation86 Whether the regulation of global protein synthesis involves changes in other components of the plant cap-binding complex, such as eIF4E, eIFiso4E, eIF4G, eIFiso4G, or eIF4A remains an outstanding question. However, the apparent lack of a plant homolog of 4E-BP (4E-binding protein), which in animals and yeast binds to eIF4E to block the interaction between eIF4E and eIF4G and repress global protein synthesis,Citation118,119 or a plant homolog of Paip2 (PABP interacting protein 2), which in animals binds to PABP and represses its poly(A) binding activity,Citation71 suggests that the regulation of interactions among the cap-associated factors and PABP in plants may rely to a greater extent on post-translational modifications than observed in other species. Future research focusing on these questions in plants will reveal those aspects common among eukaryotes but also those features that are unique to a very distinct eukaryotic kingdom.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
Work cited from the author's laboratory was funded by grants from the National Science Foundation (DBI-0820047), USDA, and the University of California Agricultural Experiment Station.
References
- Preiss TW, Hentze M. Starting the protein synthesis machine: eukaryotic translation initiation. Bioessays 2003; 25:1201-11; PMID:14635255; http://dx.doi.org/10.1002/bies.10362
- Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu Rev Biochem 2004; 73:657-704; PMID:15189156; http://dx.doi.org/10.1146/annurev.biochem.73.030403.080419
- Pestova TV, Lorsch JR, Hellen CUT. The mechanism of translation initiation in eukaryotes. In: Mathews MB, Sonenberg N, Hershey JWB ed. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press. 2007:87-128.
- Parsyan A, Svitkin Y, Shahbazian D, Gkogkas C, Lasko P, Merrick WC, Sonenberg N. mRNA helicases: the tacticians of translational control. Nat Rev Mol Cell Biol 2011; 12:235-45; PMID:21427765; http://dx.doi.org/10.1038/nrm3083
- Korneeva NL, First EA, Benoit CA, Rhoads RE. Interaction between the NH2-terminal domain of eIF4A and the central domain of eIF4G modulates RNA-stimulated ATPase activity. J Biol Chem 2005; 280:1872-81; PMID:15528191; http://dx.doi.org/10.1074/jbc.M406168200
- Rogers GW Jr., Komar AA, Merrick WC. eIF4A: the godfather of the DEAD box helicases. Prog Nucleic Acid Res Mol Biol 2002; 72:307-31; PMID:12206455; http://dx.doi.org/10.1016/S0079-6603(02)72073-4
- Goyer C, Altmann M, Lee HS, Blanc A, Deshmukh M, Woolford JL Jr., Trachsel H, Sonenberg N. TIF4631 and TIF4632: two yeast genes encoding the high-molecular-weight subunits of the cap-binding protein complex (eukaryotic initiation factor 4F) contain an RNA recognition motif-like sequence and carry out an essential function. Mol Cell Biol 1993; 13:4860-74; PMID:8336723
- Gradi A, Imataka H, Svitkin YV, Rom E, Raught B, Morino S, Sonenberg N. A novel functional human eukaryotic translation initiation factor 4G. Mol Cell Biol 1998; 18:334-42; PMID:9418880
- Gallie DR, Browning KS. eIF4G functionally differs from eIFiso4G in promoting internal initiation, cap-independent translation, and translation of structured mRNAs. J Biol Chem 2001; 276:36951-60; PMID:11483601; http://dx.doi.org/10.1074/jbc.M103869200
- Bellsolell L, Cho-Park PF, Poulin F, Sonenberg N, Burley SK. Two structurally atypical HEAT domains in the C-terminal portion of human eIF4G support binding to eIF4A and Mnk1. Structure 2006; 14:913-23; PMID:16698552; http://dx.doi.org/10.1016/j.str.2006.03.012
