Abstract
The presence of even one extra chromosome severely impairs cellular growth. This effect of aneuploidy (a term describing chromosome numbers deviating from multiples of haploid chromosome content) has been observed in many different organisms, from yeast to humans. Accordingly, abnormal karyotypes are detected in nearly 30% of spontaneously aborted embryos. The rarely surviving infants, such as with trisomy of chromosome 21, are severely handicapped. The causes remain enigmatic, although recent studies exploiting yeast and mouse models provided first glimpses of the imbalanced inner life of aneuploid cells. Using comparative genomics, transcriptomics and proteomics we have analyzed the fate of the transcripts and proteins coded on the extra chromosomes as well as the general response to aneuploidy in human cells.
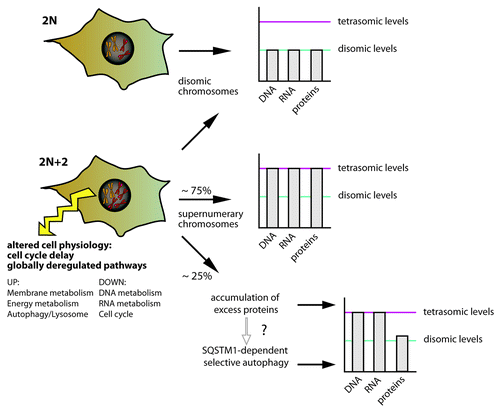
Attempts to analyze the response to aneuploidy in human cells have been hampered by the lack of isogenic diploid and aneuploid cell lines that would allow identification of changes in aneuploids in comparison to their diploid counterparts. To overcome this limitation, we introduced an extra chromosome labeled with a selection marker by micronuclei-mediated transfer to recipient human diploid and chromosomally stable cell lines HCT116 and RPE1. All analyzed tri- and tetrasomic cell lines grew significantly slower than the cognate diploid cell lines, as observed in other model systems before. To determine the fate of genes coded on the extra chromosome, we quantified the aneuploid-to-diploid changes in gene copy number (by array comparative genomic hybridization), corresponding mRNAs (by microarray analysis) and proteins (using the SILAC technology). Remarkably, whereas mRNA levels scale up with the gene copy number changes, the relative abundance of corresponding proteins was significantly lower. Direct comparison showed that approximately 25–30% of proteins coded on the tetrasomes are present at lower abundance than expected from the gene and mRNA copy numbers. Further analysis revealed that most of these proteins are subunits of macromolecular complexes and protein kinases, suggesting that their abundance is tightly controlled.
The apparent effect of aneuploidy on cell growth suggests alterations in multiple cellular pathways. Indeed, pathway enrichment analysis of both transcriptome and proteome data sets revealed multiple up- and downregulated pathways. In particular, all analyzed aneuploid cell lines show a strong downregulation of the cell cycle related pathways, DNA and RNA metabolism, splicing and ribosome biogenesis. At the same time, we observed significant upregulation of proteins associated with the endoplasmic reticulum, Golgi complex, lysosome and membrane metabolism as well as oxidative and glycolytic metabolism. We suggest that this pathway pattern represents a general cellular response to aneuploidy, as it was identified in all types of aneuploidy regardless of the type of extra chromosome and the cell line.
Remarkably, upregulation of lysosome-mediated degradation and autophagy was observed consistently in all analyzed human aneuploids. The higher demand of aneuploid cells for autophagy was further confirmed in vivo. The aneuploid cells show a 2-fold increase in LC3 foci formation as well as an increased abundance of LC3-II, a lipidated form of LC3 and hence a reliable marker for active autophagy. Interestingly, mRNA and protein levels of SQSTM1/p62 were elevated in all analyzed cell lines; correspondingly, immunofluoresence staining with SQSTM1 antibody shows elevated formation of SQSTM1 foci in aneuploid cells. This is further enhanced by inhibition of the late steps of autophagy by treatment with bafilomycin A1. SQSTM1 is a cytosolic receptor that is active in so-called selective autophagy that likely targets ubiquitinated proteins to autophagy due to its ability to bind ubiquitinated proteins via its UBA domain, and LC3 via its LIR domain. Accordingly, we observed an enhanced formation of ubiquitin-positive foci in aneuploid cells, and these foci frequently colocalized with the SQSTM1 immunofluorescence signal. Again, this observation holds true for nearly all analyzed aneuploid cell lines.
What triggers the activation of autophagy in aneuploid cells? There are several possible explanations. First, replication, transcription, translation and consequent inevitable degradation of supernumerary proteins coded on the extra chromosomes comes at a cost; the depletion of cellular energy supplies then in turn activates autophagy to provide the necessary resources. However, starvation-induced autophagy usually does not lead to elevated SQSTM1 expression. Rather, we speculate that elevated levels of proteins targeted for degradation—such as excessive subunits of protein complexes—overwhelm the proteasome and accumulate in the cytoplasm, thus causing an imbalance of protein homeostasis. This activates SQSTM1-mediated autophagy. The excessive proteins are subsequently sequestered by SQSTM1 and cleaned up by autophagy (). Finally, it remains possible that another as yet unidentified signal triggers the selective autophagy pathway in aneuploid cells.
The surplus of cellular proteins likely leads to proteotoxic stress. This could explain at least partially the growth defect of aneuploid cells. Notably, nearly 90% of solid malignant tumors show some degree of aneuploidy, yet there is no indication of growth impairment, at least as can be observed in vitro. At the same time, cancer cells often show signs of proteotoxic stress, and several inhibitors of the ubiquitin-proteasome system and autophagy are being developed and tested for cancer treatment. Identification of the strategies that allow cancer cells to overcome the protein imbalance might be an important step for developing novel therapeutic approaches that target an intrinsic feature of many cancer cells—their abnormal chromosomal content.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.