- Marcotrigiano J, Lomakin IB, Sonenberg N, Pestova TV, Hellen CU, Burley SK. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol Cell 2001; 7:193-203; PMID:11172724; http://dx.doi.org/10.1016/S1097-2765(01)00167-8
- Cheng S, Gallie DR. Competitive and noncompetitive binding of eIF4B, eIF4A, and the poly(A) binding protein to wheat translation initiation factor eIFiso4G. Biochemistry 2010; 49:8251-65; PMID:20795652; http://dx.doi.org/10.1021/bi1008529
- Cheng S, Gallie DR. Eukaryotic initiation factor 4B and the poly(A)-binding protein bind eIF4G competitively. Translation 2013; 1:1-13; http://dx.doi.org/10.4161/trla.24038
- Marintchev A, Edmonds KA, Marintcheva B, Hendrickson E, Oberer M, Suzuki C, Herdy B, Sonenberg N, Wagner G. Topology and regulation of the human eIF4A4G4H helicase complex in translation initiation. Cell 2009; 136:447-60; PMID:19203580; http://dx.doi.org/10.1016/j.cell.2009.01.014
- Lamphear BJ, Kirchweger R, Skern T, Rhoads RE. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J Biol Chem 1995; 270:21975-83; PMID:7665619; http://dx.doi.org/10.1074/jbc.270.37.21975
- Imataka H, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol Cell Biol 1997; 17:6940-7; PMID:9372926
- Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J 1998; 17:7480-9; PMID:9857202; http://dx.doi.org/10.1093/emboj17.24.7480
- Marintchev A, Wagner G. Translation initiation: structures, mechanisms and evolution. Q Rev Biophys 2004; 37:197-284; PMID:16194295; http://dx.doi.org/10.1017/S0033583505004026
- Schütz P, Bumann M, Oberholzer AE, Bieniossek C, Trachsel H, Altmann M, Baumann U. Crystal structure of the yeast eIF4A-eIF4G complex: an RNA-helicase controlled by protein-protein interactions. Proc Natl Acad Sci USA 2008; 105:9564-9; PMID:18606994; http://dx.doi.org/10.1073/pnas.0800418105
- Rajagopal V, Park EH, Hinnebusch AG, Lorsch JR. Specific domains in yeast translation initiation factor eIF4G strongly bias RNA unwinding activity of the eIF4F complex toward duplexes with 5¢-overhangs. J Biol Chem 2012; 287:20301-12; PMID:22467875; http://dx.doi.org/10.1074/jbc.M112.347278
- Oberer M, Marintchev A, Wagner G. Structural basis for the enhancement of eIF4A helicase activity by eIF4G. Genes Dev 2005; 19:2212-23; PMID:16166382; http://dx.doi.org/10.1101/gad.1335305
- Hilbert M, Kebbel F, Gubaev A, Klostermeier D. eIF4G stimulates the activity of the DEAD box protein eIF4A by a conformational guidance mechanism. Nucleic Acids Res 2011; 39:2260-70; PMID:21062831; http://dx.doi.org/10.1093/nargkq1127
- Feoktistova K, Tuvshintogs E, Do A, Fraser CS. Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity. Proc Natl Acad Sci U S A 2013; 110:13339-44; PMID:23901100; http://dx.doi.org/10.1073/pnas.1303781110
- Park EH, Walker SE, Zhou F, Lee JM, Rajagopal V, Lorsch JR, Hinnebusch AG. Yeast eukaryotic initiation factor 4B (eIF4B) enhances complex assembly between eIF4A and eIF4G in vivo. J Biol Chem 2013; 288:2340-54; PMID:23184954; http://dx.doi.org/10.1074/jbc.M112.398537
- Lomakin IB, Hellen CU, Pestova TV. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol Cell Biol 2000; 20:6019-29; PMID:10913184; http://dx.doi.org/10.1128/MCB.20.16.6019-6029.2000
- Nielsen KH, Behrens MA, He Y, Oliveira CL, Jensen LS, Hoffmann SV, Pedersen JS, Andersen GR. Synergistic activation of eIF4A by eIF4B and eIF4G. Nucleic Acids Res 2011; 39:2678-89; PMID:21113024; http://dx.doi.org/10.1093/nargkq1206
- LeFebvre AK, Korneeva NL, Trutschl M, Cvek U, Duzan RD, Bradley CA, Hershey JW, Rhoads RE. Translation initiation factor eIF4G-1 binds to eIF3 through the eIF3e subunit. J Biol Chem 2006; 281:22917-32; PMID:16766523; http://dx.doi.org/10.1074/jbc.M605418200
- Villa N, Do A, Hershey JW, Fraser CS. Human eukaryotic initiation factor 4G (eIF4G) protein binds to eIF3c, -d, and -e to promote mRNA recruitment to the ribosome. J Biol Chem 2013; 288:32932-40; PMID:24092755; http://dx.doi.org/10.1074/jbc.M113.517011
- Korneeva NL, Lamphear BJ, Hennigan FL, Rhoads RE. Mutually cooperative binding of eukaryotic translation initiation factor (eIF) 3 and eIF4A to human eIF4G-1. J Biol Chem 2000; 275:41369-76; PMID:11022043; http://dx.doi.org/10.1074/jbc.M007525200
- He H, von der Haar T, Singh CR, Ii M, Li B, Hinnebusch AG, McCarthy JE, Asano K. The yeast eukaryotic initiation factor 4G (eIF4G) HEAT domain interacts with eIF1 and eIF5 and is involved in stringent AUG selection. Mol Cell Biol 2003; 23:5431-45; PMID:12861028; http://dx.doi.org/10.1128/MCB.23.15.5431-5445.2003
- Singh CR, Watanabe R, Chowdhury W, Hiraishi H, Murai MJ, Yamamoto Y, Miles D, Ikeda Y, Asano M, Asano K. Sequential eukaryotic translation initiation factor 5 (eIF5) binding to the charged disordered segments of eIF4G and eIF2β stabilizes the 48S preinitiation complex and promotes its shift to the initiation mode. Mol Cell Biol 2012; 32:3978-89; PMID:22851688; http://dx.doi.org/10.1128/MCB.00376-12
- Yamamoto Y, Singh CR, Marintchev A, Hall NS, Hannig EM, Wagner G, Asano K. The eukaryotic initiation factor (eIF) 5 HEAT domain mediates multifactor assembly and scanning with distinct interfaces to eIF1, eIF2, eIF3, and eIF4G. Proc Natl Acad Sci USA 2005; 102:16164-9; PMID:16254050; http://dx.doi.org/10.1073/pnas.0507960102
- Tarun SZ Jr., Sachs AB. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J 1996; 15:7168-77; PMID:9003792
- Le H, Tanguay RL, Balasta ML, Wei C-C, Browning KS, Metz AM, Goss DJ, Gallie DR. Wheat germ poly(A) binding protein enhances the binding affinity of eukaryotic initiation factor 4F and (iso)4F for cap analogues. J Biol Chem 1997b; 272:16247-55; PMID:9195926; http://dx.doi.org/10.1074/jbc.272.26.16247
- Piron M, Vende P, Cohen J, Poncet D. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J 1998; 17:5811-21; PMID:9755181; http://dx.doi.org/10.1093/emboj17.19.5811
- Fraser CS, Pain VM, Morley SJ. The association of initiation factor 4F with poly(A)-binding protein is enhanced in serum-stimulated Xenopus kidney cells. J Biol Chem 1999; 274:196-204; PMID:9867830; http://dx.doi.org/10.1074/jbc.274.1.196
- Park EH, Walker SE, Lee JM, Rothenburg S, Lorsch JR, Hinnebusch AG. Multiple elements in the eIF4G1 N-terminus promote assembly of eIF4G1•PABP mRNPs in vivo. EMBO J 2011; 30:302-16; PMID:21139564; http://dx.doi.org/10.1038/emboj.2010.312
- Wei C-C, Balasta ML, Ren J, Goss DJ. Wheat germ poly(A) binding protein enhances the binding affinity of eukaryotic initiation factor 4F and (iso)4F for cap analogues. Biochemistry 1998; 37:1910-6; PMID:9485317; http://dx.doi.org/10.1021/bi9724570
- Andreou AZ, Klostermeier D. eIF4B and eIF4G jointly stimulate eIF4A ATPase and unwinding activities by modulation of the eIF4A conformational cycle. J Mol Biol 2014; 426:51-61; PMID:24080224; http://dx.doi.org/10.1016/j.jmb.2013.09.027
- Lax S, Fritz W, Browning K, Ravel J. Isolation and characterization of factors from wheat germ that exhibit eukaryotic initiation factor 4B activity and overcome 7-methylguanosine 5¢-triphosphate inhibition of polypeptide synthesis. Proc Natl Acad Sci U S A 1985; 82:330-3; PMID:3855554; http://dx.doi.org/10.1073/pnas.82.2.330
- Lax SR, Browning KS, Maia DM, Ravel JM. ATPase activities of wheat germ initiation factors 4A, 4B, and 4F. J Biol Chem 1986; 261:15632-6; PMID:2946676
- Browning KS, Lax SR, Ravel JM. Identification of two messenger RNA cap binding proteins in wheat germ. Evidence that the 28-kDa subunit of eIF-4B and the 26-kDa subunit of eIF-4F are antigenically distinct polypeptides. J Biol Chem 1987; 262:11228-32; PMID:2440886
- Browning KS, Fletcher L, Lax SR, Ravel JM. Evidence that the 59-kDa protein synthesis initiation factor from wheat germ is functionally similar to the 80-kDa initiation factor 4B from mammalian cells. J Biol Chem 1989; 264:8491-4; PMID:2722784
- Rogers GW Jr., Richter NJ, Merrick WC. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J Biol Chem 1999; 274:12236-44; PMID:10212190; http://dx.doi.org/10.1074/jbc.274.18.12236
- Rogers GW Jr., Richter NJ, Lima WF, Merrick WC. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J Biol Chem 2001; 276:30914-22; PMID:11418588; http://dx.doi.org/10.1074/jbc.M100157200
- Özeş AR, Feoktistova K, Avanzino BC, Fraser CS. Duplex unwinding and ATPase activities of the DEAD-box helicase eIF4A are coupled by eIF4G and eIF4B. J Mol Biol 2011; 412:674-87; PMID:21840318; http://dx.doi.org/10.1016/j.jmb.2011.08.004
- Trachsel H, Erni B, Schreier MH, Staehelin T. Initiation of mammalian protein synthesis. II. The assembly of the initiation complex with purified initiation factors. J Mol Biol 1977; 116:755-67; PMID:592399; http://dx.doi.org/10.1016/0022-2836(77)90269-8
- Benne R, Hershey JW. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J Biol Chem 1978; 253:3078-87; PMID:641056
- Methot N, Pickett G, Keene JD, Sonenberg N. In vitro RNA selection identifies RNA ligands that specifically bind to eukaryotic translation initiation factor 4B: the role of the RNA remotif. RNA 1996a; 2:38-50; PMID:8846295
- Altmann M, Wittmer B, Méthot N, Sonenberg N, Trachsel H. The Saccharomyces cerevisiae translation initiation factor Tif3 and its mammalian homologue, eIF-4B, have RNA annealing activity. EMBO J 1995; 14:3820-7; PMID:7543843
- Pestova TV, Hellen CU, Shatsky IN. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43 S preinitiation complexes. Mol Cell Biol 1996; 16:6859-69; PMID:8943341
- Morino S, Imataka H, Svitkin YV, Pestova TV, Sonenberg N. Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol Cell Biol 2000; 20:468-77; PMID:10611225; http://dx.doi.org/10.1128/MCB.20.2.468-477.2000
- Méthot N, Song MS, Sonenberg N. A region rich in aspartic acid, arginine, tyrosine, and glycine (DRYG) mediates eukaryotic initiation factor 4B (eIF4B) self-association and interaction with eIF3. Mol Cell Biol 1996b; 16:5328-34; PMID:8816444
- Altmann M, Müller PP, Wittmer B, Ruchti F, Lanker S, Trachsel H. A Saccharomyces cerevisiae homologue of mammalian translation initiation factor 4B contributes to RNA helicase activity. EMBO J 1993; 12:3997-4003; PMID:8404865
- Dmitriev SE, Terenin IM, Dunaevsky YE, Merrick WC, Shatsky IN. Assembly of 48S translation initiation complexes from purified components with mRNAs that have some base pairing within their 5¢ untranslated regions. Mol Cell Biol 2003; 23:8925-33; PMID:14645505; http://dx.doi.org/10.1128/MCB.23.24.8925-8933.2003
- Metz AM, Wong KC, Malmström SA, Browning KS. Eukaryotic initiation factor 4B from wheat and Arabidopsis thaliana is a member of a multigene family. Biochem Biophys Res Commun 1999; 266:314-21; PMID:10600500; http://dx.doi.org/10.1006/bbrc.1999.1814
- Cheng S, Gallie DR. Wheat eukaryotic initiation factor 4B organizes assembly of RNA and eIFiso4G, eIF4A, and poly(A)-binding protein. J Biol Chem 2006; 281:24351-64; PMID:16803875; http://dx.doi.org/10.1074/jbc.M605404200
- Rozovsky N, Butterworth AC, Moore MJ. Interactions between eIF4AI and its accessory factors eIF4B and eIF4H. RNA 2008; 14:2136-48; PMID:18719248; http://dx.doi.org/10.1261/rna.1049608
- Zhou F, Walker SE, Mitchell SF, Lorsch JR, Hinnebusch AG. Identification and characterization of functionally critical, conserved motifs in the internal repeats and N-terminal domain of yeast translation initiation factor 4B (yeIF4B). J Biol Chem 2014; 289:1704-22; PMID:24285537; http://dx.doi.org/10.1074/jbc.M113.529370
- Bushell M, Wood W, Carpenter G, Pain VM, Morley SJ, Clemens MJ. Disruption of the interaction of mammalian protein synthesis eukaryotic initiation factor 4B with the poly(A)-binding protein by caspase- and viral protease-mediated cleavages. J Biol Chem 2001; 276:23922-8; PMID:11274152; http://dx.doi.org/10.1074/jbc.M100384200
- Lanker S, Müller PP, Altmann M, Goyer C, Sonenberg N, Trachsel H. Interactions of the eIF-4F subunits in the yeast Saccharomyces cerevisiae. J Biol Chem 1992; 267:21167-71; PMID:1400427
- Neff CL, Sachs AB. Eukaryotic translation initiation factors 4G and 4A from Saccharomyces cerevisiae interact physically and functionally. Mol Cell Biol 1999; 19:5557-64; PMID:10409745
- Browning KS. The plant translational apparatus. Plant Mol Biol 1996; 32:107-44; PMID:8980477; http://dx.doi.org/10.1007/BF00039380
- Sachs AB, Davis RW, Kornberg RD. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol Cell Biol 1987; 7:3268-76; PMID:3313012
- Kühn U, Pieler T. Xenopus poly(A) binding protein: functional domains in RNA binding and protein-protein interaction. J Mol Biol 1996; 256:20-30; PMID:8609610; http://dx.doi.org/10.1006/jmbi.1996.0065
- Deo RC, Bonanno JB, Sonenberg N, Burley SK. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell 1999; 98:835-45; PMID:10499800; http://dx.doi.org/10.1016/S0092-8674(00)81517-2
- Baer BW, Kornberg RD. Repeating structure of cytoplasmic poly(A)-ribonucleoprotein. Proc Natl Acad Sci U S A 1980; 77:1890-2; PMID:6929525; http://dx.doi.org/10.1073/pnas.77.4.1890
- Kessler SH, Sachs AB. RNA recognition motif 2 of yeast Pab1p is required for its functional interaction with eukaryotic translation initiation factor 4G. Mol Cell Biol 1998; 18:51-7; PMID:9418852
- Otero LJ, Ashe MP, Sachs AB. The yeast poly(A)-binding protein Pab1p stimulates in vitro poly(A)-dependent and cap-dependent translation by distinct mechanisms. EMBO J 1999; 18:3153-63; PMID:10357826; http://dx.doi.org/10.1093/emboj18.11.3153
- Cheng S, Gallie DR. eIF4G, eIFiso4G, and eIF4B bind the poly(A)-binding protein through overlapping sites within the RNA recognition motif domains. J Biol Chem 2007; 282:25247-58; PMID:17606619; http://dx.doi.org/10.1074/jbc.M702193200
- Khaleghpour K, Svitkin YV, Craig AW, DeMaria CT, Deo RC, Burley SK, Sonenberg N. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol Cell 2001; 7:205-16; PMID:11172725; http://dx.doi.org/10.1016/S1097-2765(01)00168-X
- Roy G, De Crescenzo G, Khaleghpour K, Kahvejian A, O’Connor-McCourt M, Sonenberg N. Paip1 interacts with poly(A) binding protein through two independent binding motifs. Mol Cell Biol 2002; 22:3769-82; PMID:11997512; http://dx.doi.org/10.1128/MCB.22.11.3769-3782.2002
- Uchida N, Hoshino S, Imataka H, Sonenberg N, Katada T. A novel role of the mammalian GSPTeRF3 associating with poly(A)-binding protein in CapPoly(A)-dependent translation. J Biol Chem 2002; 277:50286-92; PMID:12381739; http://dx.doi.org/10.1074/jbc.M203029200
- Hosoda N, Kobayashi T, Uchida N, Funakoshi Y, Kikuchi Y, Hoshino S, Katada T. Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J Biol Chem 2003; 278:38287-91; PMID:12923185; http://dx.doi.org/10.1074/jbc.C300300200
- Albrecht M, Lengauer T. Survey on the PABC recognition motif PAM2. Biochem Biophys Res Commun 2004; 316:129-38; PMID:15003521; http://dx.doi.org/10.1016/j.bbrc.2004.02.024
- Gallie DR. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev 1991; 5:2108-16; PMID:1682219; http://dx.doi.org/10.1101/gad.5.11.2108
- Michel YM, Poncet D, Piron M, Kean KM, Borman AM. Cap-Poly(A) synergy in mammalian cell-free extracts. Investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J Biol Chem 2000; 275:32268-76; PMID:10922367; http://dx.doi.org/10.1074/jbc.M004304200
- Bi X, Ren J, Goss DJ. Wheat germ translation initiation factor eIF4B affects eIF4A and eIFiso4F helicase activity by increasing the ATP binding affinity of eIF4A. Biochemistry 2000; 39:5758-65; PMID:10801326; http://dx.doi.org/10.1021/bi992322p
- von Der Haar T, Ball PD, McCarthy JE. Stabilization of eukaryotic initiation factor 4E binding to the mRNA 5¢-Cap by domains of eIF4G. J Biol Chem 2000; 275:30551-5; PMID:10887196; http://dx.doi.org/10.1074/jbc.M004565200
- Kahvejian A, Svitkin YV, Sukarieh R, M’Boutchou MN, Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev 2005; 19:104-13; PMID:15630022; http://dx.doi.org/10.1101/gad.1262905
- Hinton TM, Coldwell MJ, Carpenter GA, Morley SJ, Pain VM. Functional analysis of individual binding activities of the scaffold protein eIF4G. J Biol Chem 2007; 282:1695-708; PMID:17130132; http://dx.doi.org/10.1074/jbc.M602780200
- Luo Y, Goss DJ. Homeostasis in mRNA initiation: wheat germ poly(A)-binding protein lowers the activation energy barrier to initiation complex formation. J Biol Chem 2001; 276:43083-6; PMID:11571283; http://dx.doi.org/10.1074/jbc.M104970200
- Khan MA, Goss DJ. Translation initiation factor (eIF) 4B affects the rates of binding of the mRNA m7G cap analogue to wheat germ eIFiso4F and eIFiso4F.PABP. Biochemistry 2005; 44:4510-6; PMID:15766281; http://dx.doi.org/10.1021/bi047298g
- Amrani N, Ghosh S, Mangus DA, Jacobson A. Translation factors promote the formation of two states of the closed-loop mRNP. Nature 2008; 453:1276-80; PMID:18496529; http://dx.doi.org/10.1038/nature06974
- Svitkin YV, Evdokimova VM, Brasey A, Pestova TV, Fantus D, Yanagiya A, Imataka H, Skabkin MA, Ovchinnikov LP, Merrick WC, et al. General RNA-binding proteins have a function in poly(A)-binding protein-dependent translation. EMBO J 2009; 28:58-68; PMID:19078965; http://dx.doi.org/10.1038/emboj.2008.259
- Le H, Browning KS, Gallie DR. The phosphorylation state of poly(A)-binding protein specifies its binding to poly(A) RNA and its interaction with eukaryotic initiation factor (eIF) 4F, eIFiso4F, and eIF4B. J Biol Chem 2000; 275:17452-62; PMID:10747998; http://dx.doi.org/10.1074/jbc.M001186200
- Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell 1998; 2:135-40; PMID:9702200; http://dx.doi.org/10.1016/S1097-2765(00)80122-7
- Gallie DR. The role of the initiation surveillance complex in promoting efficient protein synthesis. Biochem Soc Trans 2004; 32:585-8; PMID:15270682; http://dx.doi.org/10.1042/BST0320585
- Iizuka N, Najita L, Franzusoff A, Sarnow P. Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol Cell Biol 1994; 14:7322-30; PMID:7935446
- Tarun SZ Jr., Sachs AB. A common function for mRNA 5¢ and 3¢ ends in translation initiation in yeast. Genes Dev 1995; 9:2997-3007; PMID:7498795; http://dx.doi.org/10.1101/gad.9.23.2997
- Cheng S, Sultana S, Goss DJ, Gallie DR. Translation initiation factor 4B homodimerization, RNA binding, and interaction with Poly(A)-binding protein are enhanced by zinc. J Biol Chem 2008; 283:36140-53; PMID:18977752; http://dx.doi.org/10.1074/jbc.M807716200
- Drawbridge J, Grainger JL, Winkler MM. Identification and characterization of the poly(A)-binding proteins from the sea urchin: a quantitative analysis. Mol Cell Biol 1990; 10:3994-4006; PMID:2196442
- Gallie DR, Le H, Caldwell C, Tanguay RL, Hoang NX, Browning KS. The phosphorylation state of translation initiation factors is regulated developmentally and following heat shock in wheat. J Biol Chem 1997; 272:1046-53; PMID:8995401; http://dx.doi.org/10.1074/jbc.272.2.1046
- Schwartz DC, Parker R. Interaction of mRNA translation and mRNA degradation in Saccharomyces cerevisiae. In Sonenberg, N, Hershey, JWB., Mathews, M.B (eds), Translational Control of Gene Expression. Cold Spring Harbor press, New York, pp. 807-25, 2000.
- Le H, Chang S-C, Tanguay RL, Gallie DR. The wheat poly(A)-binding protein functionally complements pab1 in yeast. Eur J Biochem 1997a; 243:350-7; PMID:9030759; http://dx.doi.org/10.1111/j.1432-1033.1997.0350a.x
- Friend K, Brook M, Bezirci FB, Sheets MD, Gray NK, Seli E. Embryonic poly(A)-binding protein (ePAB) phosphorylation is required for Xenopus oocyte maturation. Biochem J 2012; 445:93-100; PMID:22497250; http://dx.doi.org/10.1042/BJ20120304
- Brook M, McCracken L, Reddington JP, Lu ZL, Morrice NA, Gray NK. The multifunctional poly(A)-binding protein (PABP) 1 is subject to extensive dynamic post-translational modification, which molecular modelling suggests plays an important role in co-ordinating its activities. Biochem J 2012; 441:803-12; PMID:22004688; http://dx.doi.org/10.1042/BJ20111474
- Duncan R, Hershey JW. Regulation of initiation factors during translational repression caused by serum depletion. Covalent modification. J Biol Chem 1985; 260:5493-7; PMID:3886657
- Duncan RF, Hershey JW. Protein synthesis and protein phosphorylation during heat stress, recovery, and adaptation. J Cell Biol 1989; 109:1467-81; PMID:2793930; http://dx.doi.org/10.1083/jcb.109.4.1467
- Manzella JM, Rychlik W, Rhoads RE, Hershey JW, Blackshear PJ. Insulin induction of ornithine decarboxylase. Importance of mRNA secondary structure and phosphorylation of eucaryotic initiation factors eIF-4B and eIF-4E. J Biol Chem 1991; 266:2383-9; PMID:1989989
- Hilliker A, Gao Z, Jankowsky E, Parker R. The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex. Mol Cell 2011; 43:962-72; PMID:21925384; http://dx.doi.org/10.1016/j.molcel.2011.08.008
- Tarn WY, Chang TH. The current understanding of Ded1pDDX3 homologs from yeast to human. RNA Biol 2009; 6:17-20; PMID:19106629; http://dx.doi.org/10.4161/rna.6.1.7440
- Dhote V, Sweeney TR, Kim N, Hellen CU, Pestova TV. Roles of individual domains in the function of DHX29, an essential factor required for translation of structured mammalian mRNAs. Proc Natl Acad Sci U S A 2012; 109:E3150-9; PMID:23047696; http://dx.doi.org/10.1073/pnas.1208014109
- Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Hellen CU, Pestova TV, Frank J. Structure of the mammalian ribosomal 43S preinitiation complex bound to the scanning factor DHX29. Cell 2013; 153:1108-19; PMID:23706745; http://dx.doi.org/10.1016/j.cell.2013.04.036
- Gallie DR. Cap-independent translation conferred by the 5¢-leader of tobacco etch virus is eIF4G-dependent. J Virol 2001; 75:12141-52; PMID:11711605; http://dx.doi.org/10.1128/JVI.75.24.12141-12152.2001
- Gallie DR. The 5¢-leader of tobacco mosaic virus promotes translation through enhanced recruitment of eIF4F. Nucleic Acids Res 2002; 30:3401-11; PMID:12140325; http://dx.doi.org/10.1093/nargkf457
- Mayberry LK, Allen ML, Dennis MD, Browning KS. Evidence for variation in the optimal translation initiation complex: plant eIF4B, eIF4F, and eIF(iso)4F differentially promote translation of mRNAs. Plant Physiol 2009; 150:1844-54; PMID:19493973; http://dx.doi.org/10.1104/pp.109.138438
- Pause A, Méthot N, Svitkin Y, Merrick WC, Sonenberg N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J 1994b; 13:1205-15; PMID:8131750
- Leathers V, Tanguay R, Kobayashi M, Gallie DR. A phylogenetically conserved sequence within viral 3¢ untranslated RNA pseudoknots regulates translation. Mol Cell Biol 1993; 13:5331-47; PMID:8355685
- Gallie DR, Kobayashi M. The role of the 3¢-untranslated region of non-polyadenylated plant viral mRNAs in regulating translational efficiency. Gene 1994; 142:159-65; PMID:8194747; http://dx.doi.org/10.1016/0378-1119(94)90256-9
- Gallie DR, Tanguay RL, Leathers V. The tobacco etch viral 5¢ leader and poly(A) tail are functionally synergistic regulators of translation. Gene 1995; 165:233-8; PMID:8522182; http://dx.doi.org/10.1016/0378-1119(95)00521-7
- Gallie DR, Lewis NJ, Marzluff WF. The histone 3¢-terminal stem-loop is necessary for translation in Chinese hamster ovary cells. Nucleic Acids Res 1996; 24:1954-62; PMID:8657580; http://dx.doi.org/10.1093/nar24.10.1954
- Ling J, Morley SJ, Pain VM, Marzluff WF, Gallie DR. The histone 3¢-terminal stem loop binding protein enhances translation through a functional and physical interaction with eIF4G and eIF3. Mol Cell Biol 2002; 22:7853-67; PMID:12391154; http://dx.doi.org/10.1128/MCB.22.22.7853-7867.2002
- Krab IM, Caldwell C, Gallie DR, Bol JF. Coat protein enhances translational efficiency of Alfalfa mosaic virus RNAs and interacts with the eIF4G component of initiation factor eIF4F. J Gen Virol 2005; 86:1841-9; PMID:15914864; http://dx.doi.org/10.1099/vir.0.80796-0
- Wakiyama M, Imataka H, Sonenberg N. Interaction of eIF4G with poly(A)-binding protein stimulates translation and is critical for Xenopus oocyte maturation. Curr Biol 2000; 10:1147-50; PMID:10996799; http://dx.doi.org/10.1016/S0960-9822(00)00701-6
- Prévôt D, Décimo D, Herbreteau CH, Roux F, Garin J, Darlix JL, Ohlmann T. Characterization of a novel RNA-binding region of eIF4GI critical for ribosomal scanning. EMBO J 2003; 22:1909-21; PMID:12682023; http://dx.doi.org/10.1093/embojcdg175
- Yanagiya A, Svitkin YV, Shibata S, Mikami S, Imataka H, Sonenberg N. Requirement of RNA binding of mammalian eukaryotic translation initiation factor 4GI (eIF4GI) for efficient interaction of eIF4E with the mRNA cap. Mol Cell Biol 2009; 29:1661-9; PMID:19114555; http://dx.doi.org/10.1128/MCB.01187-08
- Lin TA, Kong X, Haystead TAJ, Pause A, Belsham G, Sonenberg N, Lawrence JC Jr. PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science 1994; 266:653-6; PMID:7939721; http://dx.doi.org/10.1126/science.7939721
- Pause A, Belsham GJ, Gingras A-C, Donzé O, Lin T-A, Lawrence JC Jr., Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5¢-cap function. Nature 1994a; 371:762-7; PMID:7935836; http://dx.doi.org/10.1038371762/a0
